Abstract
Background
Clinical heterogeneity and variations in methods to identify major depressive disorder (MDD) across studies compromise replicability of research findings. This study evaluated potential implications of different MDD case definitions in a large biobank cohort.
Methods
Among Mayo Clinic Biobank participants, MDD was identified using two methods: self-report MDD in a participant questionnaire (PQ-MDD) and MDD ICD codes in the electronic health record (EHR-MDD). We examined agreement between these definitions and evaluated relationships between case agreement and participant characteristics, including MDD polygenic risk scores (PRS). Finally, we evaluated associations between different MDD case/control definitions and participant characteristics known to be related to MDD.
Results
Among 55,656 participants, 23 % were identified as PQ-MDD cases and 17 % as EHR-MDD cases, with 85 % overall agreement (61 % case agreement) between these definitions. Among participants identified as MDD cases by one method, older and male patients, and those with lower measures of morbidity at enrollment, were less likely to be identified as cases by the other method. The strength of the associations between different MDD case/control definitions and participant characteristics varied depending on whether MDD definitions used the same source of information (i.e., EHR-only, self-report only)—resulting in stronger associations—versus different sources of information (i.e., one from EHR, one from self-report)—resulting in weaker associations.
Conclusion
Our results demonstrate how the methods used to identify patients with history of MDD can affect sample characteristics and risk factor associations, highlighting the importance of considering phenotype ascertainment in the interpretation of research results.
Keywords: Mental health, Depressive disorder, Major, Electronic health records, Self report, Genetic risk score
Highlights
-
•
Major depressive disorder is heterogeneous, and studies may define it differently.
-
•
Large cohort studies can define depression using electronic records or self-report.
-
•
Different methods affect sociodemographic composition of case and control samples.
-
•
Data sources for case definitions and other variables affect association strength.
-
•
Interpretation of depression research must consider case identification methods.
1. Introduction
With a worldwide lifetime prevalence of 12 %, major depressive disorder (MDD) is a leading public health problem [1]. Furthermore, depressive disorders account for the greatest burden to health among all mental illnesses [2]. Research efforts have sought to characterize MDD in terms of risk and protective factors and response to treatment, yet the generalizability and replicability of findings have been limited by the considerable heterogeneity in both clinical presentation [3] and disease operationalization [4]. The latter might stem from different methods used to identify MDD cases and controls in research.
In recent decades, biobanks have emerged as major resources for health-related research, catalyzed by the vast amount of data they collect from various sources such as questionnaires, interviews, measurements, biospecimen donations, and—increasingly—electronic health record (EHR) linkage [5]. Disease-oriented biobanks, focused on a single disease, may identify cases using gold-standard diagnostic instruments, such as the Structured Clinical Interview for DSM-5 [6], [7]. However, gold-standard diagnostic procedures are often unavailable or not feasible to implement in more general biobanks, resulting in a wide variety of methods used to identify cases and controls. Specifically, general biobanks, aiming to support research for a diverse range of phenotypes, may prioritize high-throughput phenotyping, capitalizing on EHR linkage, or employ self-report measures [8]. However, by including a broad range of phenotypes that may not be simultaneously available in disease-oriented biobanks, general biobanks are better suited to study relationships among complex or heterogeneous traits [9], [10].
Phenotypes based on different sources of information are subject to unique biases. For example, EHRs can be subject to coding errors or bias driven by billing practices, restricted to a single medical institution or health care system, and exclude individuals who could not obtain medical care (e.g., those facing barriers in access to health care) [11]. On the other hand, self-reported measures may be affected by sociodemographic factors, disease heterogeneity, recall bias, willingness to disclose, or the specific phrasing of questions [12], [13], [14]. Given these unique complexities, it is not surprising that the genetic architecture of so-called minimally phenotyped MDD (e.g., using only participant self-report) differs from stricter definitions of MDD [15]. This is particularly important given that a growing proportion of MDD studies rely on minimal phenotyping [16]. As an alternative to linkage with a single EHR, certain general biobanks such as the National Institutes of Health All of Us® Research Program [17] allow participants to share their EHR data, effectively linking the biobank to different EHRs. Additionally, the relative ease of access to direct-to-consumer genotyping has resulted in datasets that are not associated with an EHR and are thus limited to minimal phenotyping through questionnaires (e.g., 23andMe [18]). And, while self-reported MDD may not equate to MDD defined by diagnostic interview, questionnaire-based phenotypes may be better suited to inform epidemiological studies, since they may include populations underrepresented in EHRs due to barriers to health care access [19].
Prior studies have shown that the choice of method used to identify MDD in research studies introduces substantial heterogeneity between samples [4] in addition to the already heterogeneous nature of MDD. Similar to general medical conditions such as diabetes mellitus [20], it is important to understand the yield and characteristics of the cohorts obtained by different MDD definitions across distinct sources of information. Doing so would allow researchers to account for the added variability at each stage of research, while failing to do so could lead to conflating different phenotypes as one, generating inaccurate or non-generalizable conclusions.
To evaluate the potential implications of different MDD case definitions in a large biobank cohort, we compared the level of agreement between two distinct methods of identifying MDD cases, using self-report versus structured EHR data. Additionally, we explored the effect that choice of method may have on case identification by conducting a bidirectional examination of method agreement and its associations with sociodemographic, health-related, clinical, well-being-related characteristics, and polygenic risk for MDD. Finally, we examined the relationship between different MDD case definitions and participant characteristics known to be related to MDD.
2. Materials and methods
2.1. Study design and participants
This study included data from all Mayo Clinic Biobank (MCB) participants. The design, implementation and cohort profile of the biobank have been described elsewhere [21], [22]. Briefly, MCB is an institutional biorepository that actively recruited participants from April 2009 to March 2016. Invitations were primarily sent to patients with appointments in departments providing primary care to enrich the biobank for individuals with the greatest likelihood of having comprehensive EHR data. Individuals were able to volunteer without a study invitation. Eligibility was not restricted by medical conditions and only required interested individuals to be Mayo Clinic patients, 18 years or older, current US residents, and able to provide informed consent. The final sample includes individuals with varying degrees of EHR coverage; approximately 70 % with visits during at least 3 of the 5 years prior to biobank enrollment [23]. At enrollment, participants underwent blood draw and storage; completed a participant questionnaire (PQ) related to their general health and functioning, personal and family medical history, and demographic data; and provided consent to use their EHR data for research. Follow-up was conducted both actively and passively, through follow-up surveys and data collection from the EHR. All participants provided informed consent prior to enrollment into the MCB. This study was approved by the Mayo Clinic Institutional Review Board (IRB) and the Mayo Clinic Biobank Access Committee and was conducted in accordance with the Declaration of Helsinki.
2.2. Data collection and quality control
2.2.1. Identification of participants with MDD
Identification of participants with MDD was conducted using two distinct information sources (the specific codes and questions used for each source are available in Table S1). First, PQ-MDD, based on the biobank’s enrollment participant questionnaire, was defined as any positive response to the question of being previously diagnosed with depression. Second, EHR-MDD, based on Mayo Clinic EHR up to enrollment, was defined as the presence of at least one MDD diagnostic code from a list of ICD-9/10-CM codes previously reported by Ryu et al. [23]. Participants were excluded from analyses if they self-reported a prior diagnosis of bipolar disorder or if they had at least one ICD-9/10-CM code for bipolar disorder at enrollment based on phecode maps 1.2 and 1.2b [24], [25].
2.2.2. Participant characteristics
We obtained data related to sociodemographic, health, clinical, and social and emotional well-being from participants’ EHR or questionnaire data to examine associations between these variables and the level of agreement between the two identification methods. A full list of variables and their source is available in Table S2. BMI at enrollment was calculated using height and weight information and further categorized into underweight, normal, overweight, and obese [26]. To understand the effect of cumulative comorbidity, we computed the Charlson Comorbidity Index, a severity-weighted sum of 19 conditions [27], from participants’ EHR available as of biobank enrollment. In addition, we collected information on comorbid anxiety disorders, attention-deficit/hyperactivity disorder (ADHD), and substance use disorders (SUD), which were defined by the presence of one or more relevant ICD-9/10-CM diagnostic codes (Table S2). For anxiety disorder, we also obtained self-reported information from PQ.
