Abstract
The periaxin gene (PRX) encodes two PDZ-domain proteins, L- and S-periaxin, that are required for maintenance of peripheral nerve myelin. Prx−/− mice develop a severe demyelinating peripheral neuropathy, despite apparently normal initial formation of myelin sheaths. We hypothesized that mutations in PRX could cause human peripheral myelinopathies. In accordance with this, we identified three unrelated Dejerine-Sottas neuropathy patients with recessive PRX mutations—two with compound heterozygous nonsense and frameshift mutations, and one with a homozygous frameshift mutation. We mapped PRX to 19q13.13-13.2, a region recently associated with a severe autosomal recessive demyelinating neuropathy in a Lebanese family (Delague et al. 2000) and syntenic to the location of Prx on murine chromosome 7 (Gillespie et al. 1997).
Introduction
Dejerine-Sottas neuropathy (DSN [MIM 145900]) and Charcot-Marie-Tooth disease type 1 (CMT1 [MIM 118200]) represent genetically heterogeneous inherited peripheral myelinopathies. These conditions constitute part of a spectrum of neuropathy phenotypes ranging in severity from congenital hypomyelinating neuropathy (CHN [MIM 605253]) to adult-onset hereditary neuropathy with liability to pressure palsies (HNPP [MIM 162500]) (Lupski and Garcia 2001). At least 15 genetic loci and six genes have been associated with these disorders; identified genetic causes include altered dosage of peripheral myelin protein 22 (PMP22) and mutations in one of the following genes: PMP22, the gap junction protein β1 gene (GJB1), the myelin protein zero gene (MPZ), the early growth response gene 2 (EGR2), the myotubularin-related protein 2 gene (MTMR2), or the N-myc downstream-regulated gene 1 (NDRG1) (Lupski and Garcia 2001). These genes encode proteins of diverse functions: compact myelin structural proteins (MPZ and PMP22), a noncompact myelin gap junction protein (GJB1), signal transduction proteins (NDRG1 and MTMR2), and a transcription factor for late myelin genes (EGR2). Both dominant mutant alleles (PMP22, GJB1, MPZ, and EGR2) and recessive ones (MTMR2, NDRG1, PMP22, and EGR2) have been described. Historically considered an autosomal recessive disorder (Dejerine and Sottas 1893), DSN has been associated predominantly, until this report, with de novo dominant mutations in PMP22 (Roa et al. 1993), MPZ (Hayasaka et al. 1993), or EGR2 (Timmerman et al. 1999), although rare recessive mutations in PMP22 have also been reported (Parman et al. 1999; Lupski 2000).
We hypothesized that the human orthologue of murine and rat Prx, which expresses L- and S-periaxin by alternative intron retention (Dytrych et al. 1998), is a good candidate gene for human inherited myelinopathies. In murine embryonic Schwann cells, L-periaxin initially is concentrated in the nuclei but redistributes to the plasma membrane—predominantly adaxonal with initiation of myelination, and then to the abaxonal, Schmidt-Lanterman incisure, and paranodal membranes with maturation of the myelin sheath (Scherer et al. 1995; Sherman and Brophy 2000). In addition, L-periaxin expression recapitulates this pattern following crush injury (Scherer et al. 1995). This shift in periaxin localization after the spiralization phase of myelination suggests that periaxin participates in membrane-protein interactions that are required to stabilize the mature myelin sheath. As a cytoskeleton-associated protein, L-periaxin may mediate such stabilization by facilitating integration of extracellular signaling through the cytoskeleton, which is essential for changes in Schwann cell shape and regulation of gene expression during axonal ensheathment (Fernandez-Valle et al. 1997; Tapon and Hall 1997). Such a signaling function is supported by the observation that L-periaxin contains a PDZ motif and a nuclear localization signal (Dytrych et al. 1998; Sherman and Brophy 2000). The PDZ domain, which consists of a nearly 90–amino acid protein-binding motif that interacts with the carboxy termini of plasma membrane proteins and with the cortical cytoskeleton, is named after the three proteins in which it was first identified: the postsynaptic density protein PSD-95, the Drosophila discs large tumor suppressor, and the tight junction-associated protein ZO-1 (Fanning and Anderson 1999). This domain has been implicated in the assembly of signaling complexes at sites of cell-cell contact.
