Abstract
We have studied cultured skin fibroblasts from three siblings and one unrelated individual, all of whom had fatal mitochondrial disease manifesting soon after birth. After incubation with 1 mM glucose, these four cell strains exhibited lactate/pyruvate ratios that were six times greater than those of controls. On further analysis, enzymatic activities of the pyruvate dehydrogenase complex, the 2-oxoglutarate dehydrogenase complex, NADH cytochrome c reductase, succinate dehydrogenase, and succinate cytochrome c reductase were severely deficient. In two of the siblings the enzymatic activity of cytochrome oxidase was mildly decreased (by ∼50%). Metabolite analysis performed on urine samples taken from these patients revealed high levels of glycine, leucine, valine, and isoleucine, indicating abnormalities of both the glycine-cleavage system and branched-chain α-ketoacid dehydrogenase. In contrast, the activities of fibroblast pyruvate carboxylase, mitochondrial aconitase, and citrate synthase were normal. Immunoblot analysis of selected complex III subunits (core 1, cyt c1, and iron-sulfur protein) and of the pyruvate dehydrogenase complex subunits revealed no visible changes in the levels of all examined proteins, decreasing the possibility that an import and/or assembly factor is involved. To elucidate the underlying molecular defect, analysis of microcell-mediated chromosome-fusion was performed between the present study's fibroblasts (recipients) and a panel of A9 mouse:human hybrids (donors) developed by Cuthbert et al. (1995). Complementation was observed between the recipient cells from both families and the mouse:human hybrid clone carrying human chromosome 2. These results indicate that the underlying defect in our patients is under the control of a nuclear gene, the locus of which is on chromosome 2. A 5-cM interval has been identified as potentially containing the critical region for the unknown gene. This interval maps to region 2p14-2p13.
Introduction
Inherited mitochondrial defects in which more than one enzyme is affected with low activity are relatively common in association with mtDNA that is mutated, deleted, or depleted (Zeviani et al. 1989; Brown and Wallace 1994; Bodnar et al. 1995; Grossman and Shoubridge 1996; Moraes et al. 1999). However, defects involving multiple enzymopathies, which are associated with defects in nuclear genes, are rare examples, being abnormalities in either import proteins (Jin et al. 1996, 1999; Koehler et al. 1999) or the Lon proteases responsible for mitochondrial-protein turnover (van Dijl et al. 1998).
Fatal neonatal infantile forms of the defects associated with primary lactic acidosis have been reported, including defects of the pyruvate dehydrogenase (PDH) complex (PDHC) (Robinson et al. 1987a; Brown et al. 1988; Robinson 1995; Morten et al. 1998), the NADH–coenzyme Q reductase complex (Moreadith et al. 1984; Hoppel et al. 1987), the cytochrome oxidase complex (DiMauro et al. 1988; Lombes et al. 1989, 1996), and the ubiquinol cytochrome c reductase complex (Kennaway 1988; Valnot et al. 1999). Here we report two cases in which cultured skin fibroblasts showed decreased activities of the pyruvate dehydrogenase complex, the 2-oxoglutarate dehydrogenase complex, and succinate cytochrome c reductase. Furthermore, analysis of urine metabolites indicated possible deficiencies of branched-chain α-ketoacid dehydrogenase and of the glycine-cleavage system. Here we show that this new syndrome is caused by a defect in a gene located on chromosome 2p14-2p13 (84–89 cM).
Subjects, Material, and Methods
Three Siblings
Three siblings—two males and one female (cell strain numbers 4142, 4143, and 4144)—born a few years apart to nonconsanguineous parents after uncomplicated pregnancies, presented with feeding difficulty, weakness, lethargy, and decreasing responsiveness within a few days after birth. Laboratory investigation showed metabolic acidosis with elevated blood lactate levels. Assisted ventilation was necessary because of respiratory failure. Urine organic-acid analysis performed on one of the siblings (4143) showed a highly elevated urinary lactate level, with a secondary elevation of 2-hydroxybutyrate (5.5 mEq creatinine/ml, by GC). Measurement of amino acid levels in the blood showed glycine, leucine, isoleucine, phenylalanine, alanine, valine, and taurine levels to be elevated, the most significant elevation being observed in glycine (5.4 times the normal level). All three siblings died <1 mo after birth. A limited autopsy was done on another sibling (4142), and significant findings included pulmonary oxygen toxicity, pneumonitis, eosinophilic infarction of the right and left anterior papillary muscles, focal tracheal mucosal denudation, focal intra-alveolar hemorrhage, focal renal hemorrhage, and white-matter spongiosis, all consistent with a metabolic abnormality.
Unrelated Patient
A male infant was born by spontaneous vaginal delivery at 33 wk gestation, as a product of in vitro fertilization, to a 30-year-old G4, P0, ectopic 3 mother (that is, of four pregnancies, none were live births, three were ectopic and therefore failed). The parents were first cousins. During pregnancy, the mother had gestational diabetes and pregnancy-induced hypertension with proteinuria. Four months after birth, the infant developed epileptic seizures as well as elevated levels of serum glycine (571 μM) and CSF glycine (28 μM). At 6 mo of age he was developmentally delayed, and the frequency of seizures increased. Subsequently, he developed dilated cardiomyopathy and epileptic encephalopathy. He suffered from recurrent vomiting, progressive lethargy, tachypnea, diaphoresis, respiratory distress, and hepatomegaly. At 7 mo of age he became acidotic. His initial lactate level was 11.5 mM, but it increased to 19 mM in the course of few weeks. His CSF glycine levels rose to 1,039 μM, and his condition progressively deteriorated. He died at 11 mo of age.
Cell Lines
The cell lines used in this study were obtained by skin biopsy of the forearm of patients, after informed consent was obtained. Primary fibroblast lines were established from the four patients (4142, 4143, 4144, and 11571) with compound mitochondrial-enzyme deficiency, all of whom exhibited the same biochemical phenotype. Fibroblasts were grown in Eagle's minimal essential medium (α-MEM) and 10% fetal calf serum, supplemented with glucose to a concentration of 10.5 mM. Measurements of the lactate-to-pyruvate (L/P) ratio in confluent fibroblast cultures were performed as described elsewhere (Robinson et al. 1985). The primary fibroblast cell lines were transduced with a retroviral vector expressing the E6E7 region of type 16 human papilloma virus, to extend their life span (Compton 1993), and were grown in high-glucose Dulbecco's minimal essential medium (DMEM) supplemented with 20% fetal bovine serum.
Microcell-Mediated Chromosome Transfer
A panel of human:mouse monochromosomal hybrids (Cuthbert et al. 1995) was used as the source of normal human donor chromosomes. All 22 autosomes and the X chromosome were serially transferred into one of the patient cell lines by microcell-mediated chromosome transfer (Fournier 1981). In brief, donor cells were plated in α-MEM containing 10% fetal bovine serum and hygromycin B (400 U/ml; ICN), 3 d before fusion. The medium was then changed to DMEM plus 20% fetal bovine serum, and the cells were exposed to colchicine (0.06 μg/ml) for 48 h to induce micronucleation. The cells were then collected by trypsinization and were plated on plastic “bullets” (custom made from tissue-culture plates, to fit into 50-ml centrifuge tubes) coated with cross-linked concanavalin A (Sigma). Microcells were prepared by centrifugation at 34°C–37°C and 15,000 g (SS34 rotor) in media containing cytochalasin B (10 μg/ml; Sigma), were filtered through 8-μm and 5-μm filters, were pelleted at 3,000 g on a benchtop centrifuge, and were resuspended in serum-free medium. The microcell suspension and phytohemagglutinin were added to the plate containing the recipient cells (100 μg/ml) and incubated for 20–30 min. Microcells were fused with 45% polyethylene glycol plus 10% dimethyl sulfoxide for 60 s, washed with serum-free medium, and incubated in α-MEM plus 20% fetal bovine serum. After 48–72 h, fused cells were selected in hygromycin B (100 U/ml). Colonies were picked, expanded, and analyzed 3–4 wk later.