The following MDD-related variables were collected: MDD screening, as measured by a modified version of the 2-item Patient Health Questionnaire (PHQ-2) at enrollment; MDD pharmacotherapy exposure, by quantifying the total number of unique antidepressant classes prescribed and recorded in participant EHRs prior to MCB enrollment; and recency of last MDD diagnosis, calculated as time since most recent MDD-related diagnostic code until MCB enrollment. To evaluate the depth of EHR data for participants, we calculated two measures of EHR coverage. Length of EHR was calculated as the number of years from the date of the first to last available ICD code prior to biobank enrollment. Diagnostic frequency was calculated as the number of days with at least one diagnostic code per year in participants’ EHR from first occurrence of an ICD code to the date of MCB enrollment.
2.2.3. Genetic data and polygenic risk scores
We computed polygenic risk scores (PRS) for MDD (MDD-PRS) for eligible participants. Samples were genotyped using a genotype-by-sequencing approach developed by Regeneron Genetics Center, which adds probes for GWAS scaffold SNPs to exome sequencing capture [28], and calculation of MDD-PRS for MCB participants was conducted as previously described [29]. Principal components were calculated and used to account for population substructure in the subsequent analysis. The PRS analysis was limited to only unrelated European-ancestry participants. The summary statistics were from the Psychiatric Genomics Consortium GWAS for MDD [30], which meta-analyzed three independent studies where MDD cases were identified using a mix of questionnaire-based and clinical methods. MDD-PRS was calculated using these summary statistics and the LDpred2-auto approach [31].
2.3. Statistical analyses
Descriptive statistics were used to summarize the basic sociodemographic characteristics of participants identified as MDD cases and controls by each method of identifying MDD, independent of the other method’s inclusion and exclusion criteria. For each of the two methods used to identify MDD, univariable logistic regression models were used to test the association of the different sociodemographic variables with the odds of case status.
To assess agreement between the two methods of identifying MDD, we calculated three agreement measures: overall, case, and control agreement [32], [33]. Overall agreement represents the proportion of individuals classified the same by both methods. It was calculated by dividing the number of concordant cases or controls by the total number of participants. Case agreement, also called positive agreement [32], represents how well the two methods agree on who has MDD. It was calculated by dividing the number of concordant cases by the average number of cases identified by both methods. Control agreement, also called negative agreement [32], represents how well the methods agree on who does not have MDD (the control group). It was calculated by dividing the number of concordant controls by the average number of controls identified by both methods. When assessing agreement between case definitions, we also explored the effect that increasingly restrictive EHR-MDD criteria would have on the measures of agreement by repeating the analyses after increasing the number of MDD ICD codes required to at least 2 or restricting MDD ICD code eligibility to within 5, 3, or 1 year(s) prior to biobank enrollment. We also conducted a sensitivity analysis by restricting the analyses to individuals living within Olmsted County, MN, where Mayo Clinic Rochester is located, and six surrounding counties (i.e., areas where EHR coverage is highest), hypothesizing that agreement would be greater for local participants in comparison to the overall study cohort consisting of participants with a varying degree of EHR coverage.
Then, we conducted a bidirectional examination of MDD identification method agreement and participant characteristics. Specifically, among MDD cases identified by each method, we compared the participant characteristics, including MDD-PRS, of those identified by both methods (PQ-MDD and EHR-MDD) with those of participants identified by a single method (only PQ-MDD or EHR-MDD) using linear (MDD-PRS) and logistic regression models. First, among EHR-MDD cases, we tested whether any participant characteristics were associated with also being identified as PQ-MDD cases—i.e., participants with a history of MDD recorded in their EHR that subsequently self-reported a prior diagnosis of depression versus those that did not. Second, among PQ-MDD cases, we tested whether any participant characteristics were associated with also being identified as EHR-MDD cases—i.e., participants that self-reported a prior diagnosis of depression and had a history of MDD recorded in their EHR versus those that did not. All models were adjusted for age, gender, and length of EHR except for the models testing the effects of these variables. For PRS analyses, models were adjusted for the first 5 principal components in addition to age, gender, and length of EHR.
Lastly, we considered two additional MDD case/control definitions based on a combination of the methods used to identify MDD, creating a narrower case definition based on the intersection of PQ-MDD and EHR-MDD and a broader case definition based on the union of PQ-MDD and EHR-MDD. We tested the relationship between four MDD case/control definitions and participant characteristics known to be related to MDD (i.e., age, gender, PHQ-2, anxiety disorders, and MDD-PRS). Two definitions of anxiety disorders were included, one based on EHR data, and one based on PQ information, to explore how concordance between the information source used for case definitions and the one used for participant characteristics could influence associations. We employed logistic regression models adjusted for age, gender, and EHR length to examine the associations between participant characteristics and four case/control definitions: (a) PQ-MDD cases vs. PQ-MDD controls; (b) EHR-MDD cases vs. EHR-MDD controls; (c) cases with both PQ-MDD and EHR-MDD vs. controls with neither; and (d) cases with either PQ-MDD or EHR-MDD vs. controls with neither. For PRS analyses, models were subset to European-ancestry participants and adjusted for the first 5 principal components in addition to age, gender, and length of EHR.
3. Results
Our eligible sample included 55,656 biobank participants (59 % women). Most participants identified as White (92.1 %), with 1.1 % Black, 1.0 % Asian, and 4.6 % multi-racial. More participants were identified as MDD cases by PQ-MDD (n = 12,688; 22.9 %) than by EHR-MDD (n = 9584; 17.3 %) (Table 1). Overall agreement between EHR-MDD and PQ-MDD was 84.5 %, case agreement 61.0 %, and control agreement 90.3 % (Table 2). Using increasingly restrictive EHR-MDD criteria (requiring at least 2 MDD ICD codes or restricting inclusion of EHR to within 5, 3, or 1 year prior to biobank enrollment) resulted in reductions to case agreement and marginal changes to overall and control agreement (Table S3). Our sensitivity analysis restricted to individuals from areas with the highest EHR coverage in general increased agreement, with the largest difference in case agreement (73.2 %) (Table S4).
Table 1.
Demographic characteristics across two methods to identify major depressive disorder in the Mayo Clinic Biobank.