Confirming the necessity of periaxin for maintenance of the myelin sheath, Gillespie et al. (2000) recently demonstrated that Prx−/− mice ensheath and myelinate peripheral axons apparently normally but subsequently develop a severe demyelinating neuropathy associated with allodynia (pain from non-noxious stimuli) and hyperalgesia (hypersensitivity to pain).
We report PRX mutations causing recessive DSN and thus demonstrate the necessity of periaxin for maintenance of human myelin sheaths. Furthermore, consistent with the murine model, genotype-phenotype correlation shows that these patients have a more pronounced sensory involvement than do patients with classical DSN or CMT.
Subjects and Methods
Human Subjects
All patients referred to this study by their primary physician or neurologist received informed consent approved by the Institutional Review Board of Baylor College of Medicine. We isolated DNA from the peripheral blood of each patient and established lymphoblastoid cell lines.
Human PRX cDNA Sequence
We defined the human PRX cDNA sequence corresponding to L-periaxin by sequencing two expressed-sequence tag (EST) clones (AW105547 and AW337783) from the IMAGE consortium, sequencing reverse transcriptase (RT) PCR and 5′ rapid amplification of cDNA ends (RACE) products from human femoral nerve total RNA, and sequencing 150–190 control chromosomes across all coding exons. We isolated human femoral nerve total RNA using Trizol (Life Technologies) (Chomczynski and Sacchi 1987). Prior to using the RNA for RT-PCR (One-Step RT-PCR or Superscript II RNase H Reverse Transcriptase, Life Technologies) or 5′ RACE (GeneRacer Kit, Invitrogen), we treated it with ribonuclease-free deoxyribonuclease I (Life Technologies) to remove contaminating DNA. We cloned the products of the 5′ RACE reaction into the TA vector (Invitrogen) to separate and sequence the various products.
Mapping PRX
We screened the published rat Prx cDNA sequence (Z29649) through the high-throughput genomic sequence database, using the BLAST algorithm. BAC clone CTC-492K19 (AC010271) exhibited 83% identity to the cDNA sequence. Using electronic PCR (ePCR), we identified nine chromosome 19q STSs in BAC CTC-492K19 and used these to place it on the chromosome 19 physical map. We also screened the RPCI-11 BAC library with an overgo primer probe for PRX, isolated two BACs (104E13 and 4K5) containing all coding exons of PRX, and used these to map PRX by FISH.
We performed FISH on metaphase preparations of human peripheral blood lymphocytes according to a modified procedure of Shaffer et al. (1997). In brief, 200 ng of isolated BAC (104E13 and 4K5) DNA was labeled by nick translation reaction using digoxigenin and 50 ng of chromosome 19q13.4 control cosmid probe (F13141 from Lawrence Livermore National Laboratory flow-sorted chromosome 19–specific cosmid library) using biotin (Boehringer Mannheim). Biotin was detected with fluorescein isothiocyanate-avidin (Vector Labs), and digoxigenin was detected with rhodamine–anti-digoxigenin antibodies (Sigma). Chromosomes were counterstained with DAPI diluted in Vectashield antifade (Vector Labs). Cells were viewed under a Zeiss Axioskop fluorescence microscope equipped with an appropriate filter combination. Monochromatic images were captured and pseudocolored using MacProbe 4.2.2 (Perceptive Scientific Instruments) on a Power Macintosh G4 system.