Enzymology and Spectrophotometric and Immunological Analysis
Activity of the PDH complex in both the native (unstimulated) and the dichloroacetate-activated state was determined in fibroblast extracts by the method of Sheu et al. (1981). Activity of the 2-oxoglutarate complex was determined in fibroblast extracts by the method of Hyland and Leonard (1983). Activity of the NADH cytochrome c reductase was assayed in 0.1 M potassium phosphate (KPi) buffer (pH 7.0) containing 94 μM cytochrome c and 1mM azide. To start the reaction, 20 μM NADH was added, and reduction of cytochrome c was monitored at 550 nm. Cytochrome oxidase activity was assayed in 0.1 M KPi buffer (pH 7.0). Cytochrome c was reduced with 3.2 mM ascorbate, followed by dialysis against 0.1 M KPi for 2 d, during which the buffer was changed twice. Reduced cytochrome c (94 μM) was added to start the reaction, and oxidation of cytochrome c was monitored at 550 nm. Citrate synthase assay was performed as described by Robinson et al. (1987b). Succinate dehydrogenase activity was determined in mitochondria purified from skin fibroblasts by the method of Hatefi and Stiggall (1978). Succinate cytochrome c reductase activity was determined in fibroblast extracts by the method of Yu and Yu (1982). Aconitase activity was determined in mitochondria purified from skin fibroblasts by the method of Racker (1950). Activity of the bc1 complex was determined according to the method described by Darley-Usmar et al. (1987). Western blotting was performed on mitochondrial extracts prepared by the method of Pitkänen et al. (1996), using polyclonal antibodies against (a) porcine-heart PDH-complex raised in rabbit (Ekong 1982) and (b) holo bc1 and iron-sulfur protein (ISP)–bc1 bovine heart-and-liver complex raised in rabbit (Vazquez-Acevedo et al. 1993).
Preparation of Human Liver Mitochondria
Mitochondria were isolated from frozen post-mortem human livers by a method that is a combination of those described by Pitkänen et al. (1996) and Craig and Hood (1997). Tissues were finely minced and then were gently homogenized in buffer (50 mM HEPES, pH 7.4, 70 mM sucrose, 220 mM mannitol, 1 mM EGTA and 2 mg fatty acid–free BSA/ml) (Sigma-Aldrich). After homogenization, the mixture was centrifuged at 3,000 g for 10 min. The supernatant was then centrifuged at 15,000 g for 10 min. The pellet was gently solubilized in homogenization buffer, and the centrifugation steps were repeated. The pellet was resolubilized and recentrifuged at 15,000 g for a final time. Then the mitochondrial pellet was resuspended in buffer (0.34 M sucrose, 100 mM KCl, 10 mM Tris-Cl, and 1 mM EDTA) at a concentration of 1 mg/ml.
Free Radical–Production Assay
Superoxide production was determined using the chemiluminescent dye lucigenin (10,10-dimethyl-9,9-biacridinium dinitrate; Sigma) (Pitkänen and Robinson 1996). The concentration of the working solution of lucigenin was 4.3 μM. The measurements were performed in buffer containing 70 mM sucrose, 220 mM mannitol, 2.5 mM KPi (pH 7.5), 2.5 mM MgCl2, 0.5 mM EDTA, and 0.1% BSA. Lucigenin was added to the buffer immediately before measurements were taken. All measurements were made at room temperature, with a Lumat luminometer (model 9507; Berthold). The signal from the dye was integrated over a period of 30 s after the addition of substrate (2 mM succinate), and the background signal during this same period was subtracted from the sample signal. Mitochondria were preincubated, with or without inhibitors, in the measurement buffer for 1 min prior to addition of substrate. The substrate was added using the automatic-injection system of the Lumat 9507, after determination of the background signal during a 15-s period. Measurements were carried out in the presence or absence of either antimycin A (final concentration 20 μg/ml), myxothiazol (final concentration 10 μM) (Sigma-Aldrich), or stigmatellin (final concentration 20 μM) (Fluka Biochemicals). These inhibitor concentrations were shown to fully inhibit enzyme activity (Raha et al. 2000). Ten micrograms of mitochondrial proteins per sample was used for luminometric monitoring.
Deletion and Exclusion Mapping
Oligonucleotide primers for polymorphic microsatellite markers on chromosome 2 were obtained (Research Genetics, Huntsville, Alabama and ACGT, Toronto). Deletion mapping was done on DNA isolated from hygromycin B–resistant clones obtained, after microcell fusion, from the A9-2 monochromosomal hybrid line, by a Pure Gene kit (Gentra Systems, Minneapolis). Exclusion mapping was carried out on the three affected siblings as well as on the family of the unrelated individual. This family included the affected individual, his brother, and both parents. Genomic DNA for this experiment was isolated from primary skin fibroblasts, with a total-DNA–isolation kit (Qiagen). PCR was performed using the Research Genetics protocol, AmpliTaq Gold (PE Biosystems), and γ35S-dATP. PCR products were run on 6% sequencing gels exposed overnight, with BioMax film (Kodak), and developed. PCR was also performed, as above, in the absence of γ35S-dATP. Products were run on Supergel 250TM (Helixx); gels were stained with Sybr Green (Helixx, Toronto) for 20 min, and DNA bands were visualized by UV illumination.
Results
Skin fibroblast cultures established from three siblings and one unrelated patient showed a four- to sevenfold elevation in the L/P ratio formed on incubation with 1 mM glucose for 1 h (Robinson et al. 1987a) (table 1). A very low activity of the pyruvate dehydrogenase complex was also recorded, with values in the dichloroacetate-activated state reaching, at most, 25% of the control value. Furthermore, breakdown of the activities of individual PDH-complex components revealed a decreased activity of the first subcomplex, pyruvate dehydrogenase (designated “E1”), whereas the activities of the remaining components, dihydrolipoamide transacetylase (designated “E2”) and dihydrolipoamide dehydrogenase (designated “E3”), were normal. Activities of the NADH cytochrome c reductase (complexes I + III), succinate cytochrome c reductase (complexes II + III), succinate dehydrogenase (complex II) bc1 complex (complex III), and the 2-oxoglutarate dehydrogenase complex were also decreased, reaching, at most, 30%, 35%, 45%, 60%, and 28%, respectively, of control values. The activity of cytochrome c oxidase (COX) was decreased to ∼50% of control values in two of the siblings (4143 and 4144). In contrast, activity values for pyruvate carboxylase, mitochondrial aconitase, and citrate synthase were normal. The above-described phenotype was observed in both primary and transformed cells (data not shown). Transformation did not alter enzymatic activities of the assayed complexes. Urine organic-acid analysis performed on one of the siblings (4143) showed a highly elevated urinary lactate level with a secondary elevation of 2-hyroxybutyrate (5.548 mEq/ml creatinine by GC). In all four patients, amino acid levels in the blood showed glycine, leucine, isoleucine, phenylalanine, alanine, valine, and taurine at above-normal levels, with the most significant abnormality being a fivefold elevation in glycine.