| MDD identified by participant questionnaires | MDD identified by electronic health records | |||||
|---|---|---|---|---|---|---|
| Demographic Characteristics,n(row %) |
Cases (n = 12,688) |
Controls (n = 42,542) |
ORa(95 % CI) |
Cases (n = 9584) |
Controls (n = 45,792) |
ORa(95 % CI) |
| Age at enrollment | ||||||
| 18–44 | 2875 (31.5 %) | 6250 (68.5 %) | Reference | 2136 (23.3 %) | 7017 (76.7 %) | Reference |
| 45–54 | 2469 (27.1 %) | 6658 (72.9 %) | 0.81 (0.76,0.86) | 1894 (20.7 %) | 7248 (79.3 %) | 0.86 (0.80,0.92) |
| 55–64 | 3335 (25.2 %) | 9877 (74.8 %) | 0.73 (0.69,0.78) | 2403 (18.1 %) | 10855 (81.9 %) | 0.73 (0.68,0.78) |
| 65 + | 4009 (16.9 %) | 19757 (83.1 %) | 0.44 (0.42,0.47) | 3151 (13.2 %) | 20672 (86.8 %) | 0.5 (0.47,0.53) |
| Gender Identity | ||||||
| Female | 9275 (28.6 %) | 23135 (71.4 %) | Reference | 7024 (21.6 %) | 25474 (78.4 %) | Reference |
| Male | 3413 (15.0 %) | 19407 (85.0 %) | 0.44 (0.42,0.46) | 2560 (11.2 %) | 20318 (88.8 %) | 0.46 (0.44,0.48) |
| Race | ||||||
| White | 11603 (22.8 %) | 39273 (77.2 %) | Reference | 8804 (17.3 %) | 42166 (82.7 %) | Reference |
| Black or African American | 138 (22.5 %) | 474 (77.5 %) | 0.99 (0.81,1.19) | 69 (11.2 %) | 548 (88.8 %) | 0.6 (0.47,0.77) |
| Asian | 69 (12.1 %) | 502 (87.9 %) | 0.47 (0.36,0.59) | 53 (9.20 %) | 520 (90.8 %) | 0.49 (0.36,0.64) |
| American Indian, Alaska Native, Native Hawaiian, or Pacific Islander |
29 (31.9 %) | 62 (68.1 %) | 1.58 (1.00,2.44) | 18 (19.8 %) | 73 (80.2 %) | 1.18 (0.68,1.93) |
| Multi-racial | 724 (28.6 %) | 1809 (71.4 %) | 1.35 (1.24,1.48) | 536 (20.8 %) | 2035 (79.2 %) | 1.26 (1.14,1.39) |
| Other | 57 (25.1 %) | 170 (74.9 %) | 1.13 (0.83,1.52) | 48 (21.1 %) | 180 (78.9 %) | 1.28 (0.92,1.74) |
| Missing | 68 | 252 | 56 | 270 | ||
| Ethnicity | ||||||
| Non-Hispanic | 12345 (22.9 %) | 41467 (77.1 %) | Reference | 9343 (17.3 %) | 44607 (82.7 %) | Reference |
| Hispanic | 220 (26.5 %) | 611 (73.5 %) | 1.21 (1.03,1.41) | 146 (17.5 %) | 687 (82.5 %) | 1.01 (0.84,1.21) |
| Missing | 123 | 464 | 95 | 498 | ||
| Education | ||||||
| High school or less | 1975 (22.0 %) | 6989 (78 %) | 1.07 (1.00,1.14) | 1626 (18.1 %) | 7373 (81.9 %) | 1.18 (1.10,1.26) |
| Some college | 4734 (27.1 %) | 12765 (72.9 %) | 1.40 (1.33,1.48) | 3674 (20.9 %) | 13864 (79.1 %) | 1.41 (1.33,1.50) |
| College graduate | 2888 (20.9 %) | 10937 (79.1 %) | Reference | 2189 (15.8 %) | 11674 (84.2 %) | Reference |
| Postgraduate | 2877 (20.5 %) | 11179 (79.5 %) | 0.97 (0.92,1.03) | 1915 (13.6 %) | 12166 (86.4 %) | 0.84 (0.79,0.90) |
| Missing | 214 | 672 | 180 | 715 | ||
MDD, major depressive disorder.
Odds ratios and 95 % confidence intervals for Mayo Clinic Biobank participants, comparing MDD cases vs. controls across two methods of identifying MDD.
Table 2.
Measures of agreement between two methods of identifying major depressive disorder.
| PQ-MDD | Agreement | |||||
|---|---|---|---|---|---|---|
| EHR-MDD | Cases (n = 12,506) | Controls (n = 42,444) | Excluded by PQ-BD | Overall | Case | Control |
| Cases (n = 9409) | 6689 | 2720 | 175 | 84.5 % | 61.0 % | 90.3 % |
| Controls (n = 45,541) | 5817 | 39724 | 251 | |||
| Excluded by EHR-BD | 182 | 98 | 373 | |||
Case and control counts do not include participants excluded by any method.
BD, bipolar disorder; EHR, electronic health record; MDD, major depressive disorder; PQ, participant questionnaire.
3.1. Relationship between identification method agreement and participant characteristics
Among participants with a history of MDD recorded in their EHRs (EHR-MDD cases), self-reporting a diagnosis of MDD (PQ-MDD) was less likely among those who were older (OR [95 % CI] = 0.47 [0.41–0.53] for 65 + vs. 18–44), male (OR [95 % CI] = 0.71 [0.65, 0.79] vs. female), or identified as an understudied population (e.g., Asian participants (OR [95 % CI] = 0.45 [0.26–0.81] vs. White participants). This was also the case for those with the longest time since their last recorded MDD diagnosis (OR [95 % CI] = 0.23 [0.20–0.26] for 5 + years vs. <90 days prior to the enrollment). On the other hand, participants identified as EHR-MDD cases were more likely to self-report MDD when they had a higher educational attainment (OR [95 % CI] = 1.17 [1.02–1.35] for postgraduate degree vs. college graduate), positive PHQ-2 at enrollment (OR [95 % CI] = 2.98 [2.54–3.52]), higher number of different antidepressant classes in their EHR (OR [95 % CI] = 8.98 [7.44–10.85] for 3 + vs. 0), and worse mental/emotional well-being (Table 3). Among individuals with European ancestry identified as EHR-MDD cases (n = 7474), higher MDD-PRS was associated with self-reporting MDD (OR [95 % CI] = 1.10 [1.05–1.16]).
Table 3.
Associations with self-reporting MDD among participants with a recorded history of MDD.
|
Self-reported history of depression among EHR-MDD |
|||
|---|---|---|---|
| Participant characteristics | Positive (n = 6689) | Negative (n = 2720) | aOR (95 % CI)a |
| Sociodemographic characteristics,n(row %) | |||
| Age at enrollment | |||
| 18–44 | 1643 (78.9 %) | 439 (21.1 %) | Reference |
| 45–54 | 1392 (75 %) | 465 (25 %) | 0.82 (0.71,0.95) |
| 55–64 | 1742 (73.7 %) | 621 (26.3 %) | 0.79 (0.69,0.91) |
| 65 + | 1912 (61.5 %) | 1195 (38.5 %) | 0.47 (0.41,0.53) |
| Gender Identity | |||
| Female | 5073 (73.4 %) | 1839 (26.6 %) | Reference |
| Male | 1616 (64.7 %) | 881 (35.3 %) | 0.71 (0.65,0.79) |
| Race | |||
| White | 6188 (71.5 %) | 2471 (28.5 %) | Reference |
| Black or African American | 44 (67.7 %) | 21 (32.3 %) | 0.67 (0.39,1.16) |
| Asian | 30 (57.7 %) | 22 (42.3 %) | 0.45 (0.26,0.81) |
| American Indian, Alaska Native, Native Hawaiian, or Pacific Islander |
11 (64.7 %) | 6 (35.3 %) | 0.66 (0.24,1.96) |
| Multi-racial | 356 (68.7 %) | 162 (31.3 %) | 0.95 (0.78,1.16) |
| Other | 26 (57.8 %) | 19 (42.2 %) | 0.51 (0.28,0.94) |
| Ethnicity | |||
| Non-Hispanic | 6539 (71.3 %) | 2635 (28.7 %) | Reference |
| Hispanic | 97 (68.3 %) | 45 (31.7 %) | 0.71 (0.50,1.04) |
| Education | |||
| High school or less | 1056 (66.2 %) | 540 (33.8 %) | 0.93 (0.80,1.07) |
| Some college | 2622 (72.8 %) | 981 (27.2 %) | 1.09 (0.96,1.23) |
| College graduate | 1543 (71.6 %) | 612 (28.4 %) | Reference |
| Postgraduate | 1358 (72.2 %) | 523 (27.8 %) | 1.17 (1.02,1.35) |
| Marital status | |||
| Married or marriage-like | 4579 (69.9 %) | 1972 (30.1 %) | Reference |
| Separated/Divorced | 887 (77.9 %) | 252 (22.1 %) | 1.44 (1.24,1.68) |