Mutation Screening
By aligning the human genomic sequence from BAC clone CTC-492K19 with the rat Prx cDNA, we identified all coding exons; we confirmed each exon after characterization of the human cDNAs. Using the Primer v3 program, we designed primers to amplify exons and intronic splice junctions and then screened amplified PCR products from patient genomic DNA for mutations, using the WAVE DNA-fragment analysis system (Transgenomic). In brief, by PCR we amplified the coding region of PRX from 50 ng of patient genomic DNA, using the primers listed in table 1 and Qiagen HotStarTaq. All forward primers had a −21 M13 primer tail (TGTAAAACGACGGCCAGT) and all reverse primers had a M13 reverse tail (CAGGAAACAGCTATGACC). We generated each PCR product, except that corresponding to exon 5, with the following conditions: 95°C for 15 min, 40 cycles of amplification (95°C for 30 s, 60°C for 30 s, and 72°C for 1 min), and 72°C for 7 min. For exon 5, we added 1.5 U of Qiagen HotStarTaq, following the above protocol, and then performed an additional 15 cycles of amplification. To prepare the PCR products for DHPLC analysis, we pooled the products from every two patients and denatured the products at 95°C for 5 min and reannealed them by decreasing the temperature from 95°C to 20°C over a period of 50 min. We analyzed these PCR products for heteroduplexes by means of denaturing high-performance liquid chromatography (DHPLC), using a linear acetonitrile gradient (flow rate of 0.9 ml/min, 2% slope [buffer A, 0.1 M triethylammoniumacetate; buffer B, 0.1 M triethylammoniumacetate/25% acetonitrile]; for column temperatures, see table 1); we determined optimal column temperatures empirically and identified potential heteroduplexes by visual inspection of elution chromatograms.
Table 1.
Primer Pairs Used for Amplification of the PRX Coding Region and Optimized DHPLC Column Temperatures for Each Amplicon
| Primer Name | Primer Pair | DHPLCColumnTemperature(°C) |
| Exon 4: | ||
| Forward | GTAAGCATGGCCTCCACCT | 63 |
| Reverse | CTCCTTGCTGCCCTAGTCTG | |
| Exon 5: | ||
| Forward | ACCTGTTGAGCGCCAATG | 66 |
| Reverse | CCCAAGGCAGATTCCTAACC | |
| Exon 6: | ||
| Forward | CGTGCAAGTGGGCAGAACTA | 65 |
| Reverse | TGACAAGACAGAGGGCAAGG | |
| Exon 7a: | ||
| Forward | AATACCAGGTGGGGCTCTTC | 63 |
| Reverse | CTCTAGGCAGGGAAGTGTGG | |
| Exon 7b: | ||
| Forward | AGCCGTGGGAATCCAGGT | 63 |
| Reverse | TGACACTTTGGGCAGCTCTA | |
| Exon 7c: | ||
| Forward | CAGAGGTTCGACTCCCAGAG | 62 |
| Reverse | GCCATCTCAGGCATTTTAGG | |
| Exon 7d: | ||
| Forward | CTGAGGTGAAACTCCCGAAG | 63 |
| Reverse | GCAGAGTGAGAGAGGGGACA | |
| Exon 7e: | ||
| Forward | AAGCTAGGGAGGGCAGAGTC | 63 |
| Reverse | AACTTGGGGAGAGCAAACCT | |
| Exon 7f: | ||
| Forward | CCTCAGGCAAGGTAGAGGTG | 63 |
| Reverse | GTCACGGTGGGCATCTTAAA | |
| Exon 7g: | ||
| Forward | CAGGCTACAGGGTTCAGGTG | 65 |
| Reverse | TTCTCTCTGACGGGGGACTT | |
| Exon 7h: | ||
| Forward | GTCCGCTTGCCACGTGTAG | 62 |
| Reverse | GTACAGGCACTCCTGCCAGA | |
| S-PRX C: | ||
| Forward | CCGAGCCTTACAAAGTCTCCT | NDa |
| Reverse | AGTTTGGGGCAGAGAGGAAG |
ND = not determined.
Using the Qiagen 96-PCR purification kit (Qiagen), we purified patient PCR products having an abnormal elution profile and appropriate PCR products from relatives' chromosomes and control chromosomes and sequenced them with dye-primer chemistry (PE Applied Biosystems), using an ABI377 automated sequencer (PE Applied Biosystems). We aligned the resulting sequences and evaluated mutations with the Sequencher sequence alignment program (ACGT Codes). We numbered the PRX cDNA sequence beginning with the adenine of the presumed initiating methionine and described mutations according to den Dunnen and Antonarakis (2000).
