Abstract
Leigh syndrome (LS) affects 1/40,000 newborn infants in the worldwide population and is characterized by the presence of developmental delay and lactic acidosis and by a mean life expectancy variously estimated at 3–5 years. Saguenay–Lac-Saint-Jean (SLSJ) cytochrome oxidase (COX) deficiency (LS French-Canadian type [LSFC] [MIM 220111]), an autosomal recessive form of congenital lactic acidosis, presents with developmental delay and hypotonia. It is an LS variant that is found in a geographically isolated region of Quebec and that occurs in 1/2,178 live births. Patients with LSFC show a phenotype similar to that of patients with LS, but the two groups differ in clinical presentation. We studied DNA samples from 14 patients with LSFC and from their parents, representing a total of 13 families. Because of founder effects in the SLSJ region, considerable linkage disequilibrium (LD) was expected to surround the LSFC mutation. We therefore performed a genomewide screen for LD, using 290 autosomal microsatellite markers. A single marker, D2S1356, located on 2p16, showed significant (P < 10−5) genomewide LD. Using high-resolution genetic mapping with additional markers and four additional families with LSFC, we were able to identify a common ancestral haplotype and to limit the critical region to ∼2 cM between D2S119 and D2S2174. COX7AR, a gene encoding a COX7a-related protein, had previously been mapped to this region. We determined the genomic structure and resequenced this gene in patients with LSFC and in controls but found no functional mutations. Although the LSFC gene remains to be elucidated, the present study demonstrates the feasibility of using a genomewide LD strategy to localize the critical region for a rare genetic disease in a founder population.
Introduction
The metabolic production of ATP is made possible by electron transfer and proton pumping within the inner membrane of the mitochondrion. This respiratory chain consists of five major multiprotein complexes, of which the cytochrome oxidase (COX) complex, known as “complex IV,” catalyzes the crucial reaction of oxygen reduction. COX is a multimeric, transmembrane protein of ∼200 kD that consists of 13 subunits—of which 10 are encoded by nuclear DNA and 3 are encoded by mtDNA. Inherited deficiencies of COX activity in humans have been reported to exist in multiple forms: autosomal recessive, autosomal dominant, X linked, and maternal, and the variations are presumed to depend on the gene involved (Dahl 1998). Only a few cases of COX deficiency have been shown to result from defects in the mitochondrial COX genes. Defects in three different nuclear genes that encode proteins involved in the maturation and assembly of complex IV can also lead to COX deficiencies. Each defect presents a different clinical phenotype: (1) mutations in SURF1 (Tiranti et al. 1998; Zhu et al. 1998), a gene located in chromosomal region 9q34, have been associated with Leigh syndrome (LS [MIM 256000]) (subacute necrotizing encephalomyopathy); (2) mutations in SCO2, a gene located in chromosomal region 22q13 have been associated with fatal infantile cardioencephalomyopathy (Papadopoulou et al. 1999); (3) mutations in COX10, a gene located in chromosomal region 17p13.1-q11.1, have been associated with tubulopathy and leukodystrophy (Valnot et al. 2000b); and (4) mutations in SCO1, a gene located in chromosomal region 17p11, have been associated with hepatic failure and encephalopathy (Valnot et al. 2000a). In addition, although LS can result from mutations in the SURF1 gene, it can also result from defects in pyruvate dehydrogenase, mitochondrial complex I, mitochondrial complex V, or succinate dehydrogenase (Robinson 1993; Bourgeron et al. 1995).
During the past few decades, several children in a geographically isolated region of northeastern Quebec, particularly in the Saguenay–Lac-Saint-Jean (SLSJ) region, were found to have a phenotype similar to that of LS. Children with LS French-Canadian type (LSFC) present with developmental delay and hypotonia (Morin et al. 1993); because of acute crises of metabolic acidosis and strokelike episodes, the death rate among these infants is high (Merante et al. 1993; Morin et al. 1999). At autopsy, changes typical of LS are present; and, although the course of LSFC is less fulminant than that of most cases of LS described in the literature (Montpetit et al. 1971; Pincus 1972), all the typical features—including ataxia, dystonia, optic atrophy, ophthalmoplegia, ptosis, nystagmus, tremor, and respiratory abnormalities—have been observed in various patients with LSFC.
Many tissues from patients with LSFC are deficient in COX activity. Specifically, brain and liver tissues have 10%–20% of the normal COX activity; fibroblasts and skeletal muscle have 50% of normal activity; and kidney and heart tissues have almost 100% of normal activity (Merante et al. 1993). In humans, 2 (VIa and VIIa) of the 10 nuclear-encoded subunits of COX exist in two isoforms (heart and liver, denoted by “H” and “L,” respectively), which differ both in sequence and in expression patterns. COXVIa-L and COXVIIa-L are expressed in many tissues, but COXVIa-H and COXVIIa-H are expressed mainly in heart and skeletal muscle, with low levels in smooth muscle. Because COX deficiency is most pronounced in the liver of these patients, liver tissue–mRNA samples were analyzed using northern blot analysis for subunits VIa-L and VIIa-L (Merante et al. 1993). The transcript levels appeared to be normal, but western blot analysis using an anti-holoenzyme COX antibody showed decreased amounts of COX subunits within the liver mitochondria (Merante et al. 1993). These results imply that, although the mRNAs are present at normal levels, the COX subunits fail to be efficiently assembled within the mitochondria, at least in liver cells. However, sequencing of the 10 nuclear-encoded COX subunits (the liver isoforms of VIa and VIIa) failed to identify any potential mutations (Lee et al. 1998). Consequently, it is likely that the biochemical defect in patients with SLSJ COX deficiency (i.e., LSFC) resides in a COX assembly factor or cofactor that plays a more important role in the liver and brain than in other tissues, such as kidney or heart.
The SLSJ region of Quebec is 125 miles northeast of Quebec City and was settled during the years 1838–1911, during which ∼75% of the settlers were from the Charlevoix region (Gavreau and Bourque 1988). The community of Charlevoix itself was started in 1675, with 599 founders of French descent arriving, in the course of seven generations, from Quebec City (Jette et al. 1991; Labuda et al. 1996). There has been little migration into the SLSJ region since 1870, and the community has grown from 18,000 in 1852 to 320,000 in 2000 (Labuda et al. 1996). Because of the founder effect (De Braekeleer et al. 1993; Heyer 1999), several dominant and recessive genetic disorders are more prevalent in the SLSJ region than they are elsewhere. The incidence of LSFC is 1/2,178 live births (De Braekeleer et al. 1993), and carrier rates for LSFC have been estimated to be 1/23 adults (Morin et al. 1993). Several other autosomal recessive disorders that also have a prevalence of ∼1/2,000 include cystic fibrosis, tyrosinemia type I, vitamin D–dependent rickets, polyneuropathy with corpus callosum agenesis, and autosomal spastic ataxia of Charlevoix-Saguenay (De Braekeleer et al. 1991).