| Widowed | 378 (61.3 %) | 239 (38.7 %) | 0.85 (0.71,1.02) |
| Never married | 677 (78.3 %) | 188 (21.7 %) | 1.23 (1.03,1.47) |
| Unemployed under age 65 | |||
| No | 5021 (69.0 %) | 2251 (31.0 %) | Reference |
| Yes | 1608 (78.4 %) | 442 (21.6 %) | 1.22 (1.07,1.40) |
| Health-related characteristics,n(row %) | |||
| Smoking status | |||
| Never | 3515 (70.8 %) | 1448 (29.2 %) | Reference |
| Former | 2468 (70.8 %) | 1017 (29.2 %) | 1.13 (1.02,1.24) |
| Current | 596 (75.5 %) | 193 (24.5 %) | 1.11 (0.93,1.32) |
| Alcohol use | |||
| Never | 1756 (70.4 %) | 737 (29.6 %) | Reference |
| Once a month or less | 1664 (74.4 %) | 574 (25.6 %) | 1.10 (0.96,1.25) |
| 2–4 a month | 1477 (71.4 %) | 593 (28.6 %) | 0.94 (0.83,1.08) |
| 2–3 a week | 822 (68.2 %) | 383 (31.8 %) | 0.84 (0.72,0.98) |
| 4–5 a week | 460 (69.3 %) | 204 (30.7 %) | 0.99 (0.82,1.20) |
| 6 or more a week | 472 (70.3 %) | 199 (29.7 %) | 1.09 (0.90,1.32) |
| Body mass index | |||
| Underweight | 71 (74.7 %) | 24 (25.3 %) | 1.13 (0.71,1.86) |
| Normal | 1629 (70.1 %) | 694 (29.9 %) | Reference |
| Overweight | 1952 (68.8 %) | 886 (31.2 %) | 1.07 (0.95,1.21) |
| Obese | 2663 (73.3 %) | 972 (26.7 %) | 1.28 (1.13,1.44) |
| Charlson index | |||
| 0 | 2130 (73.2 %) | 778 (26.8 %) | Reference |
| 1–2 | 1917 (73.1 %) | 706 (26.9 %) | 1.17 (1.04,1.33) |
| 3–4 | 688 (67.9 %) | 325 (32.1 %) | 1.10 (0.93,1.30) |
| 5 or more | 563 (65.8 %) | 292 (34.2 %) | 1.01 (0.85,1.21) |
| Anxiety disordersb | |||
| No | 4058 (68.4 %) | 1877 (31.6 %) | Reference |
| Yes | 2631 (75.7 %) | 843 (24.3 %) | 1.39 (1.26,1.53) |
| Attention-deficit/hyperactivity disorderb | |||
| No | 6446 (70.7 %) | 2677 (29.3 %) | Reference |
| Yes | 243 (85.0 %) | 43 (15.0 %) | 1.93 (1.40,2.72) |
| Substance use disordersb | |||
| No | 5390 (70.7 %) | 2233 (29.3 %) | Reference |
| Yes | 1299 (72.7 %) | 487 (27.3 %) | 1.07 (0.95,1.20) |
| Clinical characteristics,n(row %) | |||
| MDD Screen at enrollmentc | |||
| Negative | 5311 (68.2 %) | 2478 (31.8 %) | Reference |
| Positive | 1315 (87.3 %) | 191 (12.7 %) | 2.98 (2.54,3.51) |
| Total unique antidepressant classes | |||
| 0 | 385 (41.0 %) | 555 (59.0 %) | Reference |
| 1 | 2632 (67.8 %) | 1250 (32.2 %) | 3.27 (2.81,3.82) |
| 2 | 2073 (77.1 %) | 614 (22.9 %) | 5.45 (4.62,6.43) |
| 3 or more | 1599 (84.2 %) | 301 (15.8 %) | 8.98 (7.44,10.85) |
| Recency of last MDD episode | |||
| < 90days | 2051 (82.6 %) | 432 (17.4 %) | Reference |
| 90days-< 1 yr | 1352 (78.8 %) | 363 (21.2 %) | 0.82 (0.70,0.96) |
| 1yr-< 3 yrs | 1644 (71.8 %) | 645 (28.2 %) | 0.58 (0.50,0.67) |
| 3yrs-< 5 yrs | 712 (64.4 %) | 393 (35.6 %) | 0.41 (0.35,0.48) |
| 5 yrs+ | 930 (51.2 %) | 887 (48.8 %) | 0.23 (0.20,0.26) |
| Social and emotional well-being characteristics,n(row %) | |||
| Overall mental well-being | |||
| 0–5 (low) | 1208 (84.9 %) | 215 (15.1 %) | 2.83 (2.42,3.33) |
| 6–7 (low normal) | 1705 (78.0 %) | 480 (22.0 %) | 1.82 (1.62,2.05) |
| 8 + (high normal) | 3763 (65.1 %) | 2017 (34.9 %) | Reference |
| Overall emotional well-being | |||
| 0–5 (low) | 1808 (85.4 %) | 310 (14.6 %) | 3.39 (2.96,3.89) |
| 6–7 (low normal) | 1965 (76.2 %) | 615 (23.8 %) | 1.87 (1.67,2.08) |
| 8 + (high normal) | 2906 (61.9 %) | 1787 (38.1 %) | Reference |
| Emotional health | |||
| Excellent | 236 (47.8 %) | 258 (52.2 %) | Reference |
| Very good | 1537 (61.8 %) | 949 (38.2 %) | 1.75 (1.44,2.13) |
| Good | 2121 (69.3 %) | 939 (30.7 %) | 2.42 (1.99,2.94) |
| Fair | 1919 (81.5 %) | 435 (18.5 %) | 4.58 (3.72,5.65) |
| Poor | 809 (88.7 %) | 103 (11.3 %) | 7.59 (5.78,10.04) |
| EHR Density,n(row %) | |||
| EHR length (years) | |||
| Q1: ≤ 2.62 | 966 (80.6 %) | 232 (19.4 %) | 1.86 (1.58,2.19) |
| Q2: 2.63–8.17 | 1664 (71.0 %) | 680 (29.0 %) | 1.17 (1.04,1.31) |
| Q3: 8.18–13.3 | 1833 (70.4 %) | 772 (29.6 %) | 1.11 (0.99,1.24) |
| Q4: 13.4 + | 2226 (68.2 %) | 1036 (31.8 %) | Reference |
| Diagnostic frequency (mean visits per year) | |||
| Q1: < 2.72 | 624 (72.9 %) | 232 (27.1 %) | 1.04 (0.87,1.23) |
| Q2: 2.72–5.82 | 1331 (68.6 %) | 610 (31.4 %) | 0.85 (0.75,0.97) |
| Q3: 5.83–11.6 | 2141 (69.9 %) | 921 (30.1 %) | 0.89 (0.80,1.00) |
| Q4: 11.7 + | 2593 (73.0 %) | 957 (27.0 %) | Reference |
| MDD Polygenic Risk Score | |||
| Continuous PRS (Linear) | |||
| Participants, n (missing) | 5303 (1386) | 2171 (549) | 1.10 (1.05,1.16) |
| PRS-MDD, median (IQR) | 0.2 (−0.5:0.9) | 0.1 (−0.6:0.8) | |
aOR, adjusted odds ratio; CI, confidence interval; EHR, electronic health record; IQR, interquartile range; MDD, major depressive disorder; PQ, participant questionnaire; Q, quartile.
Odds ratios and 95 % confidence intervals for participants with EHR-MDD, comparing participants that self-reported MDD history vs. those that did not. Adjusted for age, gender, and EHR length. Tests exploring age, gender and length of EHR were unadjusted for said variable.
Comorbidities were defined as the presence of at least one diagnostic code in a participants EHR.
Screened using a modified version of the Patient Health Questoinnaire-2 in the participant questionnaire.
For participants that self-reported having been diagnosed with MDD (PQ-MDD cases), the likelihood of having a history of MDD recorded in their EHR (EHR-MDD) was similarly associated with participant characteristics, with some key distinctions (Table S5). For instance, those with higher educational attainment had lower odds of having a MDD recorded in their EHR (OR [95 % CI] = 0.88 [0.79–0.99] for postgraduate degree vs. college graduate). In addition, using the same information source to define both MDD and participant characteristics (e.g., EHR-MDD with EHR characteristics) resulted in stronger associations than when these variables were based on different sources of information (e.g., PQ-MDD with EHR characteristics). For example, having a history of 3 + antidepressant classes recorded in the EHR was more strongly associated with having a diagnosis of MDD in the EHR among participants that self-reported MDD (EHR-MDD OR [95 % CI] = 39.62 [32.86–48.00]) than with self-reporting MDD among participants with a diagnosis of MDD in the EHR (PQ-MDD OR [95 % CI] = 8.98 [7.44–10.85]). Similarly, self-reporting worse emotional health was more strongly associated with self-reporting MDD among participants with EHR-MDD (PQ-MDD OR [95 % CI] = 7.59 [5.78–10.04]) than with having a diagnosis of MDD in the EHR among participants with PQ-MDD (EHR-MDD OR [95 % CI] = 2.44 [1.95–3.05]) (Table 3, Table S5).
