Abstract
To study the genomic divergences among hominoids and to estimate the effective population size of the common ancestor of humans and chimpanzees, we selected 53 autosomal intergenic nonrepetitive DNA segments from the human genome and sequenced them in a human, a chimpanzee, a gorilla, and an orangutan. The average sequence divergence was only 1.24% ± 0.07% for the human-chimpanzee pair, 1.62% ± 0.08% for the human-gorilla pair, and 1.63% ± 0.08% for the chimpanzee-gorilla pair. These estimates, which were confirmed by additional data from GenBank, are substantially lower than previous ones, which included repetitive sequences and might have been based on less-accurate sequence data. The average sequence divergences between orangutans and humans, chimpanzees, and gorillas were 3.08% ± 0.11%, 3.12% ± 0.11%, and 3.09% ± 0.11%, respectively, which also are substantially lower than previous estimates. The sequence divergences in other regions between hominoids were estimated from extensive data in GenBank and the literature, and Alus showed the highest divergence, followed in order by Y-linked noncoding regions, pseudogenes, autosomal intergenic regions, X-linked noncoding regions, synonymous sites, introns, and nonsynonymous sites. The neighbor-joining tree derived from the concatenated sequence of the 53 segments—24,234 bp in length—supports the Homo-Pan clade with a 100% bootstrap value. However, when each segment is analyzed separately, 22 of the 53 segments (∼42%) give a tree that is incongruent with the species tree, suggesting a large effective population size (Ne) of the common ancestor of Homo and Pan. Indeed, a parsimony analysis of the 53 segments and 37 protein-coding genes leads to an estimate of Ne = 52,000 to 96,000. As this estimate is 5 to 9 times larger than the long-term effective population size of humans (∼10,000) estimated from various genetic polymorphism data, the human lineage apparently had experienced a large reduction in effective population size after its separation from the chimpanzee lineage. Our analysis assumes a molecular clock, which is in fact supported by the sequence data used. Taking the orangutan speciation date as 12 to 16 million years ago, we obtain an estimate of 4.6 to 6.2 million years for the Homo-Pan divergence and an estimate of 6.2 to 8.4 million years for the gorilla speciation date, suggesting that the gorilla lineage branched off 1.6 to 2.2 million years earlier than did the human-chimpanzee divergence.
Introduction
The degree of sequence divergence between the human (Homo sapiens) and chimpanzee (Pan troglodytes) genomes has been a subject of numerous studies (e.g., King and Wilson 1975; Sibley and Ahlquist 1987; Goodman et al. 1990), and it has been commonly thought that the two genomes differ by ∼1.6%. However, as this estimate was based mainly on DNA hybridization data (Sibley and Ahlquist 1987) and the DNA sequence data from the η-globin pseudogene region (Bailey et al. 1991), it may not represent an accurate estimate of the average divergence between the two genomes. Indeed, large variation in sequence divergence is often seen among genomic regions. For example, the last intron of the ZFY gene shows only 0.69% divergence between human and chimpanzee (Dorit et al. 1995), whereas the olfactory receptor OR1D3P pseudogene shows a divergence of 3.04% (Glusman et al. 2000). The divergences between the genomes of other hominoids are much less well studied. To have reliable estimates of the average divergences between hominoid genomes, sequence data from many genomic regions are needed.
The divergence dates between human and other hominoids also have received much attention (e.g., Sarich and Wilson 1967; Sibley and Ahlquist 1987; Takahata et al. 1995; Ruvolo 1997). However, there is still much uncertainty about these dates, especially the internodal time span between the human-chimpanzee divergence and the branch node of the gorilla lineage. For example, according to Horai et al.’s study of mitochondrial DNA sequences (1992), the divergence between human and chimpanzee occurred 4.7 ± 0.5 million years ago, whereas the gorilla lineage branched off 7.7 ± 0.7 million years ago, so the time span between the two speciation events was as long as 3 million years, ∼60% of the Homo-Pan divergence time. In contrast, in Bailey et al.’s study of the η-globin pseudogene region (1991), the internodal time span was only ∼10% of the divergence time between human and chimpanzee. Which is closer to the truth? As will be explained later, this internodal time span is useful for estimating the effective size of the ancestral population before the human-chimpanzee divergence.
There has also been considerable interest in the demographic history of hominoids (e.g., Takahata 1990; Rogers and Harpending 1992; Ruvolo 1997). Of particular interest is the effective size (Ne) of the ancestral population before the human-chimpanzee divergence (Takahata et al. 1995; Ruvolo 1997), because it may tell us whether there has been a significant reduction in population size in the human lineage since its separation from the chimpanzee lineage. Current data are not sufficient for a reliable estimate of Ne; for this purpose, a fairly large number of independent loci from the human, chimpanzee, and gorilla genomes are needed.
To study the above issues, we selected 53 autosomal intergenic nonrepetitive regions throughout the human genome and obtained the homologous sequences of each region from a human, a chimpanzee, a gorilla, and an orangutan. We chose noncoding regions because they are not directly subject to natural selection and therefore can more accurately trace the history of evolution than can coding regions. We also collected and analyzed many data sets from GenBank and the literature. Some of the conclusions derived from these extensive data are substantially different from previous ones.
Material and Methods
Selection of Regions
All of the 53 DNA segments studied were chosen to avoid coding regions. They were selected randomly, with reference to the Genome Channel. We searched for autosomal contigs without any known registered GenBank genes and scanned each contig with the GRAIL and GeneScan programs to detect potential genes. From each contig used, a segment of 2 to 20 kb was chosen, at least 5 kb away from any potential genes in both directions. Then the segment was masked using the RepeatMasker program. All repetitive elements were excluded and the remaining segment, if long enough (⩾800 bp), was used for PCR primer design and amplification.
DNA Samples
DNA samples from one Asian male human (Homo sapiens), one chimpanzee (Pan troglodytes), one gorilla (Gorilla gorilla) and one orangutan (Pongo pygmaeus) were used in this study. The protocol for taking blood samples for extracting DNA has been approved by the institutional review board of the University of Chicago.
PCR Amplification and Sequencing
Touch-down PCR (Don et al. 1991) was applied to each selected segment. Then the PCR products were purified with Wizard PCR Preps DNA Purification Resin Kit (Promega). Sequencing reactions were performed according to the ABI Prism BigDye Terminator Sequencing Kits (Applied Biosystems), modified for quarter reaction. The extension products were purified with 50% Sephadex G-50 resin (DNA grade, Pharmacia) and were run on an ABI 377 XL DNA sequencer using 4.25% gels (Sooner Scientific).
ABI DNA Sequence Analysis 3.0 was used for lane tracking and base calling. All segments were sequenced in both directions and were proofread manually with the SeqMan program of DNAStar.
Data Retrieval from GenBank
Homologous introns, pseudogenes, and coding regions of hominoids were retrieved from GenBank using the Hovergen program (Duret et al. 1994). Most of the accession numbers of coding regions were taken from Satta et al. (2000) and also from the Silver Project Home Page. For sequences with more than one haplotype, the ones with the smallest distances were chosen for analyses. The accession numbers of the chromosome 12 contigs taken from GenBank were: AC007286, AC007458, AC005294, and AC011604 (human), and AC007214 and AC006582 (chimpanzee).