Founder effects are reflected in the limited allelic diversity observed in patients with these diseases. For example, De Braekeleer et al. (1998) recently demonstrated, after complete ascertainment, that only three mutations account for 94% of all cystic fibrosis chromosomes in this population. Even more striking is the observation that a single homozygous mutation was found in 80% of patients with the hereditary tyrosinemia type I in the SLSJ region (Grompe et al. 1994). Moreover, because of the founder effect in a young population, large blocks of ancestral DNA have been shown to flank the disease mutations in this population. Specifically, recent studies have demonstrated that ancestral haplotypes spanning ∼8–10 cM are common to 40%–60% of affected chromosomes (4–6 cM being common to nearly 80%) of populations with rare diseases from this region (Labuda et al. 1996; Richter et al. 1999; Couture et al. 1999).
Because the mean life expectancy for individuals with LSFC is 3–5 years (range 0–7 years), the majority of families with LSFC have only one affected child and no other affected immediate relatives. Consequently, although we had almost complete ascertainment of patients with LSFC during a period of 7 years, DNA samples from only 14 affected children from 13 families were available for study; the lack of multiplex families prevented a traditional linkage approach to disease-gene discovery. Because of the population characteristics of the SLSJ region, a genomewide LD search was performed to identify the LSFC gene. Using analysis of the genotype data from a total of 312 autosomal microsatellite markers, we identified a single marker, located on chromosome 2p16, that was in LD with the LSFC phenotype. High-resolution mapping with 11 additional microsatellite markers limited the critical region to ∼2 cM, between D2S119 and D2S2174.
Families, Material, and Methods
Families and DNA Isolation
Patients were diagnosed with LSFC on the basis of phenotype, genetic epidemiology, lactic acidemia, pathology, and levels of COX activity (Morin et al. 1993). DNA samples were drawn from children with LSFC and from their parents, after the parents had given informed consent. The probands were ascertained among patients who came to the attention of C.C.M. or G.A.M. during the period 1990–97. The DNA samples were extracted from blood, frozen tissue, fibroblast cell cultures, or frozen lymphoblast cell lines. Some individuals with LSFC were not included in the genetic study, because their samples were not available or did not yield sufficient DNA.
The genetic material available for the genomewide screen came from 14 patients with LSFC and from 26 parents, representing 13 families (family COX007 had two affected siblings). For the high-resolution mapping, DNA samples were collected from four additional families with LSFC and were genotyped. In one of these pedigrees (COX014), DNA samples from both parents and one affected child were available; in two pedigrees (COX005 and COX006), DNA was available from only one parent and one affected child; and in one pedigree (COX022-58), only the affected child's DNA was available. Control DNA samples were collected from individuals in the SLSJ region who were not affected by LSFC and who had no children with LSFC.
Genomewide Screen and High-Resolution Mapping
The genomewide screen was performed using the DNA samples from the initial group of 13 families with LSFC; the screen was performed using 290 fluorescence-labeled microsatellite markers, which had an average intermarker distance of 12 cM. The markers were from a modified version of the Cooperative Human Linkage Center (CHLC) human screening set (Research Genetics). The reactions were set up with a robotic pipetting station (Rosys Robotic Systems) in 192-well polypropylene plates (Corning Costar) and were subjected to PCR conditions in a customized thermocycler (Intelligent Automation Systems). PCR products were pooled into panels (8–10 markers per panel) on the basis of allele size range and fluorescence labeling. Aliquots of these samples were mixed either with Tamra-labeled GENESCAN 500 and GENESCAN 2500 (Applied Biosystems) or with rhodamine-labeled MapMarkers (Bioventures), and sequencing was performed on ABI 377 sequencers (Applied Biosystems). Gels were processed, tracked, and reviewed using an automated system developed at the Whitehead Institute for Biomedical Research/MIT Center for Genome Research, as described elsewhere (Rioux et al. 1998). Allele binning and inheritance checking were performed with PEDMANAGER (developed by M. P. Reeve-Daly and M. J. Daly).
The high-resolution mapping of the chromosome 2p16 region was performed using 11 additional markers flanking marker D2S1356 and in DNA samples from 18 individuals with LSFC and from 30 of their parents. The additional markers used for the scan were selected from the Généthon sex-averaged genetic map of chromosome 2. They were GATA194B06, D2S2305, D2S2259, D2S2306, D2S2294, D2S119, D2S2298, D2S2174, D2S2291, D2S2240, and D2S2378. The original marker, D2S1356, was also regenotyped to confirm the results from the genomewide screen. Samples for PCR were set up in 96-well polypropylene plates (MJ Research) and were amplified in a thermocycler (MJ Research). The gels were electrophoresed and processed in a manner similar to that used for gels in the genomewide screen.
Statistical Analysis
The genotype data from the genomewide screen were analyzed using the transmission/disequilibrium test (TDT). Specifically, the alleles at each of the 290 markers were examined for excess transmission from heterozygous parents to children with LSFC. To assess the significance of the genomewide TDT results for marker D2S1356, permutation tests were performed using the same genotype data. For each trio, chromosomes were randomly reassigned as transmitted or untransmitted, to form a permuted data set. The percentage of permuted data sets with values as significant as that seen at D2S1356 was calculated.
Four additional families with LSFC were genotyped for marker D2S1356, but both parental genotypes were available for only one of these. Fisher’s exact test was used to determine the significance of the difference between non-238 alleles and 238 alleles, in the LSFC and control chromosomes from the same population. The control genotype data for marker D2S1356 were derived from unrelated individuals (without LSFC) who were patients at the Hôpital de la Sagamie, in the SLSJ region.
To quantify the extent of LD in the region surrounding marker D2S1356, Pexcess (δ) was computed for the genotype data from the markers included in the high-resolution mapping. Pexcess represents the strength of LD and is calculated according to the following formula: (Paffected-Pnormal)/(1-Pnormal). In our study, the Paffected value is calculated from the frequency of the common allele in all the patients with LSFC, and the Pnormal value is the frequency among untransmitted parental chromosomes.
Screening YAC Clones from the 2p16 Region
In an attempt to determine whether the previously known subunits of COX could be mapped to the region identified by use of the genome scan, we designed primers for the COX subunits and screened them with a collection of YAC clones previously mapped to 2p16 (Whitehead Institute for Biomedical Research). The primers designed from the 3′UTR of each cDNA sequence by Primer software, version 3.0 (Whitehead Institute for Biomedical Research), were specific to the 10 COX subunits (IV, Va, Vb, VIa-L, VIb, VIc, VIIa-L, VIIb, VIIc, and VIII) and three of its assembly factors (COX10, COX17, and OXA1).