3.2. Varying associations with MDD case status across four different case definitions
Four MDD case/control definitions were examined. In addition to definitions based on the methods of identifying MDD previously outlined (i.e., PQ-MDD and EHR-MDD), we considered two additional MDD case/control definitions based on a combination of methods—a narrow definition based on the intersection of PQ-MDD and EHR-MDD and a broad definition based on the union of PQ-MDD and EHR-MDD. Using the broad definition of MDD (i.e., those identified as cases by either method) resulted in 27.7 % of participants identified as MDD cases (Table S6). Using the narrow definition of MDD (i.e., those identified as cases by both methods), 12.2 % of participants were classified as MDD cases.
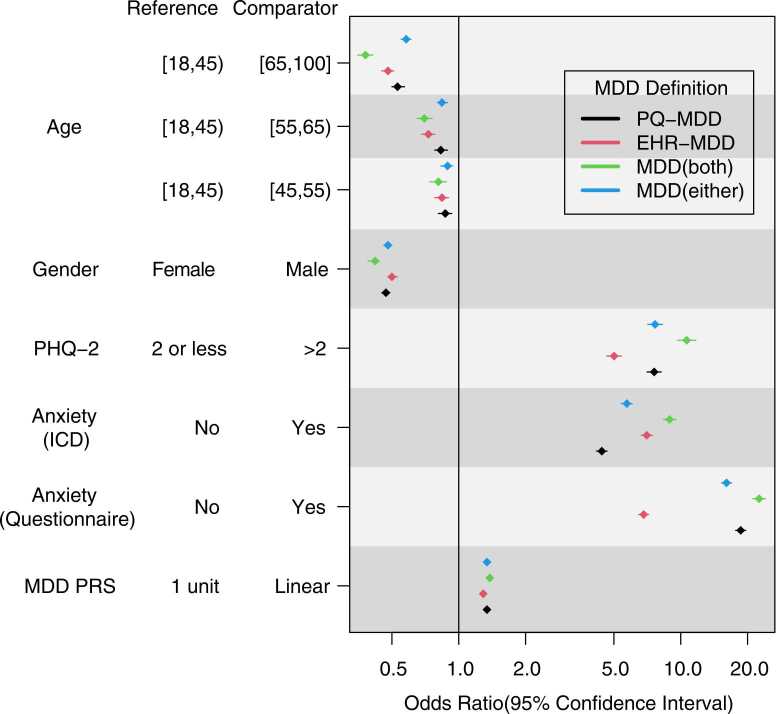
We examined how each MDD case/control definition was associated with age, gender, PHQ-2, anxiety identified in the EHR (EHR-anxiety), anxiety identified through the PQ (PQ-anxiety), and MDD-PRS (Fig. 1). Across all case/control definitions, older and male participants had a lower likelihood of being identified as MDD cases—the strongest of these associations were observed with the narrow definition (i.e., cases with both PQ-MDD and EHR-MDD vs. controls with neither) (OR [95 % CI] = 0.38 [0.35–0.41] for age 65–100 vs. 18–45 years; OR [95 % CI] = 0.42 [0.39–0.44] for male vs. female participants). While estimates suggested that MDD-PRS was more strongly associated with PQ-MDD (OR [95 % CI] = 1.34 [1.31–1.38] per unit increase in MDD-PRS) than with EHR-MDD (OR [95 % CI] = 1.29 [1.26–1.66]), the difference was not statistically significant.
Fig. 1.
Factors known to be related to MDD and their association with different MDD case/control definitionsOdds ratios comparing MDD cases vs. controls, across four different case/control definitions. Adjusted for age, gender, and EHR length. Includes two definitions for anxiety disorders, one based on EHR data and one based on self-report (PQ). PRS analyses were limited to European-ancestry participants and additionally adjusted for the first 5 genomic principal components. MDD case/control definitions: PQ-MDD, self-reported being previously diagnosed with depression at any age; EHR-MDD, presence of at least one MDD diagnostic code in EHR up to biobank enrollment; MDD (both), cases with both PQ-MDD and EHR-MDD, controls with neither; MDD (either), cases with either PQ-MDD or EHR-MDD, controls with neither.EHR, electronic health record; ICD, International Classification of Diseases; MDD, major depressive disorder; PHQ-2, 2-item Patient Health Questionnaire; PQ, participant questionnaire; PRS, polygenic risk score.
The association between history of anxiety disorders and MDD case/control definitions varied depending on whether both variables were defined using the same source of information (i.e., both derived from EHR, or both derived from self-report), or from different sources of information (i.e., one from EHR, one from self-report). When history of anxiety disorders was based on EHR, it was more strongly associated with EHR-MDD (OR [95 % CI] = 7.03 [6.44–7.44]) than with PQ-MDD (OR [95 % CI] = 4.41 [4.17–4.66]). On the other hand, when history of anxiety disorders was based on self-report (PQ), it was more strongly associated with PQ-MDD (OR [95 % CI] = 18.63 [17.68–19.63]) than with EHR-MDD (OR [95 % CI] = 6.81 [6.46–7.17]). However, irrespective of how history of anxiety disorders was defined, it was most strongly associated with the narrow case/control definition based on the intersection of both methods, i.e., cases with both PQ-MDD and EHR-MDD vs. controls with neither (OR [95 % CI] = 8.91 [8.34–9.53] for EHR-anxiety; OR [95 % CI] = 22.35 [20.98–23.82] for PQ-anxiety).
4. Discussion
In this analysis of MCB (Mayo Clinic Biobank) participants, identification of cases of MDD and controls was conducted using two methods with different information sources to compare their agreement, examine the relationship between method agreement and participant characteristics, and explore variations in MDD associations across four MDD case definitions. We explored how using EHR and questionnaire data to varying degrees in MDD case definitions, as would be expected from different large-scale studies, could affect associations with MDD. As anticipated, the prevalence of MDD differed between case identification methods and was higher when identifying MDD cases using self-report (PQ-MDD) than it was when cases were identified using participants’ EHR (EHR-MDD). The likelihood that an individual would be identified as a case by both methods (i.e., case status agreement) was differentially associated with sociodemographic, clinical, and well-being related characteristics, and varied between cases identified by each method. Furthermore, using the same information source to define both MDD and participant characteristics (e.g., EHR-based MDD with EHR-based characteristics) yielded stronger associations than when using different information sources (e.g., self-reported MDD with EHR-based characteristics). Our findings demonstrate how different methods to identify MDD can impact the selection of individuals from distinct sociodemographic groups and with different clinical characteristics, and how the collection of data across multiple information sources can influence associations observed in research studies.
In this study, we focused on several factors with a previously established relationship with MDD and explored how much the strength of their association with MDD varied across different case/control definitions. For all variables tested, the associations observed were strongest when using the narrow definition of MDD, based on the intersection of both methods used to identify MDD (i.e., cases defined as only those individuals with both PQ-MDD and EHR-MDD and controls defined as those with neither). Associations with case definitions of MDD based on a single method were highly affected by the information source used to define both the variables of interest and the case definitions. For instance, individuals with MDD frequently have comorbid anxiety disorders, and vice versa [34]. Here, the strength of the association between MDD case status and a history of anxiety disorders varied depending on the information source used to define them. Self-reported history of anxiety was much more strongly associated with self-reported MDD (PQ-MDD) than with EHR-MDD, just as EHR-based history of anxiety was more strongly associated with EHR-MDD than with PQ-MDD. This is particularly important since the method used to define MDD and variables of interest could be based on different sources of information, affecting post-hoc studies, meta-analyses, or research replication overall.