Data Analysis
The DAMBE package (Xia 2000) was used for sequence alignment, calculation of genetic distance, and phylogenetic reconstruction. All sequenced autosomal segments were analyzed separately and then were concatenated and analyzed for the overall divergence or phylogenetic reconstruction. For coding regions, the number of substitutions per synonymous site (KS) and the number of substitutions per nonsynonymous site (KA) were calculated by the method of Li (1993) in DAMBE. Introns and pseudogenes were scanned with Repeat Masker to eliminate all repeats before distance calculation. For the chromosome 12 sequences, all repeats were eliminated and then were submitted to the BLAST server to identify homologous regions between the human and chimpanzee sequences. Sequences similar to any known functional genes were excluded before alignment.
Results
Genomic Divergence
Noncoding regions
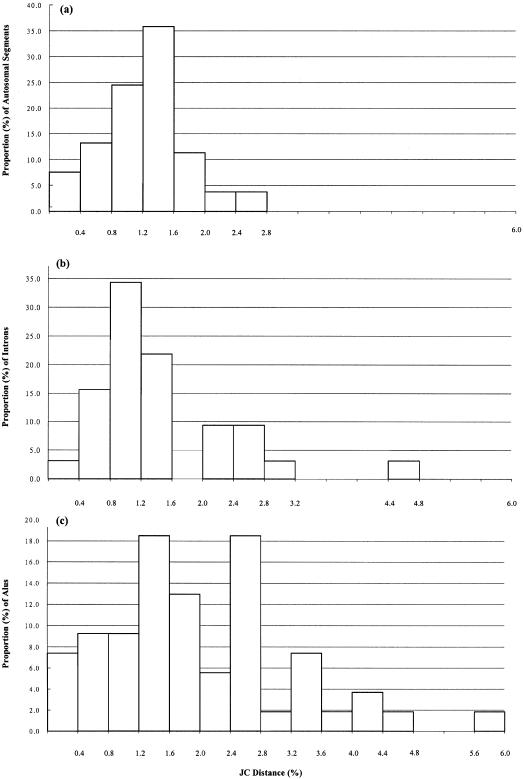
The sequence divergences among hominoids in the 53 autosomal intergenic DNA segments studied are listed in table 1. The degree of divergence varies among regions. For example, for the human-chimpanzee pair, the divergence ranges from 0% to 2.66%; figure 1 shows that the majority of the divergence values lie between 0.8% and 1.6%. The average divergences are, respectively, 1.24%, 1.62%, and 1.63% for the human-chimpanzee, human-gorilla, and chimpanzee-gorilla pairs, and they are 3.08%, 3.12%, and 3.09% for the orangutan-human, orangutan-chimpanzee, and orangutan-gorilla pairs. Therefore, the gorilla genome is substantially more different from the human genome than is the chimpanzee genome, and the orangutan genome differs by ∼3% from the human, chimpanzee, and gorilla genomes.
Table 1.
Autosomal DNA Segments Sequenced and Pairwise Divergences among Species
|
Jukes-Cantor Distancea(%) |
||||||||
| Chromosome No. | Contig No. | Length(bp) | H-C | H-G | C-G | H-O | C-O | G-O |
| Segments supporting the Homo-Pan clade: | ||||||||
| 1 | T2609 | 467 | 1.51 | 1.30 | 1.51 | 3.06 | 3.28 | 2.61 |
| 2 | T1251 | 447 | 1.58 | 3.43 | 3.67 | 6.08 | 6.33 | 6.08 |
| 7 | T2012 | 491 | 1.44 | 2.48 | 3.12 | 2.91 | 3.33 | 2.70 |
| 8 | T1364 | 479 | 1.05 | 2.55 | 2.12 | 3.42 | 2.98 | 4.08 |
| 10 | T1412 | 431 | 1.41 | 2.60 | 2.36 | 2.84 | 3.08 | 2.36 |
| 11 | T1419 | 474 | 1.28 | 1.06 | 1.92 | 3.01 | 3.90 | 3.01 |
| 12 | T1482 | 366 | 1.38 | 1.66 | 1.94 | 3.35 | 3.35 | 3.07 |
| 14 | T2963 | 350 | .00 | 1.73 | 1.73 | 2.32 | 2.32 | 2.91 |
| 15 | T2265 | 458 | .88 | 1.54 | 1.54 | 2.22 | 3.12 | 2.44 |
| 15 | T2266 | 477 | 1.27 | 2.13 | 2.78 | 1.91 | 2.56 | 3.43 |
| 16 | 598D4 | 513 | .59 | .59 | 1.18 | 1.38 | 1.98 | 1.58 |
| 17 | NT0784 | 464 | 1.30 | 1.30 | 1.52 | 4.90 | 5.13 | 4.90 |
| 17 | NT0787 | 451 | 1.34 | 1.34 | .89 | 3.17 | 2.71 | 2.25 |
| 17 | NT0801 | 489 | .62 | 1.03 | .82 | 3.35 | 3.35 | 3.35 |
| 17 | NT0812 | 336 | 1.50 | 2.42 | 2.42 | 3.97 | 4.29 | 3.04 |
| 17 | NT1574 | 359 | .84 | 1.41 | 1.69 | 2.26 | 2.55 | 1.98 |
| 17 | NT2294 | 517 | 1.17 | 2.76 | 1.96 | 3.36 | 2.76 | 2.56 |
| 17 | NT2984 | 502 | 2.22 | 3.05 | 2.43 | 3.26 | 2.84 | 2.02 |
| 17 | NT2986 | 419 | 1.20 | .96 | 1.20 | 1.93 | 2.18 | 1.45 |
| 17 | NT2988 | 468 | 1.29 | 2.17 | 1.73 | 3.05 | 3.05 | 3.05 |
| 18 | NT0864 | 373 | 1.63 | 3.29 | 2.18 | 4.99 | 3.85 | 4.42 |
| 18 | NT0866 | 443 | 1.60 | 1.37 | 1.14 | 2.29 | 1.60 | 1.37 |
| 19 | NT0953 | 479 | .63 | .84 | .84 | 2.55 | 2.55 | 2.12 |
| 20 | NT2012 | 507 | .79 | 1.39 | 1.39 | 3.84 | 3.84 | 4.05 |
| 20 | NT2018 | 535 | 1.51 | 1.70 | 1.70 | 3.64 | 3.83 | 2.08 |
| 20 | NT2020 | 449 | 1.80 | 1.35 | 1.80 | 3.18 | 3.64 | 3.18 |
| 20 | NT2064 | 513 | .39 | 1.38 | 1.58 | 2.58 | 2.78 | 3.19 |
| 20 | NT2085 | 544 | 1.49 | .74 | 1.11 | 2.24 | 2.81 | 1.86 |
| 20 | NT2352 | 511 | 1.38 | 1.18 | .98 | 2.99 | 2.79 | 2.59 |
| 20 | NT2472 | 454 | .89 | 1.33 | 1.56 | 5.01 | 5.25 | 5.72 |
| 20 | NT2560 | 522 | 1.15 | 1.54 | 1.93 | 3.52 | 3.92 | 3.13 |
| Subtotal | 31 segments | 14,288 | 1.20±.09 | 1.71±.11 | 1.75±.11 | 3.17±.15 | 3.28±.15 | 2.97±.15 |
| Segments supporting the Homo-Gorilla clade: | ||||||||
| 3 | T2659 | 557 | .90 | .90 | 1.45 | 1.82 | 2.00 | 2.37 |
| 9 | T1386 | 406 | 2.50 | 1.49 | 2.50 | 4.57 | 4.05 | 4.57 |
| 11 | T2224 | 371 | .54 | .00 | .54 | 2.19 | 1.63 | 2.19 |
| 11 | U73646 | 463 | 1.53 | 1.31 | 2.41 | 2.64 | 3.09 | 3.54 |
| 12 | T2927 | 425 | 1.67 | 1.67 | .95 | 3.86 | 2.63 | 3.12 |
| 14 | T2960 | 301 | .33 | 1.34 | 1.00 | 2.36 | 2.02 | 3.05 |
| 17 | NT2987 | 442 | 1.37 | 1.37 | 2.30 | 2.77 | 3.00 | 3.00 |
| 20 | NT1636 | 535 | 2.66 | 2.66 | 1.51 | 5.83 | 4.63 | 4.63 |