The sequences for the forward and reverse primers are as follows: COXIV, 5′-aacgagtggaagaagtgaga-3′ and 5-gtaaataggcatggagttgc-3′; COXVa, 5′-cccaaggatttattgacatt-3′ and 5′-ccattacatggcttggtact-3′; COXVb, 5′-actgagcacctgcactaaat-3′ and 5′-gagaaggagccaatgcaa-3′; COXVIa, 5′-gggaccttaagctaaccttc-3′ and 5′-ttatttaagccatctcctgc-3′; COXVIb, 5′-catctccctttcctctgtc-3′ and 5′-catgatttaggatcctggg-3′; COXVIc, 5′-ggctggtatctttcagagtg-3′ and 5′-ttcaggaacacaagtcagtg-3′; COXVIIa, 5′-ggttcagtttcattcagctc-3′ and 5′-gctttattggtggcagttac-3′; COXVIIb, 5′-aaaggaatggaggaatcagt-3′ and 5′-acagtgctttaatttggcat-3′; COXVIIc, 5′-aaggatgtttcagttcctcc-3′ and 5′-ttctagtttgatccacttcca-3′; COXVIII, 5′-ggatcatgtctattcaattcc-3′ and 5′-tttattgttacaagggggac-3′; COX10, 5′cttgacaggatgttttcgat-3′ and 5′-tgcaatgttctccacagtaa-3′; OXA1, 5′-agccttcaaccttctagctg-3′ and 5′-aaggaaaagtgcaaatgtgg-3′; and COX17, 5′-tccacatatcaaagttcgtcaaag-3′ and 5′-aggagaagaagccgctgaa-3′.
Each primer pair was tested on each of the following YAC clones, covering a region of ∼10–15 Mb around D2S1356: 929d9, 718e9, 872c7, 816c8, 908h7, 798f2, 849e9, 934e2, 748b9, 933b10, 930a1, 957b10, 913g4, 761e1, 972c5, 919c10, 955f10, 945f10, 945g11, 944d7, 899f9, 819a6, and 797a3. The PCR reactions were set up in 96-well polypropylene plates and were amplified in thermocyclers (MJ Research). The products of these reactions were electrophoresed on 1% agarose gels, and the presence or absence of bands of expected size indicated whether these genes were located within the genomic region represented by the overlapping YAC clones.
Genomic Structure of COX7AR
After determination of the LSFC critical region, by use of the genomewide and high-resolution mapping, a search through the Genemap database (National Center for Biotechnology Information) identified a cDNA sequence for a possible candidate gene called “COX7AR,” which mapped close to marker D2S119. To screen the patients' DNA for mutations, the genomic sequence for this candidate gene was elucidated. Specifically, several PCR primer pairs were designed using the published COX7AR cDNA sequence (GenBank accession number AB007618). One pair of primers was 5′-gatgcgggaagccggactct-3′ and 5′-ccagtcctctgcagcgtaag-3′, which cover the coding region and give an expected size of 392 bp on cDNA. Another pair was 5′-catgtactacaagtttagtgg-3′ and 5′-aaaaggaccttcaaaagggttt-3′, which also span the coding region and which amplify 404 bp on cDNA. A third set of primers was 5′-atctcctatctttagtgaaatct-3′ and 5′-tcatgtatttattccacagtc-3′, which span a 198-bp region in the 3′UTR. PCR assays using these primer pairs were then performed to screen pooled P1 artificial chromosome (PAC) clones.
The DNA for three positive PAC clones (RPCI-5.1126_I_4, RPCI-5.1148_J_17, and RPCI-5.1149_D_18) were isolated and used as templates in sequencing reactions. The cDNA-specific primers were used for direct sequencing of the PAC DNA, with the BigDye Terminator sequencing kit (Applied Biosystems). The sequencing reactions were purified with Sephadex G-50 medium-grade chromatography columns (Pharmacia). The samples were then electrophoresed on ABI 377 sequencers (Applied Biosystems). The sequencing gel files were processed with Bass/Grace software (Whitehead Institute for Biomedical Research), and base calling was performed using the TROUT software program (Whitehead Institute for Biomedical Research). The sequences were aligned using the Staden software package (Staden 1996). The entire intronic sequence, together with ∼1 kb of the 5′UTR and 315 bp of the 3′ end of the 3′UTR, was determined for this gene by an iterative approach of direct sequencing with primers designed from the newly obtained sequence. This genomic sequence was examined for potential transcription factor–binding sites, by TFSEARCH (version 3.1), with a threshold of 85.0 points.
Mutation Screening and Physical Mapping of COX7AR
To screen for potential mutations in the DNA of patients with LSFC, overlapping PCR assays were designed to amplify the DNA samples from three to five patients with LSFC and from three control individuals. Each PCR product was purified using the solid-phase reversible immobilization technique (Hawkins 1994), and then both strands were sequenced using the ThermoSequenase DYEnamic direct cycle-sequencing kit (Amersham). The products from the sequencing reactions were electrophoresed on ABI 377 DNA sequencing machines and were processed as described above. Three of the four single-nucleotide polymorphisms (SNPs) discovered through the use of this methodology were then genotyped in DNA samples from the remaining patients with LSFC and from their parents, by resequencing of both strands. Each sequence trace was independently reviewed by at least two individuals.
Immunoblotting of COX7RP
Mitochondria were isolated, as described elsewhere, from cultured skin fibroblasts of patients with COX deficiency (Pitkänen and Robinson 1996). Antibodies against COX7RP were generated by injecting rabbits with a peptide identical to the last 12 amino acids of the protein conjugated to keyhole limpet hemocyanin (Research Genetics). Mitochondrial proteins (100 μg of protein) were separated on 18% SDS-PAGE gels and transferred to a 0.2-mm nitrocellulose membrane. Immunoblotting procedures were performed, as described by McEachern et al. (2000), using a 1:2,000 dilution of primary antibody for all three antibodies. Antibody to beef heart COX was generated as described by Merante et al. (1993). SURF1p antibodies were generated by injecting rabbits with a SURF1/glutathione transferase–fusion protein generated in bacteria.
Results
Genomewide Screen and High-Resolution Genetic Mapping
Because the expected LD surrounding the LSFC mutation was ∼8–10 Mb, a genomewide search for LD was undertaken with 290 evenly spaced microsatellite markers. Specifically, DNA samples from 13 families, comprising 14 children with LSFC (one affected sibling pair and 12 singletons) and 26 parents, were genotyped for all markers. By use of the TDT, the alleles at each marker were examined for excess transmission. From the entire genome scan, only one marker—D2S1356, located in chromosomal region 2p16—showed significant LD with the LSFC phenotype. Table 1 shows the results from the TDT for marker D2S1356 and demonstrates that allele 238 is in strong disequilibrium with the LSFC chromosomes. Although the observed excess transmission was associated with a significant P value (1.2 × 10−5), we had tested close to 300 microsatellite markers and needed to estimate the significance of this finding after correction for multiple hypothesis testing. A conservative Bonferroni correction for performing 1,500 tests (300 markers, each with five alleles) would suggest a corrected P value of ∼.02. To determine a more accurate estimate of significance, we also performed permutation tests with the same genotype data but with the transmitted/untransmitted status assigned at random. Of 1 million permutations of our entire genomewide screen, only 2,051 (0.2%) contained a result as strong (P<.0001) as the result observed for D2S1356, which indicated that the LD for this marker was of genomewide significance.
Table 1.