Table 1.
Various Mitochondrial Enzyme Activities of Patients Suffering from the Multiple Disorder
|
Mean ± SEM Activityb [No. of Determinations] in(nmol/min/mg) |
|||||
| Enzymea | Control | Sibling 1 (4142) | Sibling 2 (4143) | Sibling 3 (4144) | Unrelated Patient (11571) |
| PDHC (active) | .98±.14 [9] | .16±.04 [6] | .22±.05 [10] | .05±.02 [9] | .21±.04 [4] |
| PDHC (DCA activated) | 1.16±.16 [9] | .13±.03 [6] | .25±.06 [10] | .04±.02 [8] | .36±.06 [4] |
| PDH E1 | 1.41±.21 [5] | .48 [1] | .63 [1] | .36±.07 [5] | N/A |
| PDH E2 | 10.68±3.89 [2] | 12.04 [1] | 12.19 [1] | 8.86±1.92 [2] | N/A |
| PDH E3 | 7.20±.53 [2] | 9.26 [1] | 7.34 [1] | 7.21±.60 [2] | N/A |
| OGDHC | 1.48±.33 [4] | 0.20±.06 [2] | .37±.05 [2] | .12±.05 [2] | 0.42±.06 [2] |
| NCR (I + III) | 6.38±.22 [5] | 1.95±.38 [4] | 1.02±.25 [4] | 1.27±.47 [3] | 1.98±.53 [3] |
| COX | 5.18±.69 [4] | 5.07±.28 [3] | 2.23±.46 [4] | 2.70±.56 [3] | 4.95±.50 [4] |
| L/P ratio | 27.1±3.5 [6] | 105.5±28.4 [4] | 157.0±20.7 [8] | 187.7±15.1 [4] | 101.2±6.5 [4] |
| Pyruvate carboxylase | 1.38±.29 [3] | 2.33 [1] | .42 [1] | 1.46 [1] | 1.17 [1] |
| Citrate synthase | 39.67±8.34 [2] | N/A | 35.34±7.30 [2] | 32.02±5.76 [2] | 42.86±6.30 [2] |
| SDH | 16.0 ± 2.4 [4] | N/A | 7.0 ±.2 [2] | N/A | 6.9 ± .2 [2] |
| Mitochondrial aconitase | 4.0 ± .6 [2] | N/A | 3.5 ± .6 [2] | N/A | 3.2 ±.5 [2] |
| bc1 complex (complex III)c | 6.9 ± 1.5 [4] | 4.5 ± .8 [2] | 3.8 ± .7 [2] | N/A | 3.9 ± .9 [3] |
| SCR | 5.94±1.01 [2] | N/A | 1.41±.33 [2] | 1.57±.36 [2] | 2.17±.71 [2] |
NCR (complexes I+III)= NADH cytochrome c reductase (complexes I + III); SDH = succinate dehydrogenase (complex II); SCR = succinate cytochrome c reductase (complexes II + III).
Determined in cultured skin fibroblasts from each patient and a group of controls. Individual determinations were made on separate cultures on different days, and multiple control cell lines were used.
Data standardized to levels of citrate synthase.
To determine the chromosomal location of the gene defect in these patients, we rescued this phenotype by functional complementation with a normal human chromosome transferred into deficient cells by microcell-mediated chromosome transfer. A panel of stable human:mouse hybrid cell lines containing human chromosomes tagged with a selectable marker (HyTK) was used as a source of donor human chromosomes (Cuthbert et al. 1995). Autosomes (with the exception of chromosomes 1, 9, 13, 15, 17, and 21) and the X chromosome were transferred, one at a time, into a patient fibroblast line (4143, one of the three siblings), and the cells were selected in hygromycin B. Three to four weeks postfusion, surviving colonies were picked, and PDH-complex activity was measured in pooled colonies obtained for each individual chromosome transfer. Transfer of chromosome 2 (A9.2 human:mouse hybrid cell line) restored PDH complex activity to control levels (table 2). This restoration was not observed with either any other autosome or the X chromosome (table 2). Transfer of chromosome 2 also restored the activity of the 2-oxoglutarate dehydrogenase complex and the NADH cytochrome c reductase in that cell line (4143).
Table 2.
Functional Complementation Analysis, with a Panel of Human Chromosomes, of Skin Fibroblasts from a Patient Suffering from Multiple Mitochondrial Enzyme Disorder
|
Mean ± SEM Activitya [No. of Determinations] of(nmol/min/mg cell protein) |
|||
| PDH Complex |
|||
| Chromosomeor Source | NADH Cytochromec Reductase (Complexes I & III) | Native | DCA Activated |
| 1 | NA | NA | NA |
| 2 | 5.98±.34 [4] | 1.12±.09 [9] | 1.5±.145 [9] |
| 3 | … | .21±.04 [2] | .24±.06 [2] |
| 4 | 1.99±.71 [2] | .23±.07 [3] | .23±.04 [3] |
| 5 | 1.87±.45 [2] | .25±.06 [2] | .19±.04 [2] |
| 6 | … | .19±.02 [2] | .29±.05 [2] |
| 7 | … | .22±.05 [3] | .20 ±.03 [3] |
| 8 | 2.08±.43 [2] | .30±.06 [2] | .31±.04 [2] |
| 9 | NA | NA | NA |
| 10 | … | .14±.07 [2] | .17±.05 [2] |
| 11 | … | .028±.06 [2] | .25±.04 [2] |
| 12 | … | .18±.03 [2] | .23±.05 [2] |
| 13 | NA | NA | NA |
| 14 | 2.24±.39 [2] | .26±.06 [3] | .21±.03 [3] |
| 15 | NA | NA | NA |
| 16 | 1.99±.22 [2] | .29±.08 [3] | .28±.07 [3] |
| 17 | NA | NA | NA |
| 18 | … | .28±.04 [2] | .20±.04 [2] |
| 19 | 2.13±.45 [2] | .23±.06 [2] | .25±.05 [2] |
| 20 | … | .26±.05 [2] | .30±.04 [2] |
| 21 | NA | NA | NA |
| 22 | 2.56±.63 [2] | .22±.05 [2] | .27±.04 [2] |
| X | … | .34±.07 [2] | .29±.03 [2] |
| Patient 4143 | 1.02±.25 [4] | .22±.05 [10] | .24±.05 [10] |
| Control | 6.56±.58 [5] | .98±.08 [9] | 1.21±.14 [9] |
Measured after microcell-mediated transfer of single HyTK-tagged normal human chromosomes into fibroblasts from patient 4143 (one of the three siblings). For multiple pooled clones recovered after selection in hygromycin B. NA = transfer of chromosome not performed. An ellipsis (…) denotes that activity was not measured.