| 20 | NT2019 | 411 | .98 | 1.97 | 1.72 | 2.98 | 2.73 | 3.49 |
| 20 | NT2568 | 488 | 1.24 | 1.24 | .62 | 2.71 | 2.08 | 2.29 |
| Subtotal | 10 segments | 4,399 | 1.42±.18 | 1.40±.18 | 1.52±.19 | 3.16±.27 | 2.83±.26 | 3.20±.27 |
| Segments supporting the Pan-Gorilla clade: | ||||||||
| 10 | T2207 | 478 | .42 | 1.27 | .63 | 3.64 | 2.99 | 3.86 |
| 10 | T2215 | 514 | 1.18 | 1.57 | 1.57 | 2.17 | 2.98 | 3.18 |
| 10 | T2891 | 542 | .93 | .74 | .93 | 2.63 | 2.63 | 2.63 |
| 12 | T2906 | 394 | .25 | 1.54 | 1.54 | 2.06 | 2.32 | 3.64 |
| 12 | T2924 | 491 | 1.64 | 2.06 | 1.44 | 2.47 | 2.89 | 2.89 |
| 17 | T0813 | 444 | .91 | 1.36 | 1.36 | 2.75 | 2.75 | 3.22 |
| 17 | 276-O15 | 434 | 1.87 | 2.34 | 2.82 | 3.06 | 3.30 | 4.02 |
| 18 | NT1506 | 461 | .44 | .65 | 1.09 | 2.20 | 2.65 | 2.87 |
| 18 | NT1584 | 482 | 2.10 | 1.26 | 1.26 | 4.27 | 4.71 | 4.05 |
| 18 | NT2558 | 446 | 1.13 | 2.51 | 1.36 | 4.15 | 3.68 | 5.10 |
| 19 | NT0946 | 320 | 1.57 | 3.18 | 2.21 | 1.89 | .94 | 2.53 |
| 20 | NT2563 | 541 | 1.67 | .92 | 1.11 | 2.05 | 2.61 | 1.86 |
| Subtotal | 12 segments | 5,547 | 1.18±.15 | 1.55±.17 | 1.40±.16 | 2.79±.23 | 2.92±.23 | 3.30±.25 |
| Overall | 53 segments | 24,234 | 1.24±.07 | 1.62±.08 | 1.63±.08 | 3.08±.11 | 3.12±.11 | 3.09±.11 |
C= chimpanzee, G= gorilla, H= human, and O= orangutan.
Figure 1.
Distributions of Jukes-Cantor distances among intergenic regions, introns, and Alus between human and chimpanzee
Table 2 shows additional data from GenBank and the literature. The ∼10-kb region on chromosome 22q11.2 shows a human-chimpanzee divergence of 1.35%, which is somewhat higher than the above estimate of 1.24%. However, the two large contigs from chromosome 12 both show a divergence of 1.2%. Thus, the estimate of 1.2% divergence between human and chimpanzee autosomal intergenic nonrepetitive regions appears to be reliable. This estimate is considerably lower than the estimate of 1.6% from the η-globin pseudogene region (Bailey et al. 1991). The 22q11.2 region shows ∼3% divergence between orangutan and chimpanzee, though a somewhat lower divergence (2.8%) between orangutan and human.
Table 2.
Jukes-Cantor Distances (%) among Noncoding Sequences
|
Jukes-Cantor Distancea(%) |
|||||||
| Sequences | Length(bp) | H-C | H-G | C-G | H-O | C-O | G-O |
| 53 autosomal segments | 24,234 | 1.24±.07 | 1.62±.08 | 1.63±.08 | 3.08±.11 | 3.12±.11 | 3.09±.11 |
| 22q11.2b | 9,772 | 1.35±.12 | …c | … | 2.83±.17 | 3.06±.18 | … |
| Chromosome 12 contig 1d | 58,593 | 1.19±.05 | … | … | … | … | … |
| Chromosome 12 contig 2d | 46,971 | 1.20±.05 | … | … | … | … | … |
| Subtotal for autosomes | 139,570 | 1.21±.03 | … | … | 3.01±.10 | 3.10±.10 | … |
| Xq13.3e | 10,097 | .92±.10 | 1.42±.12 | 1.41±.12 | 3.00±.18 | 2.99±.17 | 2.96±.17 |
| Xc36f | 14,425 | 1.32±.10 | 1.51±.10 | 1.57±.11 | … | … | … |
| Subtotal for X chromosome | 24,522 | 1.16±.07 | 1.47±.08 | 1.50±.08 | … | … | … |
| SMCY g | 4,758 | 1.68±.19 | 2.33±.22 | 2.78±.25 | 5.63±.35 | 6.02±.37 | 6.17±.37 |
Repeat elements were excluded. C = chimpanzee, G = gorilla, H = human, and O = orangutan.
Zhao et al. (2000).
Data unavailable.
Chromosome 12 sequences from GenBank. Contig 1: chimpanzee contig AC007214; Contig 2: chimpanzee contig AC006582.
Kaessmann et al. (1999).
Bohossian et al. (2000); c36 = clone 36.
Shen et al. (2000).
With respect to the X chromosome, the Xq13.3 region shows 0.92%, 1.42%, and 1.41% divergences for the human-chimpanzee, human-gorilla, and chimpanzee-gorilla pairs, respectively, whereas the Xc36 region (human X chromosome clone 36, contig NT001194) shows 1.32%, 1.51%, and 1.57% divergences for the three comparisons (table 2). The Xq13.3 region has evolved significantly more slowly than the Xc36 region in the human-chimpanzee comparison, though not in the human-gorilla and chimpanzee-gorilla comparisons. The average distances of the two X-chromosome regions are 1.16% (human-chimpanzee), 1.47% (human-gorilla), and 1.50% (chimpanzee-gorilla), which are slightly lower than the average distances for the autosomal noncoding regions studied. A slightly lower average divergence in X-linked sequences than in autosomal sequences has been noted earlier by Jaruzelska et al. (1999) and Nachman and Crowell (2000a).
For the Y chromosome, the SMCY region shows 1.68%, 2.33%, and 2.78% divergences for the human-chimpanzee, human-gorilla, and chimpanzee-gorilla comparisons, respectively. These figures are the highest among all the noncoding regions studied. The average distance on the Y chromosome between the orangutan and the human, chimpanzee, and gorilla is 5.94%—about twice the values for autosomal and X-linked noncoding regions.