Results
Mapping and Characterization of PRX
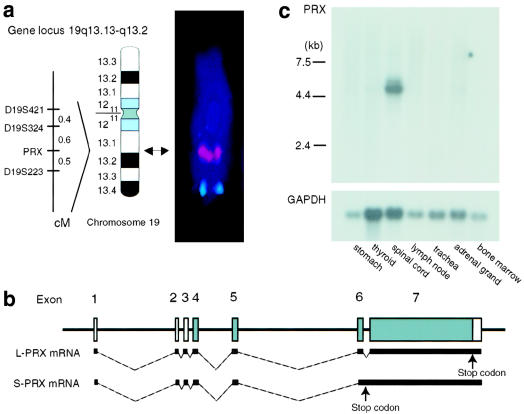
We mapped PRX in the human genome, defined its cDNA sequence and gene structure, and subsequently evaluated the tissue expression profile of PRX mRNA. By FISH and ePCR (Schuler 1997), the BAC containing PRX maps to chromosome 19q13.13-q13.2 between D19S324 and D19S223 (fig. 1a). This places PRX within a recently mapped interval for an autosomal recessive myelinopathy (Delague et al. 2000). Sequencing of RT-PCR and 5′ RACE products from femoral nerve mRNA and available EST clones defined two PRX transcripts of 4853 and 5502 bp, excluding the polyA tails. The shorter mRNA is transcribed from seven exons, and the deduced coding sequence extends from exon 4 through exon 7 (fig. 1b). The longer transcript arises by retention of intron 6 (figs. 1b, 1c, and 2); this introduces a stop codon and results in a truncated protein with an intron-encoded carboxyl terminus of 21 amino acids.
Figure 1.
Mapping of PRX and expression of PRX mRNA. a, By ePCR, BAC CTC-492K19, which contains PRX, maps between D19S324 and D19S223. We confirmed this by metaphase FISH; cohybridization with BAC RPCI-11 104E13 (red) and chromosome 19 control cosmid F13141 (green) assigned PRX to 19q13.13-q13.2 (arrow, ideogram in accord with the International System for Human Cytogenetic Nomenclature [1995]). b, Diagram showing the two PRX mRNAs resulting from alternative retention of intron 6. The large periaxin protein (L-PRX) is encoded by the shorter spliced mRNA, and the smaller periaxin protein (S-PRX) is encoded by the longer mRNA retaining intron 6. Coding regions are shaded. c, Northern analysis showing that both the 5.1- and 5.6-kb PRX mRNAs were most abundant in spinal cord, a tissue with many peripheral nerve roots.
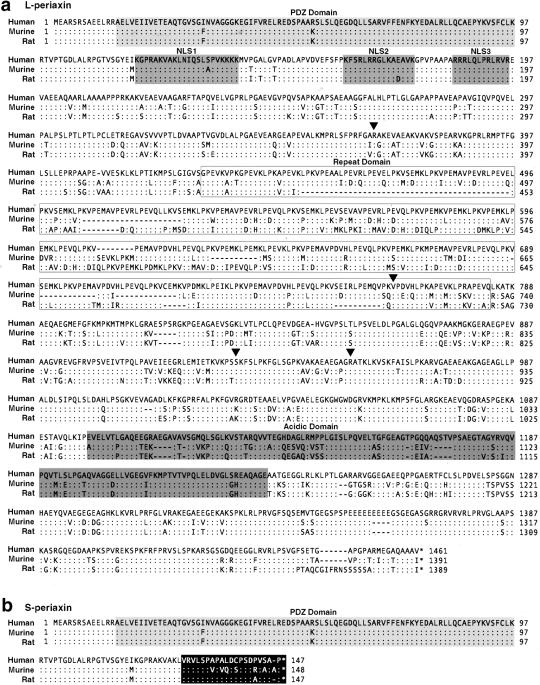
As observed in mice and rats, the amino acid sequence deduced from the shorter cDNA sequence contains a PDZ domain (amino acids 14–98), a highly basic domain (amino acids 118–194) that functions as a nuclear localization signal in mice, a repeat domain (amino acids 400–700), and an acidic domain (amino acids 1098–1235) (figs. 1b and 2) (Gillespie et al. 1994; Dytrych et al. 1998; Sherman and Brophy 2000). The amino acid sequence deduced from the longer cDNA sequence contains only the PDZ motif. Hybridization of several Clontech multitissue northern blots with a probe from exon 7 revealed expression of a 5.1-kb PRX mRNA in all tissues examined; mRNA from spinal cord, a tissue with many peripheral nerve roots, showed strongest hybridization of 5.1- and 5.6-kb bands (fig. 1c). In contrast to the nearly equal expression of each mRNA in mice (Dytrych et al. 1998), the 5.6-kb mRNA appears less abundant in humans. RT-PCR confirmed the peripheral nerve–tissue predominant expression (data not shown).