To confirm a common genotype in the selected patients, we transferred chromosome 2 into skin fibroblasts obtained from another affected sibling (4142) and from the unrelated patient (11571). Additional clones were isolated from the latter patient. As had been seen in the previous results, this chromosome restored activities of all assayed enzyme complexes (table 3).
Table 3.
Functional Complementation Analysis of Selected Enzyme Complexes (the bc1, the PDHC, and the 2-OGDHC), Pre- and Post-Chromosome 2 Transfer
|
Mean ± SEM Activity [No. of Determinations] of(nmol/min/mg cell protein) |
||||
| PDH Complex |
||||
| Cell Line | NADH Cytochrome c Reductase | Native | DCA Activated | 2-OGDHC |
| 4143 | 1.02±.25 [4] | .22±.05 [10] | .24±.05 [10] | .37±.05 [2] |
| 4143 x chr 2 | 5.98±.33 [4] | 1.12±.09 [9] | 1.50±.14 [9] | 1.04±.10 [2] |
| 4142 | 1.95±.38 [4] | .16±.04 [3] | .24±.04 [3] | .11±.09 [2] |
| 4142 x chr 2 | 6.62±1.05 [2] | .91±.09 [4] | 1.33±.18 [4] | 1.13±.16 [2] |
| 11571 | 2.28±.53 [3] | .21±.03 [4] | .36±.06 [4] | .42±.06 [2] |
| 11571 x chr 2 | 4.56±.58 [3] | 1.11±.09 [6] | 1.52±.15 [6] | 1.29±.13 [2] |
| Control | 6.56±.58 [5] | .98±.08 [9] | 1.21±.14 [9] | 1.34±.13 [4] |
We also tested PDH-complex activities in individual clones obtained from patients 4143 and 11571. Enzyme activity was restored in 23/25 independent clones derived from patient 4143 and in 30/36 clones in cells derived from patient 11571 (data not shown).
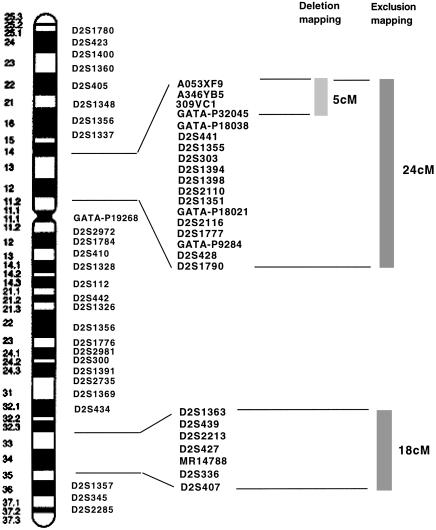
To narrow the potential locus region on chromosome 2, we genotyped three affected siblings, as well as the unrelated patient, his parents, and his unaffected brother with a set of chromosome-2 microsatellite markers. At the locus of the gene defect, affected family members, in theory, should share common alleles while being discordant with unaffected family members. This analysis resulted in isolation of two regions of interest: one on the p arm and the other on the q arm of chromosome 2 (table 4). The region on the p arm extended between markers D2S1337 and D2S1790 (∼24 cM), and the region on the q arm extended between markers D2S434 and D2S407 (∼18 cM). During the course of this experiment we had to exclude the family of the unrelated patient because the extremely high degree of consanguinity (the parents were first cousins) rendered the data noninformative.
Table 4.
Results of Exclusion-Mapping Performed on Three Siblings[Note]
|
Allele Size ina(base pairs) |
|||||
| Position(cM) | Marker | Sibling 1 | Sibling 2 | Sibling 3 | Control |
| 15 | D2S1780 | 224/232 | 220/? | 224 | … |
| 24 | D2S423 | 177/183 | 174/183 | 177 | 177/180 |
| 29 | D2S1400 | 110 | 110/114 | 110/114 | 114 |
| 42 | D2S1360 | 135/143 | 135/143 | 135/159 | 151/155 |
| 41 | D2S405 | 249 | 249 | 241/249 | 249 |
| 63 | D2S1348 | 174/180 | 159/180 | 174/180 | 171 |
| 70 | D2S1356 | ? |
202/208 |
202/226 |
202/? |
| 81 | D2S1337 | 145/147 | 145/147 | 139/145 | 143/147 |
| 90 | D2S441 | 154 | 154 | 154 | 162/174 |
| 95 | D2S1394 | 168 | 168 | 168 | 160/172 |
| 100 | D2S1777 | 201 | 201 | 201 | 198/202 |
| 107 | GATA26G08 | 137/145 | 137/145 | 137/145 | … |
| 108 | D2S1790 | 284/328 |
284/328 |
284/328 |
… |
| 120 | D2S2972 | 227/231 | 227/231 | 231/239 | 227/231 |
| 125 | D2S1784 | 205/209 | 201/205 | 205/209 | 209 |
| 130 | D2S410 | 165/173 | 163/165? | 163/173 | 165/169 |
| 138 | D2S1328 | 140/152 | 140/152 | 140 | … |
| 146 | D2S112 | 135/137 | 137/142 | 133/135 | 137 |
| 150 | D2S442 | 197/201 | 201/205 | 197/201 | 197/205 |
| 152 | D2S1326 | 242/254 | 238/246 | 242/254 | 258/266 |
| ? | D2S1353 | 144/147 | 144 | … | 144 |
| 180 | D2S1776 | 297 | 297 | 285/297 | 297 |
| 187 | D2S2981 | 233/249 | 233/249 | 241/249 | 249 |
| 189 | D2S300 | 138/142 | 138 | 142 | 142 |
| 191 | D2S1391 | 115/123 | 115/123 | 123 | 127/131 |
| 201 | D2S2735 | 248/252 | 248/252 | 236/244 | 240/244 |
| 212 | D2S1369 | 264/268 | 264/268 | 256/268 | 256 |
| 222 | D2S434 | 266/270 | 270/274 | … | 266/270 |
| 233 | D2S1363 | 186/190 |
186/190 |
178/186 |
182/190 |
| 236 | GATAE08 | 191 | 191 | 191 | … |
| 244 | D2S427 | 254 | 254 | 254 | 214/254 |
| 253 | D2S407 | 195 |
195 |
195 |
195 |
| 267 | D2S2285 | 253/259 | 253/259 | 253/259 | … |
Note.— A set of chromosome 2 markers was used for microsatellite analysis by PCR to isolate regions where the affected family members share common alleles (boxed areas).
Single values indicate homozygosity for that individual. The control individual is not related to the affected siblings. An ellipsis (…) is used when definitive designation of alleles was not possible.
Chromosomes incorporated into the genome of recipient cells by microcell transfer often undergo deletions and rearrangements (Leach et al. 1989; Newbold and Cuthbert 1996). To eliminate one of the two candidate regions obtained by exclusion mapping, as well as to obtain a narrower chromosomal interval containing the gene of interest, we used polymorphic microsatellite markers from chromosome 2. We mapped the regions that were incorporated into the genomes of complemented and noncomplemented clones isolated from patients 4143 and 11571 after hygromycin B selection. In functionally complemented clones, markers that are missing from the exogenous chromosome could be excluded from the region of interest; similarly, in noncomplemented clones, informative markers that are present could be excluded as well. Analysis of 13 complemented clones and 1 noncomplemented clone from patient 4143, and of 10 complemented and 6 noncomplemented clones from patient 11571, allowed the mapping of the gene defect to the region between markers A053XF9 and GATA-P132045 (∼5 cM) (table 5). This analysis excluded the region, on the q arm of chromosome 2, obtained by marker analysis in the three siblings. Cumulative data for both exclusion and deletion mapping are shown in figure 1. During the exclusion-mapping experiment it became apparent that neither the immortalization of the primary cells nor the chromosome-transfer procedure resulted in the loss of cellular heterozygosity, owing to the fact that, with informative sequence-tagged site (STS) markers, endogenous and exogenous alleles could be easily identified.