Results of the TDT for D2S1356
|
No. |
||||
| Allele | Transmitted | Untransmitted | χ2 | P |
| 232 | 0 | 3 | 3 | .083 |
| 235 | 0 | 8 | 8 | .005 |
| 238 | 21 | 1 | 18.18 | .00002 |
| 240 | 0 | 1 | 1 | .317 |
| 244 | 0 | 3 | 3 | .083 |
| 247 | 1 | 2 | .33 | .564 |
| 250 | 0 | 3 | 3 | .083 |
| 253 | 0 | 1 | 1 | .317 |
DNA samples from four additional probands with LSFC and from their parents were then genotyped for the D2S1356 marker. Because DNA samples were available for both parents in only one of these additional families, the full data set was examined for association, by use of a case-control design. Because the TDT results indicated that allele 238 was associated with the disease, Fisher's exact test was used to compare the proportions of all non-238 and 238 alleles in the case population (2 and 34, respectively) and in the control population (163 and 82, respectively). The results from this test demonstrated a highly significant (P<10-12) association between allele 238 of marker D2S1356 and the LSFC phenotype (table 2).
Table 2.
Results of Test of Association for D2S1356 among the Population with LSFC and among the Saguenay Control Population
|
No. of Alleles (Frequency) in |
||
| Allele Size | Population with LSFC | Control Population |
| 223 | 0 (0) | 1 (.04) |
| 232 | 0 (0) | 7 (.029) |
| 235 | 1 (.028) | 51 (.208) |
| 238 | 34 (.944) | 82 (.335) |
| 241 | 0 (0) | 8 (.033) |
| 244 | 0 (0) | 33 (.135) |
| 247 | 1 (.028) | 14 (.057) |
| 250 | 0 (0) | 38 (.155) |
| 253 | 0 (0) | 10 (.041) |
| 257 | 0 (0) | 1 (.004) |
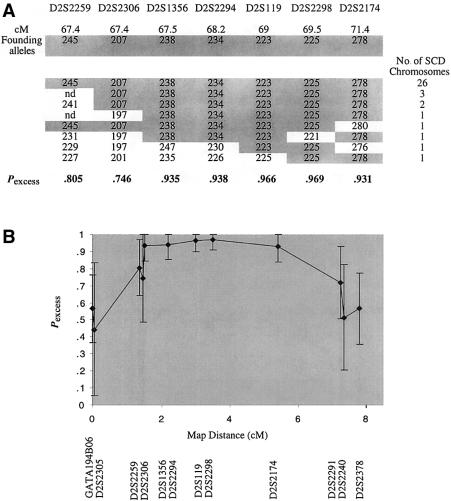
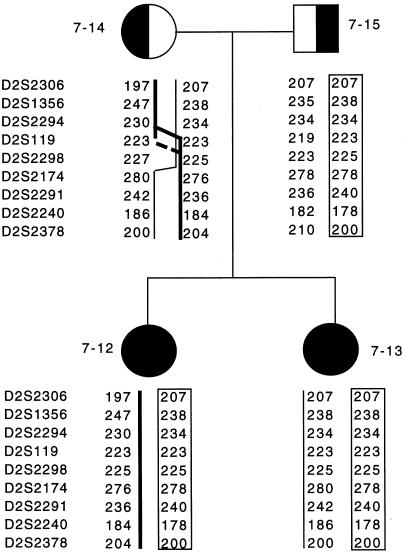
After using LD to map the LSFC gene to a chromosomal region surrounding marker D2S1356, we genotyped 11 additional microsatellite markers in the 18 LSFC samples, to identify the ancestral haplotype and to delimit the LSFC critical region. An ancestral LSFC haplotype was defined by the allele, at each marker, that was most common to the LSFC chromosomes and that showed the highest Pexcess value. Specifically, GATA194B06, D2S2305, D2S2259, D2S2306, D2S1356, D2S2294, D2S119, D2S2298, D2S2174, D2S2291, D2S2240, and D2S2378 (spanning ∼10 cM) defined the following ancestral haplotype: 288-124-245-207-238-223-225-278-240-186-198 bp (table 3). In addition, >85% (31/36) of the LSFC chromosomes were identical over a 4-cM region defined by markers D2S2306, D2S1356, D2S2294, D2S119, D2S2298, and D2S2174. This overlaps with the region of peak disequilibrium defined as Pexcess>.8 (fig. 1). Further examination of the LSFC chromosomes reveals that, apart from a likely mutation in individual COX018-43, the only marker showing identity across all LSFC chromosomes is D2S2298 (table 3). Specifically, this LSFC critical region is defined by the affected chromosomes (bearing the 225 bp allele at D252298) from families COX007 and COX021 (table 3), and figure 2 depicts the recombination event that is likely to have occurred in the maternal chromosome of family COX007, between markers D2S2294 and D2S119 or between D2S119 and D2S2298. Taken together, these results indicate that the LSFC critical region is ∼2 cM between D2S119 and D2S2174.
Table 3.
Genotypes of 36 LSFC Chromosomes in the 2p16 Region[Note]
|
COX7AR SNP |
|||||||||||||||
| Map or Proband | GATA194B06 | D2S2305 | D2S2259 | D2S2306 | 1 | 2 | 4 | D2S1356 | D2S2294 | D2S119 | D2S2298 | D2S2174 | D2S2291 | D2S2240 | D2S2378 |
| Genetic Distance(cM) |
Genetic Distance(cM) |
||||||||||||||
| Map: | |||||||||||||||
| Marshfield | 61.66 | 61.66 | 63.41 | 64.29 | 64.84 | 65.39 | 65.94 | 67.58 | 69.77 | 69.77 | 70.31 | ||||
| Généthon | 66 |
67.4 |
67.4 |
67.5 |
68.2 |
69 |
69.5 |
71.4 |
73.3 |
73.3 |
73.8 |
||||
| Allele Size |
Allele Size |
||||||||||||||
| Proband: | |||||||||||||||
| COX001-1 | 288 | 124 | 241 |
207 | A | G | nd | 238 | 234 | 223 | 225 | 278 | 240 | 186 | 198 |
| 288 |
124 | 245 | 207 | A | G | nd | 238 | 234 | 223 | 225 | 278 | 240 |
186 |
198 | |
| COX003-5 | 284 |
124 | 245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 236 |
184 |
198 |
| 288 | 124 | 245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 | 186 |
198 | |
| COX004-8 | 288 | 124 | 245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 | 178 | 198 |
| 288 |
124 |
245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 | 188 |
200 |
|
| COX005-9 | 260 |
132 |
245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 | 186 | 198 |
| 288 |
124 |
245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 | 186 | 198 | |
| COX006-10 | 284 |
124 |
245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | nd | 186 | 198 |
| 288 |
124 |
245 |
207 |
A |
G |
G |
238 |
234 |
223 | 225 | 278 |
nd |
186 |
198 |
|
| COX007-12 | 280 |
128 |
229 |
197 |
nd |
A |
A |
247 |
230 |
223 | 225 | 276 |
236 |
184 | 204 |
| 288 | 124 | 245 | 207 | nd | G | G | 238 | 234 | 223 | 225 | 278 |
240 | 178 |
200 | |
| COX007-13 | 288 | 124 | 245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 280 |
240 |
186 |
200 |
| 288 | 124 | 245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 242 |
178 |
200 |
|
| COX008-16 | nd | nd | nd | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 | nd | 198 |
| nd | nd | nd | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 | nd | 198 | |
| COX009-19 | 288 | 124 | 245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 | 186 | 198 |