Figure 2.
Comparison of human, murine, and rat L-periaxin (a) and S-periaxin (b) amino acid sequences. a, Human L-periaxin has ∼78% and ∼73% sequence identity with the murine and rat proteins, respectively. The PDZ domain, tripartite nuclear localization signal (NLS1, NLS2, and NLS3), repeat domain, and acidic domain previously characterized in mice and rats are conserved in humans. Arrowheads indicate mutations identified in patients. b, S- and L-periaxin share a common amino terminal, but retention of intron 6 in the mRNA encoding S-periaxin results in a truncated protein with 20 amino acids encoded within the intron (blackened box). Identical amino acids are indicated by a colon (:), gaps by a dash (—) and stop codons by an asterisk (*).
PRX Mutation Analysis in Neuropathy Patients
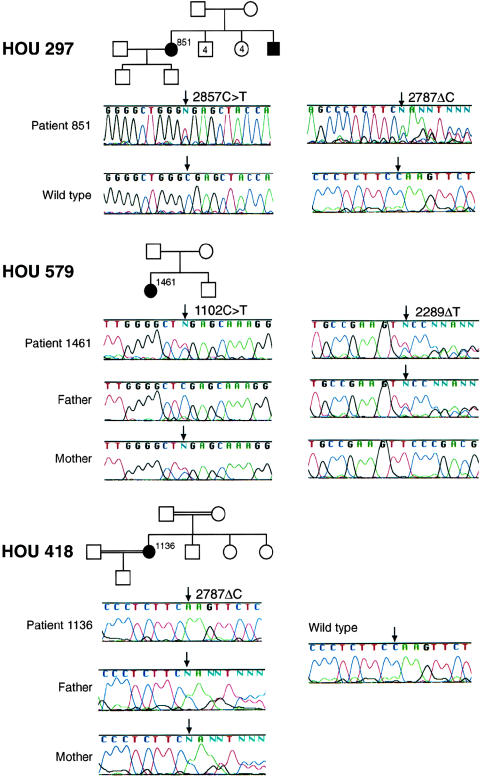
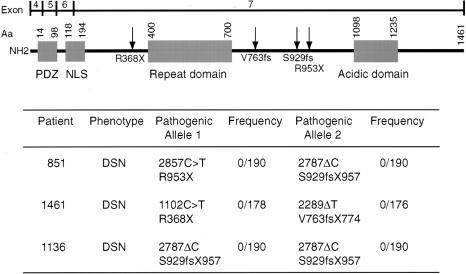
Using DHPLC, we screened each coding exon of PRX for mutations in 168 patients with peripheral neuropathy who had tested negative for mutations involving PMP22, MPZ, GJB1, EGR2, or MTMR2. We sequenced those PCR amplicons that gave an abnormal DHPLC elution profile. Patient 851 of family HOU297 is compound heterozygous for deletion 2787ΔC and transition 2857C→T. By conceptual translation, 2787ΔC causes a frameshift following amino acid S929 and terminates the protein at codon 957 (S929fsX957), whereas 2857C→T causes the nonsense mutation R953X (figs. 3 and 4). We did not observe 2787ΔC or 2857C→T in control chromosomes (fig. 4). The patient 1461 in family HOU579 is compound heterozygous for deletion 2289ΔT and a 1102C→T transition causing the nonsense mutation R368X; 2289ΔT results in a frameshift after amino acid V763 and terminates the protein at codon 774 (V763fsX774; fig. 3). The unaffected parents and son of family HOU579 are all heterozygous carriers of a PRX mutant allele (fig. 3). We did not observe 2289ΔT or 1102C→T in control chromosomes (fig. 4). Patient 1136 of family HOU418 was homozygous for deletion 2787ΔC, the same deletion observed in patient 851 of HOU297. The unaffected parents, sisters, and son of this patient are all heterozygous carriers of this deletion on one PRX allele (figs. 3 and 4); although unaware of consanguinity, both parents hailed from the same small village in Vietnam.
Figure 3.