It is quite likely that the primary phenotype for MDD may not be completely captured by a single method of identifying MDD in EHR-linked biobanks, given its heterogeneity [15], [35]. Thus, care should be taken when comparing studies that use different methods to identify MDD, particularly when sources of information vary, as this could lead to comparing different subphenotypes of MDD, or when a single study uses multiple sources of information to ascertain MDD and/or collect variables of interest. As an example, Hyde et al. (2016) failed to replicate the genetic findings of the CONVERGE study [36], [37]. However, the participant sample for the CONVERGE study was recruited from a clinical setting and included participants with at least two depressive episodes confirmed by a clinical interview [36], whereas the study by Hyde et al. (2016) included a predominantly non-clinical sample ascertained using participant self-reports [37].
In our single biobank sample, a higher number of participants were identified as MDD cases by self-report (PQ-MDD; 23 %) than by EHR (EHR-MDD; 17 %). This is in line with reports from other biobanks. In the UK Biobank, using a questionnaire-based case definition resulted in roughly one third more participants identified as MDD cases than an EHR-based method [35]. Although the UK Biobank, unlike the MCB, is population-based, it is not representative of the general population, as participation was limited (5.5 % of those invited) and greater among women, older individuals, and those living in areas with lower socioeconomic deprivation [38]. One possible explanation for the greater number of MDD cases identified by a self-report is that individuals responding to a question about previous diagnoses of depression may endorse depressive episodes for which care was sought elsewhere or not at all (i.e., censored to the EHR-based method). Alternatively, it is possible for individuals to endorse depressive symptoms for which care was sought but for which the recorded diagnosis was not MDD but rather a different disorder such as adjustment disorder, mood disorder due to another medical condition, or substance/medication-induced depressive disorder [39]. Indeed, single-item questionnaire-based case definitions of MDD may be the least specific for the phenotype of interest [16].
Prior studies have reported that sociodemographic factors and health care utilization can affect the level of agreement between self-reported and administrative health data-derived diagnosis of depression [12], [40]. Here, participants identified as EHR-MDD or PQ-MDD cases were less likely to be identified as MDD cases by the other method if they identified as men or were older. Similarly, a lower likelihood of case status agreement across identification methods was observed in those with a lower burden of illness—as inferred from EHR data on the number of unique antidepressant classes prescribed, anxiety diagnostic codes, and ADHD diagnostic codes—better social and emotional well-being scores, or lack of depressive symptoms during enrollment (i.e., negative MDD screen). These observations are in line with a previous report that found agreement between self-report and administrative data was directly associated with depression burden. Contrary to our findings, however, men and older participants had higher rates of MDD case status agreement [12]. While our study population has different demographic characteristics than the population studied by Payette et al. (e.g., age and educational attainment), the different associations observed by each study could be due to the source of the information. In our study, EHR data were limited to care received at Mayo Clinic. By contrast, Payette et al. (2020) used administrative health data from governmental health databases within a single provincial health system in Canada, which may be less prone to missing information [12]. Hence, it may also explain their higher frequency of depression ascertained by administrative health data than by self-report.
Among individuals with a recorded diagnosis of MDD in the EHR (i.e., EHR-MDD), those with the longest time elapsed since their last recorded depressive episode and those with the greatest length of EHR had much lower odds of subsequently self-reporting depression when asked about it on a questionnaire (i.e., PQ-MDD). These observations may be explained by recall bias, as both analyses were adjusted for age and the model testing the recency of last recorded depressive episode was further adjusted for EHR length. In addition, evidence suggests that recent depressive episodes are more likely to be remembered and acknowledged in a questionnaire than distant episodes [13]. At the same time, those with the greatest length of EHR would have a longer period during which to record MDD, allowing inclusion of more distant episodes. Only approximately 45 % of participants identified as EHR-MDD cases had MDD recorded in their EHR within the year prior to study enrollment.
Importantly, race was associated with MDD case status agreement independent of method, albeit differently for each method. Among those with a history of MDD recorded in their EHR (EHR-MDD), individuals identifying their race as Asian or other had a lower likelihood of subsequently self-reporting MDD than White individuals—i.e., not endorsing a prior diagnosis of MDD despite records showing otherwise. This underreporting could possibly be mediated by social factors. Previous literature highlights lower rates of mental health care among Asian individuals partially attributed to perceived need for care [41] and a possible moderation effect of social cohesion on the significantly lower likelihood of MDD diagnosis among Asian individuals when compared to other racial and ethnic groups in a population-based cohort [42]. However, further research is needed to understand these observations. On the other hand, individuals from understudied racial and ethnic groups that self-reported MDD tended to have a lower likelihood of having MDD recorded in their EHR, yet these differences were not statistically significant. Finally, educational attainment was differentially associated with MDD case status agreement. Among individuals with history of MDD in their EHR, having a postgraduate education resulted in a greater likelihood to self-report MDD. However, among those that self-reported, postgraduate education was associated with a lower likelihood of having EHR-MDD. This difference could be related to a lower burden of disease among those with greater educational attainment [43] and, thus, less care-seeking, or care being received outside of Mayo Clinic, yet further studies are needed to understand these associations.
We did not observe statistically significant differences in the strength of the association between MDD case status and MDD-PRS across distinct MDD case/control definitions. While a more in-depth exploration of the genetic architecture of PQ-MDD and EHR-MDD would allow a better understanding of this finding, such analyses were considered beyond the scope of the current study and would be underpowered with our current sample size, in certain scenarios (e.g., creating a method-specific PRS to evaluate prediction of the other method). Notwithstanding, recent large-scale studies have explored the genetic architecture of distinct MDD definitions, comparing methods of identifying MDD similar to those employed in our study. Cai et al. (2020) systematically compared genetic signals from minimally phenotyped depression, including self-report, with those from stricter definitions, including EHR-derived ICD codes, finding low specificity for minimally phenotyped definitions [15]. Furthermore, Huang et al. (2024) later reported finding significant genetic differences even between concordant symptom domains collected through different rating instruments, highlighting the high degree of variability introduced by different ascertainment methods [44]. Our findings on the low case agreement between methods of identifying MDD and the differences in characteristics associated with case agreement add to the increasing body of literature showing the phenotypic and genetic implications of MDD case definition.
Overall, our findings highlight how sociodemographic groups may not be equally represented by different methods of identifying MDD. Notwithstanding, a priori information on the sociodemographic and clinical composition of a population being studied would allow researchers to employ analytic methods to reduce potential sampling bias such as inverse probability weighted analysis [45]. Additionally, knowledge of this information may allow tailoring the identification method(s) to those with the lowest likelihood of missing cases in settings where multiple sources of information are available. For example, in a study population comprising individuals with characteristics associated with a lower likelihood of identification by both methods tested in this study (e.g., older males), a broader case definition for MDD using information from different sources (i.e., MDD cases defined as either EHR-MDD or PQ-MDD) would have a lower risk of missing cases. On the other hand, for a population composed of individuals with characteristics that are associated with a lower likelihood to endorse MDD by self-report once they have been identified as having EHR-MDD (e.g., individuals whose last recorded depressive episode occurred over 5 years ago), an EHR-based method of identifying MDD would suffice, granted sufficient length of EHR.
5. Limitations
Our study has important limitations that must be acknowledged. First, the Mayo Clinic Biobank is a relatively homogeneous cohort composed of mostly older, White individuals receiving care at Mayo Clinic. As such, our findings may not be generalizable across diverse study settings. Second, the lack of more detailed measures of depressive symptom severity or other measures of psychopathology limits our ability to explore clinical associations with the different methods to identify MDD or its case definitions. Third, code-matching for diseases of interest such as anxiety disorders, ADHD, and substance abuse, may be biased as participants without ICD codes for depression may be less likely to present or report other ICD codes—and vice versa—for non-medical reasons. Additionally, the list of ICD codes used in this study to identify depression using EHRs included non-specific diagnoses such as other depressive episodes and unspecified major depressive disorder, which might have affected its associations with the variables of interest. Fourth, the EHR of any given individual does not represent the totality of their medical history. Instead, a single individual’s EHR contains data for the time during which they received care within a given health system. As a result, health care provided outside of Mayo Clinic was not included in our data. Additionally, given that Mayo Clinic’s EHR data are available since 1995, we do not have full EHR data, especially for older individuals. On the other hand, the participant questionnaire asked individuals about any prior lifetime diagnosis of depression, extending the timeframe past that evaluated by EHR. Notwithstanding, we conducted a sensitivity analysis of the agreement measures by restricting to participants living in Olmsted County, MN, where Mayo Clinic Rochester is located, and six surrounding counties, where EHR coverage is highest. While case agreement was higher, overall and control agreement were only marginally higher (Table S4). Fifth, this study did not include a “gold standard” definition of MDD (i.e., diagnosis by structured clinical interview). As such, while we are able to compare self-report and EHR-based definitions of MDD, we cannot compare these definitions with strictly defined MDD, limiting our ability to fully understand the extent of their differences. Lastly, our study defined EHR-MDD using ICD codes only. Even though identification of MDD could be improved using more nuanced EHR data such as multiple data elements within the EHR (e.g., medication usage) or unstructured clinical data extracted through natural language processing, we decided to only use ICD codes, as they have been harmonized across different institutions and are regularly used to identify MDD in large datasets. In addition, our study did not distinguish among providers entering ICD codes (e.g., primary care vs. specialty mental health care), which may have different coding practices. However, the goal of this study was to compare minimally phenotyped MDD case definitions.