Table 5.
A Deletion Map for Two Patients with Novel Mitochondrial Syndrome
|
Results of Microsatellite PCRa |
|||||||||||||||||||||||||||||||
| Noncomplementing Clones |
Complementing Clones |
||||||||||||||||||||||||||||||
| CellLine 4143 |
Cell Line 11571 |
Cell Line 4143 |
Cell Line 11571 |
||||||||||||||||||||||||||||
| Position(cM) | Marker | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 |
| 84 | A053XF9 | NI | − | − | − | − | + | − | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | + | + | + | + | + | ? | + | + | + | + |
| 86 | A346YB5 | NI | − | − | − | − | − | − | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | + | + | + | + | ? | + | + | + | + | + |
| 88 | 309VC1 | NI | − | − | − | − | − | − | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | + | + | + | + | + | + | + | + | + | + |
| 89 | GATA-P32045 | NI |
+ |
− |
− |
− |
− |
− |
NI |
NI |
NI |
NI |
NI |
NI |
NI |
NI |
NI |
NI |
NI |
NI |
NI |
+ |
+ |
+ |
+ |
+ |
? |
+ |
+ |
+ |
+ |
| 90 | GATA-P18038 | − | + | − | − | − | − | − | − | + | + | + | + | + | + | ? | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 90 | D2S441 | − | + | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ? | + | ? | ? | + | + | + | + |
| 92 | D2S1355 | NI | + | − | − | + | − | − | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | + | + | + | + | + | ? | + | + | + | + |
| D2S303 | − | NI | NI | NI | NI | NI | NI | − | + | − | + | + | + | + | + | + | + | + | + | + | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | |
| 95 | D2S1394 | − | NI | NI | NI | NI | NI | NI | − | + | − | + | + | + | ? | ? | + | ? | + | + | + | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI |
| D2S1398 | − | + | − | − | + | − | − | − | + | − | ? | + | + | + | + | + | + | + | + | + | + | + | ? | + | + | + | − | + | + | + | |
| 98 | D2S1351 | NI | + | − | − | + | − | − | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | + | + | + | + | + | ? | − | + | ? | + |
| 100 | GATA-P18021 | − | NI | NI | NI | NI | NI | NI | − | + | − | + | + | + | + | ? | + | + | + | + | + | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI |
| D2S116 | − | + | − | − | + | − | − | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ? | − | + | + | + | |
| 107 | GATA-P9284 | − | + | − | − | + | − | − | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ? | − | + | + | + |
| 108 | D2S1790 | − | + | − | − | + | − | + | − | + | − | + | + | + | + | + | + | ? | + | + | + | + | + | + | + | + | ? | − | + | + | + |
| 138 | D2S1328 | − | + | + | − | + | − | + | |||||||||||||||||||||||
| 148 | D2S1334 | − | + | + | + | + | + | + | |||||||||||||||||||||||
| 180 | D2S1776 | − | + | + | + | + | + | + | |||||||||||||||||||||||
| 191 | D2S1391 | − | + | − | + | + | + | − | |||||||||||||||||||||||
| 233 | D2S1363 | − | + | − | + | + | − | − | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + |
| 236 | D2S439 | NI | + | − | + | + | − | − | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | + | + | + | + | + | + | − | + | + | + |
| 244 | D2S427 | NI | + | − | + | + | − | − | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | + | + | + | ? | + | + | − | − | + | + |
| 253 | D2S336 | + | + | − | + | + | − | − | − | + | − | + | − | ? | + | + | ? | + | + | + | + | + | + | + | + | + | + | − | − | + | + |
| 253 | D2S407 | NI | + | − | + | + | − | − | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | + | + | ? | + | + | + | − | − | + | + |
Plus (+) signs and minus (−) signs indicate the presence or absence, respectively, of STS markers. NI = marker not informative. ?- unable to clearly include or exclude marker. Recipient patient-derived cell lines (either 4143 or 11571) are indicated above each set of results. Only noncomplemented clones were analyzed using markers D2S1328, D2S1334, D2S1779, and D2S1391. Markers D2S2110, D2S1777, D2S428, D2S2213, and MR14788 were not informative for all analyzed clones and therefore are not included. The candidate region is boxed.
Figure 1.
Ideogram of human chromosome 2 showing results of exclusion and deletion mapping for the mitochondrial multienzyme-deficiency phenotype. Microsatellite markers that were used in the analysis are indicated. The thicker vertical lines to the right of the markers indicate regions that were obtained by either deletion or exclusion mapping.
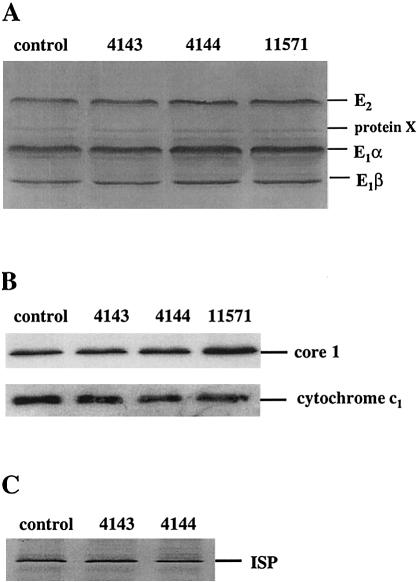
To determine whether the genetic defect shared by the affected individuals had any impact on the amount of protein expressed for the deficient complexes, immunoblotting was performed using antibodies directed against the total PDH complex, the holo bc1 complex, and the bc1 complex ISP. These immunoblots show that four components of the PDH complex and three components of the bc1 complex are expressed at levels comparable to those in control samples (fig. 2). This indicates that the disorder does not affect the import, assembly, or stability of the investigated complexes, since all protein levels investigated are quite normal despite the proven lack of activities in the enzyme complexes.
Figure 2.
Western blot analysis of endogenous expression of pyruvate dehydrogenase complex (A), core I and cytochrome c of the bc1 complex (B), and bc1 complex ISP (C). Aliquots (60 μg) of total protein from mitochondria-rich fractions were separated on 12.5% SDS/polyacrylamide gel and then were transferred to nitrocellulose. The filter was probed with antibody against affinity-purified pig heart PDH complex raised in rabbit (A), or with antibody against either holo bc1 complex, or bc1-ISP raised in rabbit (B and C). The mitochondrial extracts present on the gels are, from left to right, control cultured-skin fibroblasts (lane 1), skin fibroblasts from two affected siblings (4143 and 4144) (lanes 2 and 3), and fibroblasts from the unrelated patient (11571) (lane 4). In C the sample for the unrelated patient is not present.