Introns
The distances among hominoid introns are listed in table 3; because the orangutan data are scanty, only the distances between human, chimpanzee, and gorilla are shown. The average distances are 0.93%, 1.23%, and 1.21% for the human-chimpanzee, human-gorilla, and chimpanzee-gorilla pairs, which are slightly lower than those from the 53 intergenic regions. The human-chimpanzee data are more abundant and the total of 33 loci gives an average distance of 1.03%. Among the loci retrieved, the introns in the β-γ-globin region tend to have a higher divergence than the average. The average intron distances for these globins are 1.89%, 2.16%, and 2.06% for the human-chimpanzee, human-gorilla, and chimpanzee-gorilla pairs. High substitution rates for these genes are also observed for orangutan-human, orangutan-chimpanzee, and orangutan-gorilla pairs (the average for the three pairs is 3.85% [data not shown]).
Table 3.
Distances between Human, Chimpanzee, and Gorilla Introns
|
Jukes-Cantor Distancea(%) |
|||||
| Locus, Introns | Source or Accession Numbers | Length(bp) | H-C | H-G | C-G |
| 1q 24-25 region | Yu et al., in press | 9,556 | .54 | .99 | .96 |
| α-fetoprotein precursor, I14 | M10950, U21916, M38272 | 339 | 1.19 | 2.70 | 2.70 |
| β-1,3-galactosyltransferase, I3 | AB041413, AB041415, AB041416 | 648 | 1.25 | .00 | 1.25 |
| Complement C4, I1–4 | M14824, Z31603, Z31599 | 555 | 1.27 | 1.09 | .54 |
| Dopamine D2 receptor, I1 | AF050737, AF005639, AF005640 | 244 | 1.24 | 2.08 | 1.66 |
| ε- globin, I1–2 | U23824, AJ002051, M81363 | 975 | 1.24 | 1.24 | .62 |
| Fetal Gγ-globin, I 2 | X06490, X03109, X03111 | 856 | 2.62 | 3.22 | 3.71 |
| Ig C α heavy chain constant region, I1 | J002201, X53702, X53703 | 162 | 2.54 | 3.85 | 2.54 |
| Interstitial retinol-binding protein 3, I1 | AF003990, AF003992, AF003994 | 470 | .43 | .86 | 1.29 |
| Natriuretic protein, IA–B | AB037521, AB037522, AB037523 | 770 | 1.31 | 1.44 | 1.18 |
| Phenylalanine hydroxylase gene, I1+7 | AF003965–66b, AF003968–69, AF003971–72 | 712 | .84 | 1.42 | 1.13 |
| Protamine 1, I1 | M60331, L14591, L14587 | 91 | 2.23 | 1.11 | 1.11 |
| Protamine 2, I1 | M60332, X72968, X71336 | 161 | 4.48 | 1.89 | 3.82 |
| RNA helicase 68KD, I2–4 | AJ010931, AJ010933, AJ010932 | 574 | 1.23 | 1.05 | .88 |
| T cell receptor gv 10, I1 | X74798, X86558, X86720 | 112 | 2.75 | .91 | 1.82 |
| Transitional protein 2, I1 | HSU15422, AF215716, AF215718 | 137 | 1.47 | 2.98 | 1.47 |
| Subtotal | 16 loci | 16,362 | .93±.08 | 1.23±.09 | 1.21±.09 |
| α-2-HS glycoprotein, I1–6 | AB038689, AB038690 | 5,301 | 1.14 | … | … |
| β-globin, I1–2 | L26475, X02345 | 795 | 2.17 | … | … |
| Blue opsin, I1–4 | AF039434, AF039435, U53874 | 1,472 | 1.16 | … | … |
| Cytochrome P450, IB | M31664, AF123054 | 407 | .99 | … | … |
| Decay accelarating factor, I1 | AB003312, AB003313 | 510 | 1.19 | … | … |
| Dystrophin gene, I44 | AF085430, AF085432 | 1,450 | .83 | … | … |
| Fetal Aγ-globin, I3–4 | M91036, X03110 | 948 | 3.01 | … | … |
| Glycerol kinase, I1–2 | AF085433, AF085434, AF085435, AF085436 | 1,582 | .70 | … | … |
| Haptoglobin 1, I1–4 | M69197, M84462 | 1601 | .50 | … | … |
| Hypoxanthine phosphoribosyl-transferase, I2+8 | AF085439, AF085440, AF085441, AF085442 | 1,467 | .96 | … | … |
| Iduronate sulphate sulphatase, I2+5 | AF011889, AF085447, AF085448 | 1,498 | .07 | … | … |
| Inteleukin 2 receptor γ chain, I4+5 | AF085451, AF085452, AF085453, AF085454 | 731 | .83 | … | … |
| Lipoprotein lipase, I6+9 | M76722, Z46493, AF071092-4 | 2,163 | .93 | … | … |
| Preproinsulin, I1–2 | V00565, X61089 | 955 | 2.02 | … | … |
| Pyruvate dehydrogenase, E1 α subunit, I1–3 | AF125081, AF125077 | 2,875 | 1.19 | … | … |
| Pyruvate dehydrogenase E1 α subunit, I9–10 | AF085457, AF085459 | 1,176 | 1.20 | … | … |
| Subtotal | 16 loci | 24,931 | 1.10±.07 | … | … |
| Overall | 32 loci | 41,293 | 1.03±.04 | … | … |
H= human, C= chimpanzee, and G= gorilla.
AF003965-66: from accession number AF003965 to AF003966.
Pseudogenes
In GenBank, pseudogene sequences are less abundant than introns. For the seven loci retrieved (table 4) for human, chimpanzee, and gorilla, the average human-chimpanzee, human-gorilla, and chimpanzee-gorilla distances are 1.64%, 1.87%, and 2.14%, respectively. However, 13 additional loci for human and chimpanzee reduce the human-chimpanzee distance to 1.56% (table 4). On the other hand, for the 6 X-linked pseudogenes available for the human-chimpanzee pair, the average distance is 1.47%, which is somewhat lower than the autosomal pseudogene average, in agreement with the conclusion of Nachman and Crowell (2000a).
Table 4.
Distances between Pseudogenes
|
Jukes-Cantor Distancea(%) |
|||||
| Pseudogene | Accession Numbers | Length (bp) | H-C | H-G | C-G |
| Autosomal: | |||||
| α-1,2 fucosyltransferase | U17895, AB006612, AB006611 | 1,045 | 1.74 | 2.43 | 2.23 |
| β-globin | X02133, X02135, X02134 | 2,146 | 1.46 | 1.65 | 2.27 |
| η-globin | U01317, K02542, K02543 | 10,159 | 1.52 | 1.49 | 1.84 |
| Olfactory receptor OR1P1P | AF087927, AF101743, AF101763 | 990 | 1.94 | 3.62 | 2.26 |
| Olfactory receptor OR3A5P | AF087921, AF101735, AF101756 | 560 | 1.26 | 3.01 | 2.82 |
| Olfactory receptor OR3A4P | AF087920, AF101734, AF101756 | 806 | 1.76 | 2.92 | 2.27 |
| Olfactory receptor OR1D3P | AF087919, AF101733, | 940 | 3.04 | 2.49 | 4.27 |
| Subtotal | 7 loci | 16,646 | 1.64±.10 | 1.87±.11 | 2.14±.11 |
| γ-cytoplasmic actin | AF196978, AF196999 | 622 | 1.63 | … | … |
| α-enolase | AF196980, AF197001 | 1,034 | 1.17 | … | … |
| CAMP-dep protein kinase regulatory subunit RIα | AF196994, AF197015 | 445 | .9 | … | … |
| CAMP-dep protein kinase regulatory subunit RIα | AF196995, AF197016 | 424 | 1.43 | … | … |
| Connexin 43-kD protein | AF196981, AF197002 | 769 | 1.44 | … | … |
| Lanosterol 1,4–α-demethylase | AF196989, AF197010 | 359 | .84 | … | … |
| Cytochrome b | AF196982, AF197003 | 718 | .56 | … | … |
| α-1,3-galactosyltransferase | M60263, AF197007 | 1,048 | .77 | … | … |
| Interferon-induced 56-kD protein | AF196987, AF197008 | 713 | 1.27 | … | … |
| Malate dehydrogenase | AF196990, AF197011 | 942 | 1.07 | … | … |
| NADH dehydrogenase | AF196991, AF197012 | 1,150 | 1.14 | … | … |
| Proliferation-associated gene | AF196992, AF197013 | 881 | 1.72 | … | … |
| GPI-anchor synthesis gene | AF196993, AF197014 | 864 | 1.88 | … | … |
| Overall | 20 loci | 26,841 | 1.56±.08 | … | … |
| X-linked: | |||||
| Adaptor protein | Y09846, AF197017 | 961 | 1.79 | … | … |
| C4-sterol methyl oxidase | AF196983, AF197004 | 854 | .59 | … | … |
| Elongation factor 1- α | AF196984, AF197005 | 1,010 | 1.30 | … | … |
| Ferritin-like gene | AF196985, AF197006 | 551 | .73 | … | … |
| HTLV-1 enhancer–binding protein | U03712, AF197018 | 929 | 2.96 | … | … |
| Malate dehydrogenase | AF196990, AF197011 | 942 | 1.07 | … | … |
| Overall | 6 loci | 5,247 | 1.47±.17 | … | … |
H= human, C= chimpanzee, and G= gorilla.