6. Conclusions
In summary, our study builds upon existing literature to highlight the importance of considering the differences between methods to identify MDD, rather than advocating for a single “best” method. Our findings emphasize that the choice of method may substantially affect the sociodemographic representation and clinical makeup of participant samples. Additionally, we stress that careful consideration should be given to the information sources used by each method to identify MDD, especially when analyzing a single dataset with multiple information sources available or when comparing existing evidence from different studies.
CRediT authorship contribution statement
Jorge A. Sanchez-Ruiz: Conceptualization, Methodology, Validation, Investigation, Writing - Original Draft, Writing - Review & Editing. Nicolas A. Nuñez: Conceptualization, Validation, Writing - Original Draft, Writing - Review & Editing. Gregory D. Jenkins: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data Curation, Writing - Original Draft, Visualization, Writing - Review & Editing. Brandon J. Coombes: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data Curation, Writing - Review & Editing. Lauren A. Lepow: Conceptualization, Validation, Writing - Review & Editing. Braja Gopal Patra: Conceptualization, Validation, Writing - Review & Editing. Mark Olfson: Conceptualization, Funding acquisition, Validation, Writing - Review & Editing. J. John Mann: Conceptualization, Funding acquisition, Validation, Writing - Review & Editing. Myrna M. Weissman: Conceptualization, Validation, Writing - Review & Editing. Jyotishman Pathak: Conceptualization, Funding acquisition, Validation, Writing - Review & Editing. Alexander Charney: Conceptualization, Funding acquisition, Validation, Writing - Review & Editing. Joanna M. Biernacka: Conceptualization, Methodology, Validation, Funding acquisition, Investigation, Resources, Supervision, Writing - Original Draft, Writing - Review & Editing. Euijung Ryu: Conceptualization, Methodology, Validation, Investigation, Resources, Supervision, Writing - Original Draft, Writing - Review & Editing.
Funding
This study was supported by the National Institute of Mental Health grants R01MH121924, R01MH121923, R01MH121922, and R01MH121921. The Mayo Clinic Biobank and generation of genetic data were supported in part by Mayo Clinic Center for Individualized Medicine. We acknowledge Regeneron Genetics Center* for generating the genetic data for Mayo Clinic Biobank participants and their contributions to this manuscript. We also acknowledge the Mayo Clinic Biobank research teams as well as the patients who consented to participate in these research programs.
*Regeneron Genetics Center members and affiliations are available in Supplemental information.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Dr. Weissman has received funding from NIMH and Columbia University Institute for Developmental Sciences, receives book royalties from Perseus Press and Oxford Press, and serves on the editorial board of the Journal of Mood & Anxiety Disorders. None of these represent a conflict of interest. Dr. Mann receives royalties for commercial use of the C-SSRS from the Research Foundation for Mental Hygiene and from Columbia University for the Columbia Pathways App. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
A member of the Editorial Board is an author of this article. Editorial Board members are not involved in decisions about papers which they have written themselves or have been written by family members or colleagues or which relate to products or services in which the editor has an interest. Any such submission is subject to all of the journal’s usual procedures, with peer review handled independently of the relevant editor and their research groups.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.xjmad.2025.100136.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Kessler R.C., Ormel J., Petukhova M., McLaughlin K.A., Green J.G., Russo L.J., et al. Development of Lifetime Comorbidity in the World Health Organization World Mental Health Surveys. Arch Gen Psychiatry. 2011;68(1):90–100. doi: 10.1001/archgenpsychiatry.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GBD 2019 Mental Disorders Collaborators Global, regional, and national burden of 12 mental disorders in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Psychiatry. 2022;9(2):137–150. doi: 10.1016/S2215-0366(21)00395-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malhi G.S., Mann J.J. Depression. Lancet. 2018;392(10161):2299–2312. doi: 10.1016/S0140-6736(18)31948-2. [DOI] [PubMed] [Google Scholar]
- 4.Cai N., Choi K.W., Fried E.I. Reviewing the genetics of heterogeneity in depression: operationalizations, manifestations and etiologies. Hum Mol Genet. 2020;29(R1):R10–R18. doi: 10.1093/hmg/ddaa115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beesley L.J., Salvatore M., Fritsche L.G., Pandit A., Rao A., Brummett C., et al. The emerging landscape of health research based on biobanks linked to electronic health records: Existing resources, statistical challenges, and potential opportunities. Stat Med. 2020;39(6):773–800. doi: 10.1002/sim.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.First M.B., Williams J.B.W., Karg R.S., Spitzer R.L. American Psychiatric Association Publishing; Arlington, VA: 2016. SCID-5-CV: Structured Clinical Interview for DSM-5 Disorders: clinician version; p. 95. (p) [Google Scholar]
- 7.Frye M.A., McElroy S.L., Fuentes M., Sutor B., Schak K.M., Galardy C.W., et al. Development of a bipolar disorder biobank: differential phenotyping for subsequent biomarker analyses. Int J Bipolar Disord. 2015;3(1):30. doi: 10.1186/s40345-015-0030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linder J.E., Bastarache L., Hughey J.J., Peterson J.F. The role of electronic health records in advancing genomic medicine. Annu Rev Genom Hum Genet. 2021;22(1):219–238. doi: 10.1146/annurev-genom-121120-125204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glicksberg B.S., Johnson K.W., Dudley J.T. The next generation of precision medicine: observational studies, electronic health records, biobanks and continuous monitoring. Hum Mol Genet. 2018;27(R1):R56–R62. doi: 10.1093/hmg/ddy114. [DOI] [PubMed] [Google Scholar]
- 10.Wolford B.N., Willer C.J., Surakka I. Electronic health records: the next wave of complex disease genetics. Hum Mol Genet. 2018;27(R1):R14–R21. doi: 10.1093/hmg/ddy081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gianfrancesco M.A., Goldstein N.D. A narrative review on the validity of electronic health record-based research in epidemiology. BMC Med Res Method. 2021;21(1):234. doi: 10.1186/s12874-021-01416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Payette Y., Moura C.S. de, Boileau C., Bernatsky S., Noisel N. Is there an agreement between self-reported medical diagnosis in the CARTaGENE cohort and the Québec administrative health databases? IJPDS [Internet]. 2020 Mar 26 [cited 2023 Jan 17];5(1). Available from: 〈https://ijpds.org/article/view/1155〉. [DOI] [PMC free article] [PubMed]
- 13.Short M.E., Goetzel R.Z., Pei X., Tabrizi M.J., Ozminkowski R.J., Gibson T.B., et al. How Accurate Are Self-reports? Analysis of Self-reported Health Care Utilization and Absence When Compared with Administrative Data. J Occup Environ Med. 2009;51(7):786–796. doi: 10.1097/JOM.0b013e3181a86671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolinsky F.D., Miller T.R., An H., Geweke J.F., Wallace R.B., Wright K.B., et al. Hospital episodes and physician visits: the concordance between self-reports and medicare claims. Med Care. 2007;45(4):300–307. doi: 10.1097/01.mlr.0000254576.26353.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai N., Revez J.A., Adams M.J., Andlauer T.F.M., Breen G., Byrne E.M., et al. Minimal phenotyping yields genome-wide association signals of low specificity for major depression. Nat Genet. 2020;52(4):437–447. doi: 10.1038/s41588-020-0594-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flint J. The genetic basis of major depressive disorder. Mol Psychiatry [Internet]. 2023 Jan 26 [cited 2023 Feb 16]; Available from: 〈https://www.nature.com/articles/s41380-023-01957-9〉. [DOI] [PMC free article] [PubMed]