Superoxide can be generated from two potential sites within the bc1 complex (Raha et al. 2000). Inhibition with antimycin A prevents transfer of electrons from cytochrome b to Qi, thereby increasing the concentration of Qo semiquinone. This increase in the Qo semiquinone species results in an increase in the free-radical species, since superoxide is formed by donation of an electron from Qo to molecular oxygen (Raha et al. 2000). The antimycin A–dependent increase in production of superoxide was observed for the control human liver mitochondria (table 6). However, liver mitochondria obtained from the autopsied liver of one of the siblings failed to produce any significant increase in free-radical production after addition of antimycin A. Moreover, a statistically significant decrease in free-radical production was observed (table 6). Addition of either myxothiazol (100 μM) or stigmatellin (20 μM) to the liver mitochondria from either the patient or the control produced only a marginal (i.e., statistically nonsignificant) increase in the production of superoxides, above basal levels.
Table 6.
Effect of Inhibitor-Dependent Superoxide Production in Control and Multi-Complex Deficient Liver Mitochondria
|
Mean ± SEM SuperoxideFormationa [No. of Determinations](rel V/min) |
||
| Inhibitor | Control | Sibling 1 |
| No inhibitor | 343 ± 49 [4] | 349 ± 24 [4] |
| Antimycin A (20 μg/ml) | 1,402 ± 123 [4] | 107 ± 46 [8] |
| Stigmatellin (20 μM) | 380 ± 33 [4] | 408 ± 61 [4] |
| Myxothiazol (10 μM) | 413 ± 67 [4] | 431 ± 71 [4] |
Monitored using lucigenin luminescence after addition of succinate (5 mM) to mitochondrial suspension in the presence or absence of either antimycin A (see Subjects, Materials, and Methods section), stigmatellin, or myxothiazol. Values were standardized to the activity of citrate synthase, a mitochondrial matrix marker.
Discussion
Investigations of the cultured skin fibroblasts derived from the three siblings (4142, 4143, and 4144) and from the unrelated patient (11571) showed an increased L/P ratio in the cells, typical of a severe respiratory-chain problem (Robinson et al. 1985, 1990). Deficient activities of NADH cytochrome c reductase, the pyruvate dehydrogenase complex, the 2-oxoglutarate dehydrogenase complex, and succinate cytochrome c reductase suggested the presence of a defect that was decreasing the activity of several mitochondrial enzymes. This was further confirmed by metabolite analysis of the blood, pointing to a deficiency of both the branched-chain α-keto acid dehydrogenase complex and the glycine-cleavage system. However, the activities of pyruvate carboxylase, mitochondrial aconitase, lipoamide dehydrogenase (i.e., E3), and citrate synthase were normal. Immunoblot analysis performed on mitochondrial extracts derived from the four patient cells (fig. 2) showed that the pattern of the selected subunits (the PDH complex and the bc1 complex) was similar to that of the control cell line. This observation suggests that the examined polypeptides are correctly imported into and/or folded within the mitochondria and are assembled into complexes. Previous studies have indicated that patients with a defect in mitochondrial import machinery present with a lack of immunodetectable proteins within the organelle (Jin et al. 1999). Similarly, deficiencies in factors required for assembly of multisubunit complexes produce, at best, only partially assembled complexes (Brasseur et al. 1997; Petruzzella et al. 1998). This would indicate that the above-described genetic defect occurs at the posttranslational—and, probably, postassembly—level.
In both families there was no previous history of individuals on the maternal side who had symptoms associated with mitochondrial myopathy. Although mtDNA was not analyzed for pathogenic mutations, the fact that the majority of the affected enzyme complexes have no mitochondrially encoded components makes mitochondrial aberrations an unlikely cause.
In trying to pinpoint a genetic defect that would explain all of the above-described results, we decided to conduct functional complementation analysis using microcell-mediated chromosome transfer. Our results clearly show that introduction of a normal chromosome 2 into the three cell lines with multiple mitochondrial enzyme deficiency can revert their phenotype and restore enzyme activities. Evidence that phenotypic changes were not the mere consequence of manipulations associated with the chromosome-transfer protocols but were truly controlled by chromosome 2–specific DNA sequences comes from three observations. First, similar changes were not seen when chromosomes other than chromosome 2 were transferred into the same recipient cells. Second, genotype analysis of chromosome 2 markers for the three siblings gave two regions, one at 2p and one at 2q, in which all three siblings had common alleles. Third, only a small (5 cM) portion of the exogenous chromosome 2p in the hybrid cell line resulted in restoration of affected enzyme activities, further attesting to the association between these phenotypic properties and specific DNA sequences. This region, when absent in hybrid cell lines after chromosome 2 transfer, produced clones that still exhibited the mutant phenotype (one clone from patient 4143 and six clones from patient 11571; see the Results section).
Our ultimate goal in this study was to better define the chromosomal region(s) associated with the disorder that causes multiple mitochondrial enzyme malfunctions, in order to facilitate the eventual isolation of the specific gene(s) controlling this disorder. This was achieved with subclones of chromosome 2 hybrids that did not regain their affected enzyme activities. Examination of the integrity of the exogenous chromosome 2 in these clones showed that portions of the chromosome had been lost in each case. Similarly, examination of the integrity of the exogenous chromosome 2 clones that reverted to the mutant phenotype showed that, in these revertants, portions of it had been lost in each case. The smallest deletion that could be defined by this approach extended from A053XF9 to GATA-P132045 (table 5 and fig. 1). Most of the known polymorphisms within this region were noninformative, because the corresponding alleles on the exogenous chromosome were indistinguishable from alleles on the endogenous copies. This complicated attempts at further narrowing this candidate region.
Results from the measurement of bc1 complex (i.e., complex III) superoxide production (a) indicate that there is only a residual transfer of electrons from ubiquinone to the iron-sulfur center in the deficient cell line and (b) confirm a deficiency in the overall functioning of that complex. This reasoning is based on the observation that inhibition of electron flow between coenzyme b562 and Q°i, by antimycin, failed to produce any significant increase in reactive-species production (Raha et al. 2000). How can these observations be compatible with the Western-blot data suggesting that the ISP is fully assembled into the bc1 complex? Kapazoglou et al. (2000) showed that, in a triple-substitution mutant, C107S/H109R/C112S, the replacement of conserved residues involved in the ligation of the [2Fe-2S] center of the Rieske protein allowed the latter, in isolated chloroplasts, to assemble into the cytochrome bf complex, which nonetheless was an inactive complex. The cytochrome bf complex and the mitochondrial bc1 complex share a high degree of topological and functional similarity, especially with regard to the Rieske protein (Soriano et al. 1999). Therefore, a lack of incorporation of this [2Fe-2S] center may be a reasonable explanation for the activity loss in these patients.
In the attempt to determine the nature of the genetic defect underlying the above-described disorder, the identification of a common factor or process involved in all the affected enzyme complexes is all-important. An interesting biochemical characteristic of this disorder is the fact that, although the activities of the isolated respiratory enzymes II and III retain quite high intrinsic values (45%–60%), the combined activities of complexes I+III and complexes II+III are more dramatically suppressed (table 2). These results could suggest a defect in the mitochondrial coenzyme Q content and/or function, since both system I+III and system II+III require it as an electron carrier. Similar biochemical findings were reported, in a case of intrinsic muscle coenzyme Q deficiency, by Boitier et al. (1998). However, this hypothesis cannot explain the observed deficiencies of either the mitochondrial dehydrogenases (PDHC, 2-oxoglutarate dehydrogenase complex [OGDHC], and branched chain α-ketoacid dehydrogenase complex [BCKDHC]) or the glycine-cleavage system.