Chromatograms of PRX alterations identified in three families. Families HOU297, HOU579, and HOU418 exhibit autosomal recessive inheritance. Blackened symbols indicate DSN. Patient 851, from family HOU297, is compound heterozygous for mutations S929fsX957 and R953X; her older unaffected son is heterozygous for R953X (data not shown). Patient 1461, from family HOU579, is compound heterozygous for mutations V763fsX774 and R368X; her unaffected brother is heterozygous for V763fsX774 (data not shown). Patient 1136 from family HOU418 has the homozygous mutation S929fsX957; her two unaffected sisters and her son are heterozygous for this mutation (data not shown).
Figure 4.
Mutations identified in PRX. The location of mutations within L-periaxin is indicated in the diagram at the top by the arrows. The clinical phenotype of each patient, their mutations, and the frequency of their mutations in North American control chromosomes are listed below.
Other PRX sequence variants identified in patients and controls are shown in table 2. Most of these likely represent benign polymorphic variants, but whether the alleles identified in only one control chromosome represent rare polymorphisms or a recessive carrier state remains to be determined.
Table 2.
Alterations Occurring in North American Control Chromosomes or Unaffected Family Members
| Alteration | Frequency in Control Chromosomesa | |
| 3775G→A | E1259K | 0/190b |
| 1216G→A | A406T | 1/178 |
| 4075-4077Δ | E1359del | 1/150 |
| 1483G→C | E495Q | 2/184 |
| 3394A→G | R1132G | 6/182 |
| 3248C→G | P1083R | 24/182 |
| 2763A→G | I921M | 37/190 |
| 2645C→T | A882V | 45/190 |
| 306C→T | T102T | ND |
| 1491C→G | P497P | ND |
| 2655T→C | P885P | ND |
ND = not determined.
Observed in an unaffected sibling.
Phenotype of Patients with PRX Loss-of-Function Mutations
The clinical features of peripheral neuropathy in patients with autosomal recessive PRX mutations are comparable to those observed in the 19q13 linked family and the homozygous knockout mice (table 3) (Delague et al. 2000; Gillespie et al. 2000). In each patient, objective findings include markedly reduced nerve-conduction velocities and onion-bulb formation (OBFs) on neuropathology. These patients have a more severe sensory component than is usually seen with typical DSN or CMT1.
Table 3.
Clinical Features of Patients with Myelinopathy Secondary to PRX Mutations[Note]
|
Family/Patienta |
|||||
| Characteristic | HOU297/851 | HOU579/1461 | HOU418/1136 | CMT4F/19q13.1-q13.3a | Prx−/− Miceb |
| Current age (years) | 46 | 6 | 31 | ||
| Sex | Female | Female | Female | ||
| Age at onset (years) | <7 | 1.5 | 1 | Early childhood | 4–6 weeks |
| Inheritance pattern | Autosomal recessive | Autosomal recessive | Autosomal recessive | Autosomal recessive | Autosomal recessive |
| Motor involvement | Distal dominant, severe | Distal dominant, severe | Distal dominant, severe | Distal dominant, severe | Severe weakness |
| Sensory loss | Severe | Severe | Severe | Severe | Severe |
| Sensory ataxia | No | Yes | Yes | Yes | Unsteady gait |
| Dysesthesia | None | None | Yes | Yes | Yes |
| Foot deformity | Pes cavus | None | Pes cavus | Pes cavus, Pes equinovarus | Not described |
| Motor nerve conduction velocity | 3 m/sec | Undetectable | 2.1 m/sec in median nerve | Undetectable | Severely delayed |
| Peripheral nerve histopathology | NA | Hypomyelination, dysmyelination, OBF | NA | Severe loss of MF, OBF | Demyelination, thick and thin myelin sheaths, loss of MF, OBF |
Note.— NA = not available; MF = myelinated fibers; OBF = onion-bulb formation.
Delague et al. (2000).
Gillespie et al. (2000).
Discussion
Consistent with the Prx−/− mice, these three families establish that putative loss-of-function mutations in PRX cause autosomal recessive DSN (fig. 4). The nonsense and frameshift mutations delete the carboxyl portion of L-periaxin, including the acidic domain. The function of this portion of L-periaxin has not been defined, although acidic domains commonly mediate protein-protein interactions. Loss of this domain, therefore, might inhibit binding of L-periaxin to the cytoskeleton or might preclude L-periaxin from interacting with proteins essential for transmission of extracellular signals.