- 17.The All of Us Research Program Investigators. The “All of Us” Research Program. N Engl J Med. 2019;381(7):668–676. [DOI] [PMC free article] [PubMed]
- 18.Eriksson N., Macpherson J.M., Tung J.Y., Hon L.S., Naughton B., Saxonov S., et al. Web-based, participant-driven studies yield novel genetic associations for common traits. gibson g, editor. PLoS Genet. 2010;6(6) doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakravarthy R., Stallings S.C., Williams M., Hollister M., Davidson M., Canedo J., et al. Factors influencing precision medicine knowledge and attitudes. PLoS One. 2020;15(11) doi: 10.1371/journal.pone.0234833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richesson R.L., Rusincovitch S.A., Wixted D., Batch B.C., Feinglos M.N., Miranda M.L., et al. A comparison of phenotype definitions for diabetes mellitus. J Am Med Inf Assoc. 2013;20(e2):e319–e326. doi: 10.1136/amiajnl-2013-001952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson J.E., Ryu E., Johnson K.J., Koenig B.A., Maschke K.J., Morrisette J.A., et al. The mayo clinic biobank: a building block for individualized medicine. Mayo Clin Proc. 2013;88(9):952–962. doi: 10.1016/j.mayocp.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olson J.E., Ryu E., Hathcock M.A., Gupta R., Bublitz J.T., Takahashi P.Y., et al. Characteristics and utilisation of the Mayo Clinic Biobank, a clinic-based prospective collection in the USA: cohort profile. BMJ Open. 2019;9(11) doi: 10.1136/bmjopen-2019-032707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryu E., Jenkins G.D., Wang Y., Olfson M., Talati A., Lepow L., et al. The importance of social activity to risk of major depression in older adults. Psychol Med. 2021:1–9. doi: 10.1017/S0033291721004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denny J.C., Bastarache L., Ritchie M.D., Carroll R.J., Zink R., Mosley J.D., et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–1111. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu P., Gifford A., Meng X., Li X., Campbell H., Varley T., et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med Inf. 2019;7(4) doi: 10.2196/14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen M.D., Ryan D.H., Apovian C.M., Ard J.D., Comuzzie A.G., Donato K.A., et al. 2013 AHA/ACC/TOS guideline for the management of overweight and obesity in adults: a report of the American College of Cardiology/American Heart Association task force on practice guidelines and the obesity society. Circulation. 2014;129(25__2) doi: 10.1016/j.jacc.2013.11.004. 〈https://www.ahajournals.org/doi/10.1161/01.cir.0000437739.71477.ee〉 cited 2024 Sep 5]; [DOI] [PubMed] [Google Scholar]
- 27.Charlson M.E., Pompei P., Ales K.L., MacKenzie C.R. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 28.Gelfman S., Moscati A., Huergo S.M., Wang R., Rajagopal V., Parikshak N., et al. A large meta-analysis identifies genes associated with anterior uveitis. Nat Commun. 2023;14(1):7300. doi: 10.1038/s41467-023-43036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coombes B.J., Landi I., Choi K.W., Singh K., Fennessy B., Jenkins G.D., et al. The genetic contribution to the comorbidity of depression and anxiety: a multi-site electronic health records study of almost 178 000 people. Psychol Med. 2023:1–7. doi: 10.1017/S0033291723000983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard D.M., Adams M.J., Clarke T.K., Hafferty J.D., Gibson J., Shirali M., et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22(3):343–352. doi: 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Privé F., Arbel J., Vilhjálmsson B.J. In: Schwartz R., editor. Vol. 36. 2021. LDpred2: better, faster, stronger; pp. 5424–5431. (Bioinformatics). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbara A.M., Loeb M., Dolovich L., Brazil K., Russell M.L. Patient self-report and medical records: measuring agreement for binary data. Can Fam Physician. 2011;57(6):737–738. [PMC free article] [PubMed] [Google Scholar]
- 33.Cicchetti D.V., Feinstein A.R. High agreement but low kappa: II. resolving the paradoxes. J Clin Epidemiol. 1990;43(6):551–558. doi: 10.1016/0895-4356(90)90159-m. [DOI] [PubMed] [Google Scholar]
- 34.Lamers F., Van Oppen P., Comijs H.C., Smit J.H., Spinhoven P., Van Balkom A.J.L.M., et al. Comorbidity patterns of anxiety and depressive disorders in a large cohort study: the netherlands study of depression and anxiety (NESDA) J Clin Psychiatry. 2011 15;72(03):341–348. doi: 10.4088/JCP.10m06176blu. [DOI] [PubMed] [Google Scholar]
- 35.Glanville K.P., Coleman J.R.I., Howard D.M., Pain O., Hanscombe K.B., Jermy B., et al. Multiple measures of depression to enhance validity of major depressive disorder in the UK Biobank. BJPsych Open. 2021;7(2) doi: 10.1192/bjo.2020.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CONVERGE consortium. Cai N., Bigdeli T.B., Kretzschmar W., Li Y., Liang J., et al. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523(7562):588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyde C.L., Nagle M.W., Tian C., Chen X., Paciga S.A., Wendland J.R., et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48(9):1031–1036. doi: 10.1038/ng.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fry A., Littlejohns T.J., Sudlow C., Doherty N., Adamska L., Sprosen T., et al. Comparison of sociodemographic and health-related characteristics of UK biobank participants with those of the general population. Am J Epidemiol. 2017;186(9):1026–1034. doi: 10.1093/aje/kwx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.American Psychiatric Association . American Psychiatric Association Publishing; Washington, D.C.: 2022. Diagnostic and statistical manual of mental disorders. 5th ed., text rev. [Google Scholar]
- 40.Edwards J., Thind A., Stranges S., Chiu M., Anderson K.K. Concordance between health administrative data and survey-derived diagnoses for mood and anxiety disorders. Acta Psychiatr Scand. 2020;141(4):385–395. doi: 10.1111/acps.13143. [DOI] [PubMed] [Google Scholar]
- 41.Yang K.G., Rodgers C.R.R., Lee E., Lê Cook B. Disparities in mental health care utilization and perceived need among Asian Americans: 2012–2016. PS. 2020;71(1):21–27. doi: 10.1176/appi.ps.201900126. [DOI] [PubMed] [Google Scholar]
- 42.Kammer-Kerwick M., Cox K., Purohit I., Watkins S.C. In: Lautarescu A., editor. Vol. 1. 2024. The role of social determinants of health in mental health: an examination of the moderating effects of race, ethnicity, and gender on depression through the all of us research program dataset. (PLOS Ment Health). [Google Scholar]
- 43.van der Veen D.C., van Zelst W.H., Schoevers R.A., Comijs H.C., Voshaar R.C.O. Comorbid anxiety disorders in late-life depression: results of a cohort study. Int Psychogeriatr. 2015;27(7):1157–1165. doi: 10.1017/S1041610214002312. [DOI] [PubMed] [Google Scholar]
- 44.Huang L., Tang S., Rietkerk J., Appadurai V., Krebs M.D., Schork A.J., et al. Polygenic analyses show important differences between major depressive disorder symptoms measured using various instruments. Biol Psychiatry. 2024;95(12):1110–1121. doi: 10.1016/j.biopsych.2023.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schoeler T., Speed D., Porcu E., Pirastu N., Pingault J.B., Kutalik Z. Participation bias in the UK Biobank distorts genetic associations and downstream analyses. Nat Hum Behav. 2023;7(7):1216–1227. doi: 10.1038/s41562-023-01579-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material