A more plausible explanation is provided by the possibility that the unknown factor is somehow involved in the metabolism of sulfur in the mitochondria and/or the supply of sulfur to the affected complexes. Sulfur is an essential part of lipoic acid, which is a cofactor shared by the PDH complex, the 2-oxoglutarate dehydrogenase complex, the branched-chain α-keto acid dehydrogenase complex, and the glycine-cleavage system. Sulfur is also a part of respiratory complexes II and III, in the form of the iron-sulfur centers, and cysteine desulfurase and sulfur transferase are required for assembly. In contrast, assembled sulfur-containing active centers are not a part of either the pyruvate carboxylase complex or the citrate synthase complex.
Given the second hypothesis, how can we explain the fact that mitochondrial aconitase—an iron-sulfur center–containing enzyme—is not affected? In the past, it has been reported that, in respiratory-chain abnormalities involving iron-sulfur clusters, mitochondrial aconitase activity was also depressed (Hall et al. 1993; Rotig et al. 1997). However, no link was reported between these disorders and the activities of mitochondrial dehydrogenases. Furthermore, in the case of Friedreich ataxia, in which mutated frataxin triggers aconitase and mitochondrial iron-sulfur respiratory-enzyme deficiency, the defect stems from abnormal iron homeostasis rather than from sulfur homeostasis. Therefore, it is very possible that the mechanisms of sulfur insertion into iron-sulfur centers for respiratory-chain complexes and lipoic acid are different from the mechanisms required for aconitase.
The nearly completed Human Genome Project should provide sequence information about the isolated chromosome 2 interval where the locus for the unknown gene is most likely located. At the time of submission, extensive searches throughout this region, which contains ⩾30 predicted genes, have yielded no obvious candidates. Future work will determine whether the gene product involved proves to have either a sulfur transferase activity or a cysteine desulfurase activity. Examination of data from the Human Genome Project that are particular to this locus reveals that it is a relatively gene-rich region with a mix of well-characterized genes, such as malate dehydrogenase, and other less–well-defined genes. After extensive analysis, we could find, in the region of chromosome 2p14-2p13, no indication of either sulfur transferase genes or genes involved in ubiquinone synthesis.
Acknowledgment
We thank the Medical Research Council of Canada for the support of this investigation.
References
- Bodnar AG, Cooper JM, Leonard JV, Schapira AH (1995) Respiratory-deficient human fibroblasts exhibiting defective mitochondrial DNA replication. Biochem J 305:817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitier E, Degoul F, Desguerre I, Charpentier C, Francois D, Ponsot G, Diry M, Rustin P, Marsac C (1998) A case of mitochondrial encephalomyopathy associated with a muscle coenzyme Q10 deficiency. J Neurol Sci 156:41–46 [DOI] [PubMed] [Google Scholar]
- Brasseur G, Tron G, Dujardin G, Slonimski PP, Brivet-Chevillotte P (1997) The nuclear ABC1 gene is essential for the correct conformation and functioning of the cytochrome bc1 complex and the neighbouring complexes II and IV in the mitochondrial respiratory chain. Eur J Biochem 246:103–111 [DOI] [PubMed] [Google Scholar]
- Brown GK, Haan EA, Kirby DM, Scholem RD, Wraith JE, Rogers JG, Danks DM (1988) “Cerebral” lactic acidosis: defects in pyruvate metabolism with profound brain damage and minimal systemic acidosis. Eur J Pediatr 147:10–14 [DOI] [PubMed] [Google Scholar]
- Brown MD, Wallace DC (1994) Molecular basis of mitochondrial DNA disease. J Bioenerg Biomembr 26:273–289 [DOI] [PubMed] [Google Scholar]
- Compton, T (1993) An immortalized human fibroblast cell line is permissive for human cytomegalovirus infection. J Virol 67:3644–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EE, Hood DA (1997) Influence of aging on protein import into cardiac mitochondria. Am J Physiol 272:H2983–H2988 [DOI] [PubMed] [Google Scholar]
- Cuthbert AP, Trott DA, Ekong RM, Jezzard S, England NL, Themis M, Todd CM, Newbold RF (1995) Construction and characterization of a highly stable human:rodent monochromosomal hybrid panel for genetic complementation and genome mapping studies. Cytogenet Cell Genet 71:68–76 [DOI] [PubMed] [Google Scholar]
- Darley-Usmar VM, Rickwood D, Wilson MT (eds) (1987) Mitochondria: a practical approach. IRL Press, Oxford [Google Scholar]
- DiMauro S, Zeviani M, Servidei S, Prelle A, Miranda AF, Bonilla E, Schon EA (1988) Biochemical and molecular aspects of cytochrome c oxidase deficiency. In: DiDonato S, Mamoli A, Rowland LP (eds) Advances in neurology. Vol 48, Raven Press, New York, pp 93–105 [PubMed] [Google Scholar]
- Ekong J (1982) Studies of the deficiencies of the pyruvate dehydrogenase complex. MSc thesis, University of Toronto, Toronto [Google Scholar]
- Fournier RE (1981) A general high-efficiency procedure for production of microcell hybrids. Proc Natl Acad Sci USA 78:6349–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman LI, Shoubridge EA (1996) Mitochondrial genetics and human disease. Bioessays 18:983–991 [DOI] [PubMed] [Google Scholar]
- Hall RE, Henriksson KG, Lewis SF, Haller RG, Kennaway NG (1993) Mitochondrial myopathy with succinate dehydrogenase and aconitase deficiency: abnormalities of several iron-sulfur proteins. J Clin Invest 92:2660–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatefi Y, Stiggall DL (1978) Preparation and properties of succinate: ubiquinone oxidoreductase (complex II). Methods Enzymol 53:21–27 [DOI] [PubMed] [Google Scholar]
- Hoppel CL, Kerr DS, Dahms B, Roessmann U (1987) Deficiency of the reduced nicotinamide adenine dinucleotide dehydrogenase component of complex I of mitochondrial electron transport. J Clin Invest 80:71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland K, Leonard JV (1983) Revised assays for the investigation of congenital lactic acidosis using 14C keto acids, eliminating problems associated with spontaneous decarboxylation. Clin Chim Acta 133:177–187 [DOI] [PubMed] [Google Scholar]
- Jin H, Kendall E, Freeman TC, Roberts RG, Vetrie DL (1999) The human family of deafness/dystonia peptide (DDP) related mitochondrial import proteins. Genomics 61:259–267 [DOI] [PubMed] [Google Scholar]
- Jin H, May M, Tranebjaerg L, Kendall E, Fontan G, Jackson J, Subramony SH, Arena F, Lubs H, Smith S, Stevenson R, Schwartz C, Vetrie D (1996) A novel X-linked gene, DDP, shows mutations in families with deafness (DFN-1), dystonia, mental deficiency and blindness. Nat Genet 14:177–180 [DOI] [PubMed] [Google Scholar]