Alus
In a chromosome 12 region for the human and chimpanzee pair, 54 homologous Alu sequences are available (GenBank accession numbers AC007214, AC006582, AC005294, AC007458, AC007286, and AC011604). The individual distances are highly variable (fig. 1c), but the majority of them are higher than the distances in the intergenic regions (fig. 1a). The average distance is 2%.
Coding regions
The KS and KA values and the amino acid distances (p) among hominoids are shown in table 5. These values fluctuate greatly among genes, presumably because of stochastic effects, variation in mutation rate, and variation in selection pressure. For example, the KS values between human and chimpanzee range from 0.00% to 4.02%, and the KA values from 0.00% to 3.68%. The average KS values are 1.11% for human-chimpanzee, 1.48% for human-gorilla, and 1.64% for chimpanzee-gorilla. The KS values between orangutan and human, chimpanzee, and gorilla are 2.98%, 3.05%, and 2.95%, respectively. The average KA values are 0.80%, 0.93%, 0.90%, 1.96%, 1.93, and 1.77% for the human-chimpanzee, human-gorilla, chimpanzee-gorilla, human-orangutan, chimpanzee-orangutan, and gorilla-orangutan pairs, respectively. On the other hand, the average amino acid divergences for the six pairs above are 1.34%, 1.58%, 1.65%, 3.60%, 3.63%, and 3.45%, which are slightly higher than the respective intergenic distances.
Table 5.
Coding-Region Distances between Human, Chimpanzee, Gorilla, and Orangutan
|
KS (%)/KA (%)/AA Distance (%)a |
|||||||
| Locus | Length(bp) | H-C | H-G | C-G | H-O | C-O | G-O |
| Atrophin-related protein DRPLA | 858 | .00/.51/.00 | 1.92/.87/1.41 | 1.92/1.02/1.41 | 2.71/.72/2.11 | 2.71/.88/2.11 | 2.26/1.24/2.11 |
| Brain natriuretic protein | 1,140 | .73/1.51/1.32 | 1.35/1.33/1.84 | .53/1.32/1.57 | 3.54/3.07/4.74 | 2.87/2.79/5.25 | 3.39/2.53/4.99 |
| BRCA 1 | 3,423 | .29/1.03/2.19 | .86/.93/2.02 | .74/.59/1.23 | 2.03/1.85/3.77 | 1.90/1.41/2.89 | 2.49/1.41/2.89 |
| CC chemokine receptor 5 | 1,056 | 1.08/.56/.57 | 1.75/.74/1.14 | 2.30/.41/.57 | 3.44/.59/.85 | 2.85/.26/.28 | 3.53/.15/.28 |
| Complement C5 α receptor | 1,023 | .53/.43/.88 | 3.21/.40/.88 | 3.23/.83/1.76 | 4.49/1.84/3.82 | 4.79/2.40/4.71 | 7.04/1.80/3.82 |
| Connexin-36 | 709 | 2.15/.00/.00 | 2.15/.00/.00 | .80/.00/.00 | 3.63/.00/.00 | 2.89/.00/.00 | 2.89/.00/.00 |
| Cytochrome oxidase subunit 4 | 435 | .00/.65/.00 | .00/1.02/.69 | .00/.37/.69 | .64/2.51/3.47 | .64/1.85/3.47 | .64/2.23/4.17 |
| Eosinophil cationic protein | 483 | .85/.83/1.88 | 1.09/1.00/1.88 | 2.22/1.16/2.50 | 1.42/7.53/13.75 | 2.01/7.89/13.75 | 2.76/7.90/14.38 |
| Eosinophil-derived neurotoxin | 486 | 2.65/.24/.62 | 3.26/.32/.62 | 3.72/.48/1.24 | 7.96/3.20/6.83 | 7.55/3.46/7.45 | 8.79/3.48/7.45 |
| α-1,2-fucosyl-transferase | 1,047 | 1.85/2.63/4.35 | 1.43/1.81/2.32 | 2.27/2.03/4.35 | 3.13/2.78/5.51 | 3.62/3.39/7.54 | 3.73/2.26/5.80 |
| ε-globin | 444 | .65/.00/.00 | .65/.00/.00 | .00/.00/.00 | 1.31/.62/1.36 | 1.98/.62/1.36 | 1.98/.62/1.36 |
| γ-globin | 888 | 1.85/.00/.00 | .00/.35/.68 | 1.85/.35/.68 | 2.27/.21/2.72 | 2.29/1.21/2.72 | 2.26/1.21/2.72 |
| 5-hydroxy-tryptamine receptor 1A | 1,260 | .34/.22/.72 | .22/.45/.24 | .57/.25/.48 | .44/1.20/.72 | .85/.82/.95 | .67/1.01/.48 |
| 5-hydroxy-tryptamine receptor 1F | 1,116 | .00/.29/.00 | .24/.72/.81 | .24/.43/.81 | .49/.64/.81 | .49/.35/.81 | .73/.49/1.62 |
| 5-hydroxy-tryptamine receptor 2A | 732 | 3.22/.21/.41 | 2.72/.00/.00 | 2.58/.21/.41 | 2.59/.16/.41 | 2.99/.37/.82 | 1.97/.16/.41 |
| Ig α heavy chain constant region | 1,059 | 3.03/2.87/5.14 | 1.38/2.13/2.57 | 3.13/3.62/6.57 | 4.69/5.21/8.29 | 5.61/6.22/1.57 | 4.37/5.27/8.86 |
| Igκ constant region | 252 | .00/.00/.00 | 3.60/2.36/4.76 | 3.60/2.36/4.76 | 4.85/7.88/15.48 | 4.85/7.88/15.48 | 6.15/8.05/15.48 |
| Interleukin-8 receptor type A | 1,089 | .25/.64/1.14 | 1.25/1.40/2.57 | .99/1.03/2.29 | 1.25/1.40/2.57 | .99/1.03/2.29 | .00/.00/.00 |
| Interleukin-8 receptor type B | 1,056 | .56/.82/.57 | 2.60/1.06/2.55 | 2.60/1.30/2.55 | 3.30/1.53/4.26 | 3.24/1.95/4.26 | 2.88/1.87/3.98 |
| Leptin | 431 | 1.71/.75/.69 | 1.31/.37/1.38 | 1.71/.66/.69 | .91/.36/2.76 | 1.45/.84/3.42 | .92/.52/4.14 |
| Lysozyme | 447 | 2.85/.00/.00 | 1.26/.35/.68 | .92/.42/.68 | 1.91/.97/2.03 | 1.28/.97/2.03 | .46/1.42/2.70 |
| β2-microglobin | 369 | .78/.70/1.68 | .78/.70/1.68 | .00/.00/.00 | 1.59/2.14/5.04 | .78/.75/3.36 | .78/.75/3.36 |
| β-nerve growth factor | 721 | 2.48/.60/3.17 | 2.92/.60/2.38 | 1.24/.43/1.85 | 4.13/.82/6.08 | 2.42/.65/5.29 | 2.86/.65/5.03 |