PRX mutations are a significant cause of apparently sporadic and autosomal recessive DSN. Of 20 unrelated DSN patients in our cohort, 3 inherited two recessive mutant PRX alleles; by comparison, 4, 3, and 2 DSN patients of the 20 had de novo heterozygous causative mutations in MPZ, PMP22, and EGR2, respectively. Moreover, because HOU297, HOU579, and HOU418 are respectively of North American Hispanic, Northern European (English-German-Polish), and Vietnamese ethnicities, we suggest that PRX mutations are a significant cause of DSN in most populations. These two observations imply that identification of PRX mutations will be important for the diagnosis and recurrence-risk counseling of DSN patients and their families.
We previously hypothesized that, because mutations of the transcription factor EGR2 cause myelinopathies, mutation of genes regulated by EGR2 might also result in myelinopathies (Warner et al. 1998). J. Milbrandt and coworkers have shown, consistent with this claim, that EGR2 regulates PRX expression (personal communication). This observation suggests that other genes regulated by EGR2 may also have mutations causing CMT1 or related myelinopathies and raises the possibility that the expression of proteins interacting with L-periaxin may also be regulated by EGR2.
The association of mutations in PRX with peripheral neuropathy not only identifies another genetic cause for the CMT1 spectrum of myelinopathies but also provides further insights into the molecular mechanisms for these diseases. The interaction among L-periaxin, the cytoskeleton, and a membrane complex is reminiscent of the interactions among the proteins of the dystrophin-sarcoglycan complex (Cohen and Campbell 2000) and the signaling complexes organized by other PDZ domain proteins (Montell 2000). We hypothesize that mutations in cytoskeletal and membrane proteins interacting with L-periaxin may also cause CMT or related neuropathies.
Acknowledgments
We thank the families described for their cooperation. H.T. is a recipient of a postdoctoral fellowship from the Charcot-Marie-Tooth Association. This study was supported in part by grants from the National Institute of Diabetes, Digestive, and Kidney Diseases, NIH (K08 DK02738, to C.F.B.), and from the National Institute of Neurological Disorders and Stroke (R01 NS27042) and the Muscular Dystrophy Association (to J.R.L.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Chromosome 19 physical map, http://greengenes.llnl.gov//genome/html/chrom_map.html
- Electronic PCR, http://www.ncbi.nlm.nih.gov/genome/sts/epcr.cgi
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank (for human PRX mRNA sequence encoding S-periaxin [AF321192] and human PRX mRNA sequence encoding L-periaxin [AF321191])
- HUGO Gene Nomenclature Committee, http://www.gene.ucl.ac.uk/nomenclature/ (for registered gene name PRX)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CMT1 [MIM 118200], DSN [MIM 145900], CHN [MIM 605253], and HNPP [MIM 162500])
- Primer v3 program, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi
References
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159 [DOI] [PubMed] [Google Scholar]
- Cohen RD, Campbell KP (2000) Molecular basis of muscular dystrophies. Muscle Nerve 23:1456–1471 [DOI] [PubMed] [Google Scholar]
- Dejerine J, Sottas J (1893) Sur la névrite interstitielle hypertrophique et progressive de l'enfance. Comp Rend Seanc Soc Biol 45:63–96 [Google Scholar]
- Delague V, Bareil C, Tuffery S, Bouvagnet P, Chouery E, Koussa S, Maisonobe T, Loiselet J, Megarbane A, Claustres M (2000) Mapping of a new locus for autosomal recessive demyelinating Charcot-Marie-Tooth disease to 19q13.1-13.3 in a large consanguineous Lebanese family: exclusion of MAG as a candidate gene. Am J Hum Genet 67:236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE (2000) Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 15:7–12 [DOI] [PubMed] [Google Scholar]