- Kapazoglou A, Mould RM, Gray JC (2000) Assembly of the Rieske iron-sulphur protein into the cytochrome bf complex in thylakoid of isolated pea chloroplasts. Eur J Biochem 267:352–360 [DOI] [PubMed] [Google Scholar]
- Kennaway NG (1988) Defects in the cytochrome bc1 complex in mitochondrial diseases. J Bioenerg Biomembr 20:325–352 [DOI] [PubMed] [Google Scholar]
- Koehler CM, Leuenberger D, Merchant S, Renold A, Junne T, Schatz G (1999) Human deafness dystonia syndrome is a mitochondrial disease. Proc Natl Acad Sci USA 96:2141–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach RJ, Thayer MJ, Schafer AJ, Fournier RE (1989) Physical mapping of human chromosome 17 using fragment-containing microcell hybrids. Genomics 5:167–176 [DOI] [PubMed] [Google Scholar]
- Lombes A, Mendell JR, Nakase H, Barohn RJ, Bonilla E, Zeviani M, Yates AJ, Omerza J, Gales TL, Nakahara K (1989) Myoclonic epilepsy and ragged-red fibers with cytochrome oxidase deficiency: neuropathology, biochemistry, and molecular genetics. Ann Neurol 26:20–33 [DOI] [PubMed] [Google Scholar]
- Lombes A, Romero NB, Touati G, Frachon P, Cheval MA, Giraud M, Simon D, Ogier de Baulny H (1996) Clinical and molecular heterogeneity of cytochrome c oxidase deficiency in the newborn. J Inherit Metab Dis 19:286–295 [DOI] [PubMed] [Google Scholar]
- Moraes CT, Kenyon L, Hao H (1999) Mechanisms of human mitochondrial DNA maintenance: the determining role of primary sequence and length over function. Mol Biol Cell 10:3345–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreadith RW, Batshaw ML, Ohnishi T, Kerr D, Knowx B, Jackson D, Hruban R, Olson J, Reynafarse B, Lehninger AL (1984) Deficiency of the iron-sulfur clusters of mitochondrial reduced nicotinamide-adenine dinucleotide-ubiquinone oxidoreductase (complex I) in an infant with congenital lactic acidosis. J Clin Invest 74:685–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morten KJ, Caky M, Matthews PM (1998) Mechanisms of expression of pyruvate dehydrogenase deficiency caused by an E1 α subunit mutation. Neurology 51:1324–1330 [DOI] [PubMed] [Google Scholar]
- Newbold RF, Cuthbert AP (1996) Mapping human senescence genes using interspecific monochromosome transfer. In: Fresheny RI, Freshney MG (eds) Culture of immortalized cells. Wiley-Liss, New York, pp 53–75 [Google Scholar]
- Petruzzella V, Tiranti V, Fernandez P, Ianna P, Carrozzo R, Zeviani M (1998) Identification and characterization of human cDNAs specific to BCS1, PET112, SCO1, COX15, and COX11, five genes involved in the formation and function of the mitochondrial respiratory chain. Genomics 54:494–504 [DOI] [PubMed] [Google Scholar]
- Pitkänen S, Raha S, Robinson BH (1996) Diagnosis of complex I deficiency in patients with lactic acidemia using skin fibroblast cultures. Biochem Mol Med 59:134–137 [DOI] [PubMed] [Google Scholar]
- Pitkänen S, Robinson BH (1996) Mitochondrial complex I deficiency leads to increased production of superoxide radicals and induction of superoxide dismutase. J Clin Invest 98:345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racker E (1950) Spectrophotometric measurements of the enzymatic formation of fumaric and cis-aconitic acids. Biochem Biophys Acta 4:211–214 [DOI] [PubMed] [Google Scholar]
- Raha S, McEachern GE, Myint AT, Robinson BH (2000) Superoxides from mitochondrial complex III: the role of manganese superoxide dismutase. Free Rad Biol Med 29:170–180 [DOI] [PubMed] [Google Scholar]
- Robinson BH (1995) Lactic acidemia (disorders of pyruvate carboxylase, pyruvate dehydrogenase). In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease. 7th ed. McGraw-Hill, New York, pp 1479–1492 [Google Scholar]
- Robinson BH, DeMeirlier L, Glerum DM, Sherwood WG, Becker L (1987a) Clinical presentation of patients with mitochondria respiratory chain defects NADH coenzyme Q reductase and cytochrome oxidase: clues to the pathogenesis of Leigh’s Disease. J Pediatr 110:216–222 [DOI] [PubMed] [Google Scholar]
- Robinson BH, Glerum DM, Chow W, Petrova-Benedict R, Lightowlers R, Capaldi R (1990) The use of skin fibroblast cultures in the detection of respiratory chain defects in patients with lacticacidemia. Pediatr Res 28:549–555 [DOI] [PubMed] [Google Scholar]
- Robinson BH, MacKay N, Goodyer P, Lancaster G (1985) Defective intramitochondrial NADH oxidation in skin fibroblasts from an infant with fatal neonatal lactica acidemia. Am J Hum Genet 37:938–946 [PMC free article] [PubMed] [Google Scholar]
- Robinson JB, Brent LG, Sumegi B, Srere PA (1987b) An enzymatic approach to the study of the Krebs tricarboxylic acid cycle. In: Darley-Usmar VM, Rickwood D, Wilson MT (eds) Mitochondria: a practical approach. IRL Press, Oxford, pp 153–170 [Google Scholar]
- Rotig A, de Lonlay P, Chretien D, Foury F, Koenig M, Sidi D, Munnich A, Rustin P (1997) Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat Genet 17:215–217 [DOI] [PubMed] [Google Scholar]
- Sheu KRF, Ju CW, Utter MF (1981) Pyruvate dehydrogenase complex in normal and deficient fibroblasts. J Clin Invest 67:1463–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano GM, Ponamarev MV, Carrell CJ, Xia D, Smith JL, Cramer WA (1999) Comparison of the cytochrome bc1 complex with the anticipated structure of the cytochrome b6f complex: le plus ca change le plus c'est la meme chose. J Bioenerg Biomembr 31:201–213 [DOI] [PubMed] [Google Scholar]
- Valnot I, Kassis J, Chretien D, de Lonlay P, Parfait B, Munnich A, Kachaner J, Rustin P, Rotig A (1999) A mitochondrial cytochrome b mutation but no mutations of nuclearly encoded subunits in ubiquinol cytochrome c reductase (complex III) deficiency. Hum Genet 104:460–466 [DOI] [PubMed] [Google Scholar]
- van Dijl JM, Kutejova E, Suda K, Perecko D, Schatz G, Suzuki CK (1998) The ATPase and protease domains of yeast mitochondrial Lon: roles in proteolysis and respiration-dependent growth. Proc Natl Acad Sci USA 95:10584–10589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Acevedo M, Antaramian A, Corona N, Gonzalez-Halphen D (1993) Subunit structures of purified beef mitochondrial cytochrome bc1 complex from liver and heart. J Bioenerg Biomembr 25:401–410 [DOI] [PubMed] [Google Scholar]
- Yu L, Yu CA (1982) Quantitative resolution of succinate-cytochrome c reductase into succinate-ubiquinone and ubiquinol-cytochrome c reductases. J Biol Chem 257:2016–2021 [PubMed] [Google Scholar]
- Zeviani M, Bonilla E, DeVivo DC, DiMauro S (1989) Mitochondrial diseases. Neurol Clin 7: 123–156 [PubMed] [Google Scholar]