| N-formyl peptide receptor | 1,044 | 1.68/.42/.87 | 2.26/1.04/2.31 | 2.26/1.22/2.60 | 3.38/1.31/2.89 | 2.81/1.19/2.60 | 3.97/1.58/3.47 |
| N-formyl peptide receptor-like 2 | 1,047 | 1.22/.48/1.15 | 2.76/1.19/2.58 | 3.34/.90/2.01 | 5.41/2.15/4.87 | 6.61/1.87/4.30 | 2.95/1.51/3.44 |
| Low-affinity N-formyl peptide receptor | 1,044 | 1.36/.51/.86 | 1.09/.53/.86 | 2.48/.77/1.72 | 4.82/1.72/3.16 | 6.19/2.00/4.02 | 5.41/1.48/2.87 |
| Olfactory receptor 93 | 987 | 1.17/1.56/3.40 | 3.08/2.14/4.32 | 3.07/1.12/2.46 | 5.39/3.25/5.86 | 5.09/2.22/2.46 | 5.80/2.14/4.62 |
| Homeobox protein OTX 1 | 729 | .00/.00/.00 | .37/.58/.00 | .37/.58/.00 | .74/1.02/.41 | .74/1.02/.41 | .37/.37/.41 |
| Homeobox protein OTX 2 | 342 | .40/.24/.00 | .00/.00/.00 | .40/.24/.00 | .00/.48/.00 | .40/.71/.00 | .00/.48/.00 |
| Protamine 1 | 156 | 4.02/3.68/10.81 | 1.30/3.31/8.11 | 2.57/2.59/8.11 | 6.76/9.95/18.92 | 11.46/11.00/24.32 | 8.31/1.46/21.62 |
| Protamine 2 | 309 | 2.37/3.64/6.86 | 1.57/3.86/6.86 | .79/2.08/3.92 | 6.24/6.14/11.76 | 6.22/5.51/10.78 | 5.35/6.98/12.75 |
| RNase k6 | 453 | 1.28/.34/.67 | 1.95/.28/.67 | .64/.60/1.33 | 5.38/.85/1.33 | 4.02/1.20/2.00 | 4.69/1.11/2.00 |
| Voltage-gated Na+channel α subunit | 1,092 | 1.13/.00/.00 | .75/.00/.00 | .76/.00/.00 | 1.13/.14/.27 | 1.02/.14/.27 | .76/.14/.27 |
| T cell receptor gv10 | 357 | .83/1.30/2.52 | .83/.86/1.68 | 1.71/.43/.84 | 6.53/4.58/9.24 | 7.60/4.11/8.40 | 5.62/3.65/7.56 |
| Urate oxidase | 162 | .00/.95/.00 | 3.24/.00/1.92 | 3.24/.95/1.92 | .93/3.82/5.88 | .93/4.83/6.00 | 4.13/3.82/7.84 |
| Zinc finger protein 75 | 273 | .00/.00/.00 | .00/.00/.00 | .00/.00/.00 | 4.44/6.26/17.78 | 4.44/6.26/17.78 | 4.44/6.26/17.78 |
| Zinc finger protein 80 | 822 | 2.24/1.41/3.30 | 1.44/2.27/4.76 | 2.61/2.84/6.23 | 3.94/1.85/4.03 | 5.19/2.62/5.86 | 2.81/2.70/5.86 |
| Overall | 29,342 | 1.11/.80/1.34 | 1.48/.93/1.58 | 1.64/.90/1.65 | 2.98/1.96/3.60 | 3.05/1.93/3.63 | 2.95/1.77/3.45 |
KS = number of substitutions per synonymous site; KA = number of substitutions per nonsynonymous site; AA distance = amino acid difference per residue site. C= chimpanzee, G= gorilla, H= human, and O= orangutan.
Comparison of different regions
Figure 1 shows the distributions of distances between human and chimpanzee for the autosomal intergenic regions, introns, and Alus. The distributions are approximately normal for the intergenic regions and introns. About 60% of the intergenic distances and ∼56% of the intron distances fall in between 0.8% to 1.6%. The Alu distribution is more dispersed and only 50% of the distances fall in between 0.4% and 2.0%, but the majority of the Alus have diverged more than the intergenic regions and introns.
On average, the Alu sequences give the largest distances, followed in order by pseudogenes, intergenic regions, synonymous sites, introns, and nonsynonymous sites. For example, for the human-chimpanzee pair, the average distances are 2% (Alus), 1.56% (pseudogenes), 1.24% (autosomal intergenic regions), 1.11% (synonymous), 1.03% (introns), and 0.80% (nonsynonymous). This order is consistent with Li’s (1997) conclusion.
Phylogeny
When the 53 autosomal segments are considered together (concatenated), the neighbor-joining tree (Saitou and Nei 1987) supports the Homo-Pan clade with a 100% bootstrap value (see the topology in fig. 2). When each segment is considered individually, 31 segments support the Homo-Pan clade, 10 support the Homo-Gorilla clade, and 12 support the Pan-Gorilla clade. From these data, we can compute the likelihood ratio of the Homo-Pan clade to the trichotomy (null) hypothesis (Wu 1991):
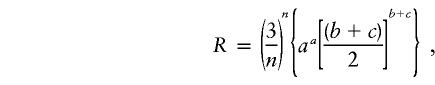 |
where a, b, and c are the numbers of loci supporting topology A (Homo-Pan), topology B (Homo-Gorilla), and topology C (Pan-Gorilla), respectively, and n is the total number of loci studied. For n=53, a=31, b=10, and c=12, we have R=1,105.8, which is much larger than the threshold value (17.2 in table 3 of Wu 1991), so the probability for accepting the Homo-Pan clade is practically 1. In addition, the Homo-Pan clade is also supported by the coding region data set (table 5). Thus, in agreement with the studies of Ruvolo (1997) and Satta et al. (2000), there is very strong support for the Homo-Pan clade.
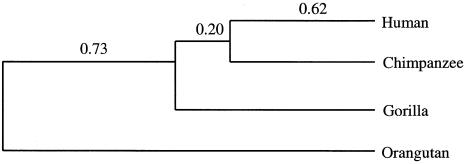
Figure 2.
Phylogeny of hominoids. The branch lengths (Jukes-Cantor distances) are computed under the assumption of rate constancy and used for estimating divergence dates.