- Dytrych L, Sherman DL, Gillespie CS, Brophy PJ (1998) Two PDZ domain proteins encoded by the murine periaxin gene are the result of alternative intron retention and are differentially targeted in Schwann cells. J Biol Chem 273:5794–5800 [DOI] [PubMed] [Google Scholar]
- Fanning AS, Anderson JM (1999) Protein modules as organizers of membrane structure. Curr Opin Cell Biol 11:432–439 [DOI] [PubMed] [Google Scholar]
- Fernandez-Valle C, Gorman D, Gomez AM, Bunge MB (1997) Actin plays a role in both changes in cell shape and gene-expression associated with Schwann cell myelination. J Neurosci 17:241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie CS, Lee M, Fantes JF, Brophy PJ (1997) The gene encoding the Schwann cell protein periaxin localizes on mouse chromosome 7 (Prx). Genomics 41:297–298 [DOI] [PubMed] [Google Scholar]
- Gillespie CS, Sherman DL, Blair GE, Brophy PJ (1994) Periaxin, a novel protein of myelinating Schwann cells with a possible role in axonal ensheathment. Neuron 12:497–508 [DOI] [PubMed] [Google Scholar]
- Gillespie CS, Sherman DL, Fleetwood-Walker SM, Cottrell DF, Tait S, Garry EM, Wallace VC, Ure J, Griffiths IR, Smith A, Brophy PJ (2000) Peripheral demyelination and neuropathic pain behavior in periaxin-deficient mice. Neuron 26:523–531 [DOI] [PubMed] [Google Scholar]
- Hayasaka K, Himoro M, Sawaishi Y, Nanao K, Takahashi T, Takada G, Nicholson GA, Ouvrier RA, Tachi N (1993) De novo mutation of the myelin P0 gene in Dejerine-Sottas disease (hereditary motor and sensory neuropathy type III). Nat Genet 5:266–268 [DOI] [PubMed] [Google Scholar]
- Lupski JR (2000) Recessive Charcot-Marie-Tooth disease. Ann Neurol 47:6–8 [PubMed] [Google Scholar]
- Lupski JR, Garcia CA (2001) Charcot-Marie-Tooth peripheral neuropathies and related disorders. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Vogelstein B, Childs B (eds) The metabolic and molecular bases of inherited disease. McGraw-Hill, New York, pp 5759–5788 [Google Scholar]
- Montell C (2000) A PDZ protein ushers in new links. Nat Genet 26:6–7 [DOI] [PubMed] [Google Scholar]
- Parman Y, Plante-Bordeneuve V, Guiochon-Mantel A, Eraksoy M, Said G (1999) Recessive inheritance of a new point mutation of the PMP22 gene in Dejerine-Sottas disease. Ann Neurol 45:518–522 [DOI] [PubMed] [Google Scholar]
- Roa BB, Dyck PJ, Marks HG, Chance PF, Lupski JR (1993) Dejerine-Sottas syndrome associated with point mutation in the peripheral myelin protein 22 (PMP22) gene. Nat Genet 5:269–273 [DOI] [PubMed] [Google Scholar]
- Scherer SS, Xu Y-t, Bannerman PGC, Sherman DL, Brophy PJ (1995) Periaxin expression in myelinating Schwann cells: modulation by axon-glial interactions and polarized localization during development. Development 121:4265–4273 [DOI] [PubMed] [Google Scholar]
- Schuler GD (1997) Sequence mapping by electronic PCR. Genome Res 7:541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Kennedy GM, Spikes AS, Lupski JR (1997) Diagnosis of CMT1A duplications and HNPP deletions by interphase FISH: implications for testing in the cytogenetics laboratory. Am J Med Genet 69:325–331 [PubMed] [Google Scholar]
- Sherman DL, Brophy PJ (2000) A tripartite nuclear localization signal in the PDZ-domain protein L-periaxin. J Biol Chem 275:4537–4540 [DOI] [PubMed] [Google Scholar]
- Tapon N, Hall A (1997) Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr Opin Cell Biol 9:86–92 [DOI] [PubMed] [Google Scholar]
- Timmerman V, De Jonghe P, Ceuterick C, De Vriendt E, Lofgren A, Nelis E, Warner LE, Lupski JR, Martin JJ, Van Broeckhoven C (1999) Novel missense mutation in the early growth response 2 gene associated with Dejerine-Sottas syndrome phenotype. Neurology 52:1827–1832 [DOI] [PubMed] [Google Scholar]
- Warner LE, Mancias P, Butler IJ, McDonald CM, Keppen L, Koob KG, Lupski JR (1998) Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat Genet 18:382–384 [DOI] [PubMed] [Google Scholar]