| 288 |
124 |
245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 | 186 | 198 | |
| COX011-25 | 260 | 128 | 245 | 207 | A | G | nd | 238 | 234 | 223 | 225 | 278 | 240 | 186 | 198 |
| 260 |
132 |
245 | 207 | A | G | nd | 238 | 234 | 223 | 225 | 278 | 240 | 186 | 198 | |
| COX012-27 | 288 | 124 | 245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 |
186 |
198 |
| 288 |
124 |
245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 236 |
174 |
200 |
|
| COX013-29 | 260 |
132 |
245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 | 186 |
198 |
| 288 | 124 | 245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 | 178 |
200 |
|
| COX014-32 | 288 | 124 | 245 | 207 | nd | nd | nd | 238 | 234 | 223 | 225 | 278 | nd | 186 | 198 |
| 288 |
124 |
245 | 207 | nd | nd | nd | 238 | 234 | 223 | 225 | 278 | nd |
186 | 204 | |
| COX016-38 | 260 |
132 |
245 | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 236 | 186 |
214 |
| 288 |
124 |
245 |
207 |
A |
G |
G |
238 | 234 | 223 | 225 |
278 | 242 |
174 |
204 | |
| COX018-43 | 264 | 132 |
231 |
197 |
T |
A |
A |
238 | 234 | 223 | 221 |
278 | 240 | 186 | 192 |
| 260 |
124 | 245 |
207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 | 186 | 214 |
|
| COX020-55 | 288 | 124 | 241 |
207 | A | G | nd | 238 | 234 | 223 | 225 | 278 | 240 | 186 | 198 |
| 288 |
124 |
245 |
207 |
A | G |
nd |
238 |
234 |
223 |
225 | 278 | 240 | 186 |
198 |
|
| COX021-57 | 263 | 130 |
227 |
201 |
A | A |
A |
235 |
226 |
225 |
225 | 278 | 240 |
182 |
204 |
| 291 | 124 |
245 |
207 |
A | G |
G |
238 | 234 | 223 | 225 | 278 | 236 |
186 |
198 | |
| COX022-58 | 264 |
130 |
nd |
197 |
A | A |
A |
238 | 234 | 223 | 225 | 278 | 240 | 184 |
198 |
| 288 | 124 | nd | 207 | A | G | G | 238 | 234 | 223 | 225 | 278 | 240 | 186 | 198 | |
Note.— Boxes denote alleles not shared by the LSFC ancestral haplotype; nd = no data.
Figure 1.
Haplotypes of patients with LSFC, for the microsatellite markers that gave significant Pexcess values. A, Ancestral haplotype, as defined by the markers (shared by 26 [72%] of 36 individuals). B, Pexcess values for all the markers, with their Généthon map distances.
Figure 2.
Pedigree and haplotypes of members of family COX007, for some of the markers used in the high-density mapping of LSFC. The inherited paternal chromosome is framed in the right column, for father and both daughters. Straight lines indicate the maternal chromosomes, and the points of recombination are depicted in the maternal genotype. The thicker vertical lines indicate the maternal alleles inherited by patient 7-12, and the thinner vertical lines indicate the maternal alleles inherited by patient 7-13. The two patients share alleles only for D2S119 and D2S2298.
Candidate Genes within the LSFC Critical Region
The genomic locations of several genes that encode the COX subunits and assembly factors were unknown, and we were interested in determining whether any were located within the LSFC critical region. None of the 10 COX subunits (IV, Va, Vb, VIa-L, VIb, VIc, VIIa-L, VIIb, VIIc, or VIII) and none of the three assembly factors (COX10, COX17, or OXA1) mapped to any of the 23 MEGAYAC clones that cover this region (data not shown). In contrast, a search of the National Center for Biotechnology Information (NCBI) public databases for genes located in the 2p16 region identified a cDNA sequence that was predicted to encode a protein similar to COXVIIa. The protein is known as “COX7-related protein” (COX7RP [GenBank accession number AB007618]), and its corresponding gene is named “COX7AR.” COX7AR had been mapped using radiation hybrids with two different markers, stSG3793 and A007B18, on GeneMap '98 (NCBI). Marker stSG3793 mapped to an interval between markers D2S119 and D2S337, and A007B18 mapped to between D2S177 and D2S119. Although the function of COX7RP is unknown, COX7AR appeared to be a possible candidate gene for LSFC, because its C-terminal region has a high amino acid–sequence identity with both liver and heart isoforms of COXVIIa and is well conserved between humans and mice (Watanabe et al. 1998).
Genomic Structure of COX7AR
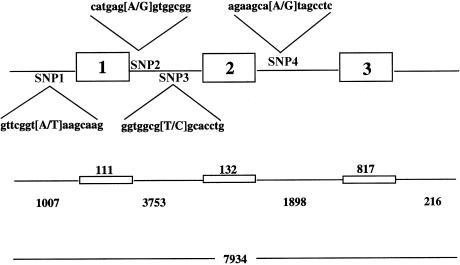
To screen patients' DNA samples for an LSFC mutation, the genomic structure of COX7AR was determined. This was accomplished by isolation of large insert genomic DNA clones containing COX7AR and by use of these as templates in sequencing experiments. Three PAC1 clones (1126_I_4, 1148_J_17, and 1149_D_18) were isolated. The sequencing experiments revealed that COX7AR consists of three exons—of 111, 132, and 817 bases—that are separated by introns of 3,753 and 1,898 bases (fig. 3). Exon 1 contains a 5′UTR of 39 bases, and exon 3 contains a 3′UTR of 723 bases. Examination of slightly more than 1 kb of sequence upstream from the start codon revealed consensus CCAT boxes at nucleotide positions −127 and −397, as well as several potential binding sites for Sp1, Oct-1, HNF-3b, and NF-κB transcription factors.
Figure 3.
Genomic structure of COX7AR. The genomic sequence was determined for a region of 7934 bp. This region contained the entire COX7AR gene, consisting of three exons (of 111, 132, and 817 bp, respectively), two introns (of 3753 and 1898 bp, respectively), and the 5′ upstream region of 1007 bp. The four SNPs found in the SLSJ population are referred to here as SNP1, SNP2, SNP3, and SNP4.