Molecular Clock
The sequence data are also useful for testing the molecular clock (rate constancy) hypothesis among the hominoids. For this purpose we can use Wu and Li’s (1985) relative rate test. This test provides the mean and standard error (SE) of the rate difference between two lineages, using a third (outgroup) lineage as a reference; when the mean/SE ratio is ⩾2, the difference is significant at the 5% level. For the 31 intergenic segments that support the Homo-Pan clade (table 1), the gorilla can be used as a reference to test the rate difference between the human and chimpanzee lineages. Since the average distances for the 31 segments are 1.71% and 1.75% for the human-gorilla pair and the chimpanzee-gorilla pair, respectively, the difference between the two distances is clearly not significant and the molecular clock holds. If the orangutan is used as the reference, the distances for the orangutan-human, orangutan-chimpanzee, and orangutan-gorilla pairs are 3.17%, 3.28%, and 2.97%, respectively, and the only significant difference among the three values is between the second and the third (0.31% ± 0.12%, computed from Wu and Li’s formula), implying a significantly slower rate in the gorilla lineage than the chimpanzee lineage. However, this is the only significant difference among all the comparisons in table 3. Indeed, when the 53 segments are considered together and the orangutan is used as the reference, rate constancy is found to hold for the three lineages, because the three distances are very similar (3.08%, 3.12%, and 3.09%; see table 1).
Table 2 provides more data for testing the molecular-clock hypothesis. For the 22q11.2 region, the distance between chimpanzee and orangutan is significantly larger than that between human and orangutan (3.06%-2.83%=0.23%; SE = 0.11%). However, when all the autosomal sequence data are considered together, the difference becomes nonsignificant. For the Y chromosome region (SMCY), the human lineage has evolved significant more slowly than the chimpanzee and gorilla lineages. For the coding regions shown in table 5, the synonymous distances between orangutan and the other three species are 2.98%, 3.05%, and 2.95%, and the nonsynonymous distances are 1.96%, 1.93%, and 1.77%, none of which (except for the difference between 1.96% and 1.77%) deviates significantly from an equal rate of evolution among the human, chimpanzee, and gorilla lineages. So, overall rate constancy holds well, except that the SMCY region has evolved more slowly in the human lineage than in the chimpanzee and gorilla lineage.
Divergence Times
We are interested in estimating the date for the Homo-Pan divergence (THC) and the date for the gorilla divergence (TG). The sequence data from the 53 autosomal intergenic segments in table 1 and the synonymous distances in table 5 are suitable for this purpose because, for these two data sets, rate constancy seems to hold among the human, chimpanzee, and gorilla lineages (see above). Assuming rate constancy, we compute the branch lengths in figure 2 for the data set of 53 intergenic regions. Assuming that the speciation time (T) of Pongo is 12 to 16 million years ago (Goodman et al. 1998; D. Pilbeam, personal communication), we obtain the Homo-Pan divergence time as THC = (0.62/1.55)T = 4.8 to 6.4 million years and the gorilla divergence time as TG = (0.82/1.55)T = 6.3 to 8.5 million years.
The internodal time span (TIN) between the gorilla speciation and the Homo-Pan common ancestor is 0.20/0.62 = 32% of the divergence time between the human and chimpanzee lineage or TIN = (6.3 to 8.5)−(4.8 to 6.4) = 1.5 to 2.1 million years. For the synonymous distances in table 5, the estimates become THC = 4.5 to 5.9 million years, TG = 6.3 to 8.3 million years, and TIN = 1.8 to 2.4 million years. These estimates are very similar to those from the 53 intergenic regions. Taking average of the two sets of estimates, we obtain THC = 4.6 to 6.2 million years, TG = 6.2 to 8.4 million years, and TIN = 1.6 to 2.2 million years.
Effective Size of the Ancestral Population
When three species are fairly closely related to each other, the tree obtained from a set of DNA sequence data (known as the gene tree) may not be congruent with the true tree that represents the two speciation events (known as the species tree). Hudson (1983) and Nei (1986) showed that the probability for the gene tree obtained from a set of sequence data to be congruent with the species tree is given by
where t=TIN is the internodal time span between the two speciation events and is expressed in units of 2Ne generations, where Ne is the effective size of the population in the time span between the two speciation events. Formula (1) implicitly assumes that an incongruent gene tree can arise because of the sharing of an ancestral polymorphism between species 1 (or 2) and species 3, though species 1 and 2 are more closely related to each other. It depends on Ne, because the smaller the Ne, the faster the decrease with time in the probability for species 1 (or 2) to share a polymorphism with species 3. Applying Wu’s (1991) maximum-likelihood estimation procedure to a set of data from independent loci, we can equate P with the proportion of loci that support the species tree. For example, if a loci among the n loci studied support the species tree, then the maximum-likelihood estimate of P is a/n. If t and P are both known, one can estimate Ne.
Because formula (1) assumes that an incongruent tree arose due to sharing of ancestral polymorphism between “wrong” species, a parsimony analysis is more appropriate than a distance analysis. Of the 53 intergenic segments, 24 segments support the Homo-Pan clade, 7 support the Homo-Gorilla clade, 2 support the Pan-Gorilla clade, and 20 segments give no resolution (i.e., they do not support any of three alternative trees). For the coding loci listed in table 5, the corresponding numbers are 12, 3, 4, and 16. In this analysis, P1 and P2 are pooled together as one locus because they are linked, and so are ε-globin and γ-globin; therefore, there are only 35 “independent loci” instead of 37. Taking the two sets of data together and excluding loci that give no resolution, we have a=24+12=36, n=33+19=52, and P=36/52=69%. From this value and formula (1) we estimate t as t=-ln[(3/2)(1-P)]=0.766(2Negenerations). We estimated above that the internodal time span is t = TIN = 1.6 to 2.2 million years. Assuming a generation time of g=15 to 20 years, we obtain Ne=t/(2×0.766g)≈52,000 to 96,000. If we use the neighbor-joining method, we obtain a considerably larger estimate of Ne. For example, for the 53 intergenic segments, we obtain a=31, n=53, P=.59 and Ne = 84,000 to 150,000.
Discussion
Genomic Divergences
We have seen that among the types of sequences included in this study, Alus have, on average, evolved at the highest rate. This is because Alu sequences are not subject to functional constraints, and they contain many CpG dinucleotides, which have a mutation rate about 10 times higher than the genomic average, because of the strong tendency for the C in CpG to mutate to T (Labuda and Striker 1989; Nachman and Crowell 2000a). In fact, there is a 62% correlation between the rate of substitution in an Alu (fig. 1) and the number of changes at CpG dinucleotide sites in the sequence (data not shown). This is a good example supporting the ideas that the mutation rate in a region may depend on its sequence context and that, when the functional constraints in a sequence are removed, the sequence may evolve at a higher rate than the genomic average.
We noted that pseudogenes show the second-highest rate among the types of autosomal sequences included in this study. Like Alus, a pseudogene may also contain more CpG dinucleotides than the average for noncoding regions, though not at a frequency as high as that in Alus. To see if this is, in fact, the case, we computed the CpG frequencies in the 53 intergenic segments included in table 1 and in the 37 genes included in table 5 (table 6). The frequency of CpG in a sequence is computed as the number of CpGs in the sequence, divided by the length of the sequence, minus 1, and the expected frequency of CpG is computed as fCfG, where fC and fG are, respectively, the frequencies of C and G in the sequence. We note that the observed frequency of CpG is much lower in the 53 (noncoding) segments (0.69%) than in the gene sequences (2.77%). The difference is highly significant, even when the expected frequencies are taken into account (table 6). A pseudogene may also contain some other sequence contexts that can confer a higher-than-average mutation rate.