Mutation Screening and Physical Mapping of COX7AR
Overlapping PCR assays, spanning nearly 8 kb of genomic sequence, were designed for resequencing of COX7AR in DNA samples from five patients with LSFC and from three control individuals. Resequencing of both strands of DNA in these samples revealed four nucleotide positions where the sequence differed between patients and controls. The four SNPs were located at positions −516, 145, 1331, and 4277 and are referred to here as “SNP1,” “SNP2,” “SNP3,” and “SNP4,” respectively (fig. 3). Three of the four SNPs (1, 2, and 4) were examined by resequencing in the remaining DNA samples from patients with LSFC (table 3) and from their parents (data not shown). None of these SNPs appear likely to be the disease-causing mutation, because none are predicted to change the function or expression of COX7RP; they do not change predicted amino acid sequence and are not located wihin the intron/exon boundaries or cis-acting elements. In addition, two individuals with LSFC (COX007-12 and COX018-43) are heterozygous for these SNPs, and 10 of the parents were homozygous for the ancestral haplotype defined by these SNPs. Moreover, these SNPs are not part of the conserved haplotype, a finding that places COX7AR outside the LSFC critical region and that probably excludes the possibility that the LSFC mutation is actually within COX7AR but remains undetected.
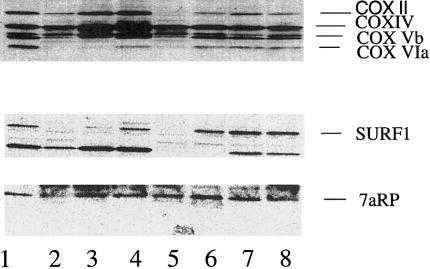
Immunoblotting of mitochondrial proteins, using an antibody generated against COX7RP, showed that levels of expression in control subjects were similar to those (1) in patients with COX deficiency due to mutations in SURF1, (2) patients with COX deficiency and LS of unknown origin, and (3) three patients with LSFC (fig. 4). Parallel blotting analysis done with anti-SURF1 antibody and anti-COX antibody shows low expression of COX subunits in several patients and low expression of SURF1 in patients with SURF1 mutations.
Figure 4.
Immunoblotting of mitochondria from fibroblast cultures. Mitochondria (100 μg) were prepared from skin fibroblast cultures, were subjected to PAGE separation, and were transferred to nitrocellulose. Parallel prepared blots were then probed with antibodies to beef heart COX (top), SURF1 (middle), and COX7RP (7aRP) (bottom). The anti-COX antibody recognizes subunits II, IV, Vb, and VIa of the human COX complex. Lane 1, Control. Lanes 2, 3, and 5, Patients with LS and SURF1 mutation. Lane 4, Patients with LS COX deficiency and normal SURF1. Lanes 6, 7, and 8, Patients with LSFC-COX.
Discussion
Isolated COX deficiencies represent a common cause of mitochondrial respiratory-chain defects in humans. Only a few rare cases of mutations in the mitochondrial-encoded subunits have been identified. In contrast, three genes that encode proteins involved in the assembly of the COX subunits have recently been shown to be mutated in patients with various COX deficiencies. Specifically, these are the SURF1, SCO2, and COX10 genes, located in chromosomal regions 9q34, 22q13, and 17p13.1-q11.1, respectively (Tiranti et al. 1998; Zhu et al. 1998; Papadopoulou et al. 1999; Valnot et al. 2000a). Less than a decade ago, a distinct autosomal recessive COX deficiency was characterized that was specific to the SLSJ region of Quebec (Merante et al. 1993; Morin et al. 1993). In an effort to identify the gene defect that causes this deficiency, samples were collected from families with LSFC. Given the devastating effect of COX deficiency on a child's life expectancy, we were able to collect samples only from families that had one affected child and from a single family that had an affected sibling pair. This limitation made a linkage study impossible. The known characteristics of the SLSJ population, however, made it likely that LSFC was caused by a single major mutation present on a common haplotype extending over a distance of many centimorgans. We therefore undertook a genomewide screen for LD and successfully identified an allele of a single marker (allele 238 of marker D2S1356), on chromosomal 2p16, that was in significant disequilibrium with the LSFC phenotype. Although the identity of the LSFC gene has yet to be determined, its chromosomal position clearly indicates that it is distinct from the genes involved in the previously described COX deficiencies.
Moreover, because none of the 10 nuclear-encoded COX subunits map to this region, it is very likely that the LSFC gene product is involved in COX assembly, as has been observed for the other nuclear-encoded COX deficiencies (Tiranti et al. 1998; Zhu et al. 1998; Papadopoulou et al. 1999; Valnot et al. 2000a, 2000b). Specifically, SCO1 and SCO2 are involved with copper transfer, COX10 is the gene for heme farnesyl transferase, and SURF1 has been shown to be involved with the subunit-assembly process. To date, ∼40 genes are associated with COX assembly in the yeast Saccharomyces cerevisiae model (Glerum 1999), and 10 have been identified—and have been recently mapped—in humans. Some of the potential processes of the LSFC gene are as follows: subunit synthesis, assembly, and degradation; chaperone roles; heme transport and synthesis; transport and placement of the metals copper, zinc, magnesium and iron. It may also reduce activities in COX by affecting the binding to either cytochrome c or oxygen or by affecting electron transfer or stability. Somewhat different from what is seen in the other COX deficiencies, however, is the fact that LSFC-COX displays some tissue specificity. The gene responsible for LSFC-COX is likely to be an assembly factor that affects the enzyme in the liver and brain tissues more than in the heart or kidney.
High-resolution genetic mapping, using 11 additional markers that spanned ∼10 cM in the region surrounding marker D2S1356, identified a haplotype likely to represent the chromosome of a common founder. More specifically, a single haplotype defined by markers D2S2306, D2S1356, D2S2294, D2S119, D2S2298, and D2S2174 (alleles 207, 238, 234, 223, 225, and 278, respectively), extending over a region of ∼4 cM, is present in >80% of LSFC chromosomes and almost certainly contains the LSFC gene. Such a large extent of disequilibrium surrounding the disease locus was expected in view of the unequal contribution, ∼12–15 generations ago, of a small number of founders of this population (Heyer 1995). Additionally, this finding was supported by experimental results from previous studies of rare diseases within the SLSJ region (Labuda et al. 1996; Richter et al. 1999). On examination of family COX007, the identification of a maternal recombination event narrowed the LSFC critical region to the ∼3-cM interval between markers D2S2294 and D2S2174. In addition, the observation that the individual with LSFC from family 21 (COX21-57) is homozygous only for markers D2S2298 and D2S2174 further reduces the critical region, to the ∼2-cM interval between D2S119 and D2S174. Patient COX18-43 is heterozygous at marker D2S2298; however, because of the extensive match with the ancestral haplotype for ∼1.5 cM on either side of D2S2298, we believe that the heterozygosity in this patient is most likely the result of a mutational event at the marker (Brinkmann et al. 1998).
No known COX subunits or assembly factors mapped to the region identified by the LD and high-resolution mapping. After we performed these experiments, other research groups mapped COX subunits and more assembly factors: SCO1, COX11, COX15, paraplegin, and pet112-L, on chromosomal regions 17p12-13, 17q22, 10q24, 10p14, and 4q27-q28, respectively (Casari et al. 1998; Petruzzella et al. 1998). In agreement with the results of our screening, none of the subunits map to 2p16. A search of the NCBI public databases identified a potential candidate gene called “COX7AR.” This gene had been mapped, by use of radiation hybrids, to an interval that overlapped the LSFC region. Its gene product was called “COX7A-related protein,” because it had extensive amino acid–sequence identity with the two isoforms of the mammalian COXVIIa family (Watanabe et al. 1998). Although the biochemical role of COX7RP has yet to be determined, its mouse orthologue (called “SIG81”) was shown to localize to the mitochondria (Segade et al. 1996). Determination of the genomic structure of the COX7AR gene made it possible to screen the patients' DNA for potential mutations. Given that all the sequence differences between patients and controls were located within COX7AR introns, none were likely to be the LSFC mutation. Furthermore, genotyping of these SNPs in samples from all the patients and parents indicated that this gene must be located outside the critical region, clearly demonstrating that the LSFC gene has yet to be identified. Finally, immunoblots using mitochondria from both patients with COX deficiency due to SURF1 mutations and from patients with SLSJ-COX deficiency showed normal levels of COX7ARp while clearly showing decreases of COX subunits, indicative of poor assembly of the mature COX complex. This result, along with the normal exon sequence of the 7ARP gene, strongly suggests that this gene is not connected with SLSJ-COX deficiency. At the time this article was submitted for publication, no human genome sequence covering the markers D2S119 and D2S2298 had been listed. Once the human reference sequence for this region is available from the Human Genome Project, it will undoubtedly become easier to identify and localize genes within the LSFC critical region.
LD has previously been used as a powerful tool for the high-resolution mapping of disease genes in founder populations, such as the diastrophic dysplasia gene in Finland (Hastbacka et al. 1992) and the genes for pseudo–vitamin D–deficiency rickets and for autosomal recessive spastic ataxia in the SLSJ region (Richter et al. 1999). However, these loci had first been identified by traditional linkage analysis. The current study is one of the first examples of a disease-gene locus identified solely by the use of a whole-genome LD-mapping approach. This approach has recently been proposed as a means of using a dense set of SNPs to localize susceptibility genes for common traits. Because the experimental data are insufficient, it is not yet clear whether this approach is applicable to common alleles in the general population. In isolated populations, however, where there has been a narrow bottleneck or where the frequency of the variant is <5%, the LD is expected to extend over a greater region (Kruglyak 1999). Taken together, our results suggest that whole-genome LD searches would be feasible for other rare traits and may also be possible for some common diseases in the SLSJ region.
Acknowledgments
We thank the patients and their families for their cooperation and support. Special thanks are due to the family of Pierre Lavoie for his world record–breaking cycle event for fundraising. The authors would also like to thank Daniel Gaudet for his contribution of the control genotype data for marker D2S1356. This work was supported by the Garrod Association of Canada, Association de l'Acidose Lactique (Québec), Medical Research Council (MRC) of Canada, Canadian Genetics Disease Network (support to B.H.R., G.M., and T.J.H.), and by a research contract from Bristol-Myers Squibb Company, Millennium Pharmaceuticals, Inc., and Affymetrix, Inc. T.J.H. is a recipient of a Clinician Scientist Award from MRC.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://www.marshmed.org/genetics
- CHLC, http://lpg.nci.nih.gov/CHLC/ (for fluorescence-labeled microsatellite markers)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for accession number AB007618)
- GeneMap'98, http://www.ncbi.nlm.nih.gov/genemap98/ (for cDNA sequence of a possible candidate gene, COX7AR [accession number AB007618])
- Généthon, http://www.genethon.fr/genethon_en.html (for additional markers flanking marker D2S1356)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim (for LS [MIM 256000] and LSFC [MIM 220111])
- PEDMANAGER, ftp://ftp-genome.wi.mit.edu/distribution/software/pedmanager/
- TFSEARCH, http://pdap1.trc.rwcp.or.jp/research/db/TFSEARCH.html
- Whitehead Institute for Biomedical Research/MIT Center for Genome Research: http://www-genome.wi.mit.edu (for YAC map information and GENEHUNTER 2.0)
References
- Bourgeron T, Rustin P, Chretien D, Birch-Machin M, Borgeois M, Viegas-Péquignot E, Munnich A, Rötig A (1995) Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat Genet 11:144–149 [DOI] [PubMed] [Google Scholar]
- Brinkmann B, Klintschar M, Neuhuber F, Huhne J, Burkhard R (1998) Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am J Hum Genet 62:1408–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, Durr A, Fontaine B, Ballabio A (1998) Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 93:973–983 [DOI] [PubMed] [Google Scholar]
- Couture P, Morisette J, Gaudet D, Vohl M, Gagne C, Bergeron J, Sepres J-P, Simard J (1999) Fine mapping of low-density lipoprotein receptor gene by genetic linkage on chromosome 19p13.1-p13.3 and study of the founder effect of four French Canadian low-density lipoprotein receptor gene mutations. Atherosclerosis 143:145–151 [DOI] [PubMed] [Google Scholar]
- Dahl H (1998) Getting to the nucleus of mitochondrial disorders: identification of respiratory chain–enzyme genes causing Leigh syndrome. Am J Hum Genet 63:1594–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Braekeleer M (1991) Hereditary disorders in Saguenay–Lac-St-Jean (Quebec, Canada). Hum Hered 41:141–146 [DOI] [PubMed] [Google Scholar]
- De Braekeleer M, Mari C, Verlingue C, Allard C, Leblanc JP, Simard F, Aubin G, Ferec C (1998) Complete identification of cystic fibrosis transmembrane conductance regulator mutations in the CF population of Saguenay–Lac-Saint-Jean (Quebec, Canada). Clin Genet 53:44–46 [DOI] [PubMed] [Google Scholar]
- De Braekeleer MGF, Mathieu J, Roy M, Bouchard J-P, Morgan K (1993) Genetic epidemiology of autosomal recessive spastic ataxia of Charlevoix-Saguenay in northeastern Quebec. Genet Epidemiol 10:17–25 [DOI] [PubMed] [Google Scholar]
- Gavreau D, Bourque M (1988) Mouvements migratoires et familles: le peuplement du Saguenay avant 1911. Rev Hist Am Fr 42:167–192 [Google Scholar]
- Glerum DM, Beers J, Tzagoloff A, Punter F, Adams D (1999) Getting copper into mitochondria. In: Sarkar B (ed) Metals and genetics. Kluwer Academic/Plenum, New York, pp 237–254 [Google Scholar]
- Grompe M, St-Louis M, Demers S, Al-Dhalimy M, Leclerc B, Tanguay R (1994) A single mutation of the fumarylacetoacetate hydrolase gene in French Canadians with hereditary tyrosinemia type I. N Engl J Med 331:353–357 [DOI] [PubMed] [Google Scholar]
- Hastbacka J, de la Chapelle A, Kaitla I, Sistonen P, Weaver A, Lander E (1992) Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nat Genet 2:204–211 [DOI] [PubMed] [Google Scholar]
- Hawkins TL, O'Connor-Morin T, Roy A, Santillan C (1994) DNA purification and isolation using a solid phase. Nucleic Acids Res 22:4543–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer ETM (1995) Variability of the genetic contribution of Quebec population founders associated to some deleterious genes. Am J Hum Genet 56:970–978 [PMC free article] [PubMed] [Google Scholar]
- ——— (1999) One founder/one gene hypothesis in a new expanding population: Saguenay (Quebec, Canada). Hum Biol 71:99–109 [PubMed] [Google Scholar]
- Jette R, Gauvreau D, Guerien M (1991) Aux origins d'une region: le peuplement foundateur de Charlevoix avant 1850. In: Bouchard G, De Braekeleer M (eds) Histoire d'un genome. Presses de l'Université du Québec, Québec, pp 75–106 [Google Scholar]
- Kruglyak L (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 22:139–144 [DOI] [PubMed] [Google Scholar]
- Labuda M, Labuda D, Korab-Laskowska M, Cole D, Zietkiewicz E, Weissenbach J, Popwska E, Pronicka E, Root A, Glorieux F (1996) Linkage disequilibrium analysis in young populations: pseudo–vitamin D–deficiency rickets and the founder effect in French Canadians. Am J Hum Genet 59:633–643 [PMC free article] [PubMed] [Google Scholar]
- Lee N, Morin C, Mitchell G, Robinson B (1998) Saguenay Lac Saint Jean cytochrome oxidase deficiency: sequence analysis of nuclear encoded COX subunits, chromosomal localization and a sequence anomaly in subunit VIc. Biochim Biophys Acta 1406:1–4 [DOI] [PubMed] [Google Scholar]
- McEachern G, Kassovska-Bratinova S, Raha S, Tarnopolsky MA, Turnbull J, Bourgeois J, Robinson BH (2000) Manganese superoxide dismutase levels are elevated in a proportion of amyotrophic lateral sclerosis patient cell lines. Biochem Biophys Res Commun 273:359–363 [DOI] [PubMed] [Google Scholar]
- Merante F, Petrova-Benedict R, MacKay N, Mitchell G, Lambert M, Morin C, De Braekeleer M, Laframboise, R, Gagne R, Robinson BH (1993) A biochemically distinct form of cytochrome oxidase (COX) deficiency in the Saguenay-Lac-Saint-Jean region of Quebec. Am J Hum Gen 53:481–48793318848 [Google Scholar]
- Montpetit V, Anderman F, Carpenter S, Fawcett JS, Zborowska-Sluis D, Gibersun HR (1971) Subacute necrotizing encephalomyopathy: a review and a study of two families. Brain 94:1–30 [DOI] [PubMed] [Google Scholar]
- Morin C, Dube J, Robinson B, Lacroix J, Michaud J, De Breakeleer M, Geoffroy G, Lortie A, Blanchette C, Lambert M, Mitchell G (1999) Stroke-like episodes in autosomal recessive cytochrome oxidase deficiency. Ann Neurol 45:389–392 [PubMed] [Google Scholar]
- Morin C, Mitchell G, Larochelle J, Lambert M, Ogier H, Robinson BH, De Braekeleer M (1993) Clinical, metabolic, and genetic aspects of cytochrome c oxidase deficiency in Saguenay-Lac-Saint-Jean. Am J Hum Genet 53:488–4968392291 [Google Scholar]
- Papadopoulou L, Sue C, Davidson M, Tanji K, Nishino I, Sadlock J, Krishna S, Walker W, Selby J, Glerum DM, Van Coster R, Lyon G, Scalais E, Lebel R, Kapllan P, Shanske S, De Vivo D, Bonilla E, Hirano M, DiMauro S (1999) Fatal infantile cardiocephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat Genet 23:333–337 [DOI] [PubMed] [Google Scholar]
- Petruzzella V, Tiranti V, Fernandez P, Ianna P, Carrozzo R, Zeviani M (1998) Identification and characterization of human cDNAs specific to BCS1, PET112, SCO1, COX15, and COX11, five genes involved in the formation and function of the mitochondrial respiratory chain. Genomics 54:494–504 [DOI] [PubMed] [Google Scholar]
- Pincus J (1972) Subacute necrotizing encephalomyopathy (Leigh's disease): a consideration of clinical features and etiology. Dev Med Child Neurol 14:87–101 [DOI] [PubMed] [Google Scholar]
- Pitkänen S, Robinson BH (1996) Mitochondrial complex I deficiency leads to increased production of superoxide radicals and induction of superoxide dismutase. J Clin Invest 98:345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A, Rioux J, Bouchard J-P, Mercier J, Mathieu J, Ge B, Poirier J, Julien D, Gyapay G, Weiseenbach J, Hudson T, Melancon S, Morgan K (1999) Location score and haplotype analyses of the locus for autosomal recessive spastic ataxia of Charlevoix-Saguenay, in chromosome region 13q11. Am J Hum Genet 64:768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux J, Stone V, Daly M, Cargill M, Green T, Nguyen H, Nutman T, Zimmerman P, Tucker M, Hudson T, Goldstein A, Lander E, Lin A (1998) Familial eosinophilia maps to the cytokine gene cluster on human chromosomal region 5q31-q33. Am J Hum Genet 63:1086–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BH (1993) Lacticacidemia. Biochim Biophys Acta 1182:231–244 [DOI] [PubMed] [Google Scholar]
- Segade F, Hurle B, Claudio E, Ramos S, Lazo PS (1996) Identification of an additional member of the cytochrome c oxidase subunit VIIa family of proteins. J Biol Chem 271:12343–12349 [DOI] [PubMed] [Google Scholar]
- Staden R (1996) The Staden sequence analysis package. Mol Biotechnol 5:233–241 [DOI] [PubMed] [Google Scholar]
- Tiranti V, Hoertnagel K, Carrozzo R, Galimberti C, Munaro M, Granatiero M, Zelante L, Gasparini P, Marzella R, Rocchi M, Bayona-Bafaluy MP, Enriquez J, Uziel G, Bertini E, Dionisi-Vici C, Franco B, Meitinger T, Zeviano M (1998) Mutations of SURF-1 in Leigh's disease associated with cytochrome c oxidase deficiency. Am J Hum Genet 63:1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valnot I, Osmond S, Gigarel N, Mehaye B, Amiel J, Cormier-Daire V, Munnich A, Bonnefont JP, Rustin P, Rötig A (2000a) Mutations of the SCO1 gene in mitochondrial cytochrome c oxidase deficiency with neonatal-onset hepatic failure and encephalopathy. Am J Hum Genet 67:1104–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valnot I, von Leist-Retzow J-C, Barrientos A, Gorbatyuk M, Taanman J, Mehaye B, Rustin P, Tzagoloff A, Munnich A, Rötig A (2000b) A mutation in the human heme A: farnesyl transferase gene (COX10) causes cytochrome oxidase deficiency. Hum Mol Genet 9:1245–1249 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Inoue S, Hiroi H, Orimo A, Kawashima H, Muramatsu M (1998) Isolation of estrogen-responsive genes with a CpG island library. Mol Cell Biol 18:442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Yao J, Johns T, Fu K, DeBie I, Macmillan D, Cuthbert AP, Newbold RF, Wang J, Chevrette M, Brown, G, Brown, R, Shoubridge, EA (1998) SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet 20:337–343 [DOI] [PubMed] [Google Scholar]