Table 6.
Frequencies of CpG Dinucleotide in Coding and Noncoding Regions[Note]
|
CpG Frequency |
|||
| Region | No. of CpG Dinucleotides Observed | Observed | Expected |
| 37 genes | 781 | .0277 | .0674 |
| 53 noncoding segments | 169 | .0069 | .0426 |
Note.— χ2=41.3, P=.0001.
The above observations suggest that the mutation rate in a functional region often may be higher than the average mutation rate in its nearby introns because of its sequence context, which has been maintained by functional constraints. Thus, the observation of a slightly higher substitution rate at synonymous sites than in introns might be due, in part, to a slightly higher rate of mutation in coding regions than in introns, though it probably also indicates slightly stronger functional constraints in introns than at synonymous sites. The observation that both introns and synonymous sites have on average evolved more slowly than intergenic regions suggests that both introns and synonymous sites are subject to some functional constraints.
For the above reasons, intergenic regions are more suitable than pseudogenes, introns, and synonymous and nonsynonymous sites for estimation of the degree of sequence divergence between hominoid genomes. In fact, the extensive data from intergenic regions suggest that the human and chimpanzee genomes differ by only ∼1.2%, rather than the 1.6% divergence estimated from the η- globin pseudogene region. The η-globin pseudogene region also shows 3.7%, 4.9%, and 4.4% divergences for the orangutan-human, orangutan-chimpanzee, and orangutan-gorilla pairs, which are considerably higher than the ∼3% divergence for these species pairs estimated from the 53 intergenic regions in this study. However, it should be emphasized that our aim is to estimate the genomic divergences among the hominoids in unique noncoding regions. The divergences in repetitive sequences among these genomes might be substantially higher because of a higher mutation rate and frequent deletion and insertion events. In particular, as Alus have a substantially higher rate of mutation than the average genome (see above), the high content (∼10%) of Alus in the hominoid genome should have accelerated the divergence between the human and chimpanzee genomes. This might partly explain the higher estimates of genomic divergence between the human and chimpanzee genomes in the literature. Although the DNA hybridization study by Sibley and Alhquist (1987) tried to exclude rapidly reassociating DNA, this procedure is unlikely to delete all repetitive elements, because such elements are very abundant and are highly dispersed in the hominoid genome.
Molecular Clocks and Divergence Dates
There has been strong evidence supporting the hominoid slowdown hypothesis (Goodman 1961), which postulates that the rate of molecular evolution has become slower in the hominoid (apes and humans) lineage since its separation from the Old World monkey lineage (for a review, see Li 1997). For some regions, a further slowdown has occurred in the human and chimpanzee lineages, in comparison to the rate in the gorilla lineage. These include the η-globin pseudogene region (Bailey et al. 1991; Graur and Li 2000), the Xq13.3 region (table 2), the last intron of the ZFX gene (Jaruzelska et al. 1999), and introns 7 and 44 of the Duchene muscular dystrophy gene (Nachman and Crowell 2000b). However, for some regions, the rate in the gorilla lineage is significantly slower than those in the human and chimpanzee lineages. For example, as noted above, the average rate for the first 31 segments in table 1 is significantly lower in the gorilla lineage than in the chimpanzee lineage, and the average nonsynonymous rate for the genes in table 5 is significantly lower in the gorilla lineage than in the human lineage. So, there is no strong trend toward a slowdown in the human or chimpanzee lineage. In fact, the data in tables 1 and 5 show that the molecular-clock hypothesis, on average, holds well among the human, chimpanzee, and gorilla lineages for the intergenic regions and the synonymous sites. This observation suggests that the generation time effect is weak and often may not be discernable. This suggestion seems reasonable, in view of the fact that the generation time in the human lineage was only slightly longer than those in the chimpanzee and gorilla lineages (see later). For Y-linked sequences, the mutation rate is higher, and the generation time effect is easier to detect. In fact, the SMCY region is seen to have evolved faster in the chimpanzee lineage than in the human lineage (see table 2).
As the molecular-clock hypothesis seems to hold well for the 53 intergenic regions and for the synonymous sites used in this study, these regions are suitable for estimating the divergence dates among the human, chimpanzee, and gorilla lineages. The only uncertain aspect is the date of the orangutan speciation event. If our assumption of 12 to 16 million years is close to the true date, then our estimates should be reliable. Our estimates are similar to those of Goodman et al. (1998) of 7 million and 6 million years ago for the gorilla branching node and the human-chimpanzee divergence, respectively. In any event, our data suggest that the internodal time span between the human-chimpanzee divergence and the gorilla speciation event is about one-third of the divergence time between the human and chimpanzee lineages. This estimate is independent of the calibration of the molecular clock and is in between the estimates of 60% from the mitochondrial data and 10% from the η-globin pseudogene sequence data.
Population-Size Estimation
Generation time
The generation time is an important factor in our estimation of the ancestral population size. According to Nowak and Paradiso (1983), both sexes of chimpanzees reach puberty at age 7 years. But females usually do not give birth until they are 13 years old, and males are not totally integrated into the social hierarchy until they are 15 years old. Reproductive capability in females can last at least until age 40 years (Nowak and Paradiso 1983), and chimpanzees can live to age 50 or even 60 years. This is concordant with Reynolds and Reynolds’s observation (1965) that wild chimpanzees could live to age >40 years and remain healthy. On the other hand, gorillas reach sexual maturity at age 8 years for females and 11 years for males (Nowak and Paradiso 1983). However, females in the wild usually give birth for the first time at age ∼10 years and live to age 30 to 40 years in the wild. In summary, chimpanzees and gorillas start to reproduce a couple of years earlier than humans and have a somewhat shorter lifespan than humans. Although the generation time in a modern human society can be >30 years (Tremblay and Vézina 2000; Sigurdadóttir et al. 2000), the generation time in the long history of human evolution is commonly taken to be 20 years (e.g., Nei and Graur 1984). For this reason and in light of the data cited above, we assume a generation time of 15 to 20 years for the common ancestor of chimpanzees and humans.
Ancestral population size
We have followed Ruvolo (1997) in the use of multiple data sets to estimate the effective size (Ne) of the ancestral population before the human-chimpanzee divergence. Because of limited data availability, Ruvolo used only 14 independent coding loci. Among them, 11 loci supported the Homo-Pan clade, and Ruvolo obtained an estimate of Ne = 35,000 to 65,000. Her estimate is considerably lower than ours. There are two possible reasons for the difference. First, the number of loci used in Ruvolo’s study was small, so the estimate had a large standard error. Second, as Ruvolo (1997) pointed out, the 14 loci included one mitochondrial locus, one X-linked locus, and one Y-linked locus, all of which tend to give a lower estimate of Ne; this is because the effective population sizes for the mtDNA, a Y-linked locus, and an X-linked locus are only Ne/4, Ne/4, and 3Ne/4, respectively, instead of Ne for an autosomal locus. If we exclude these three loci, the proportion of loci supporting the Homo-Pan clade decreases from 11/14=0.79 to 9/11=0.73, which is not significantly different from our value of 0.69.
Note that formula (1) assumes no new mutations. This assumption tends to overestimate P, because new mutations in the internodal time span would produce shared polymorphisms between species 1 and 2. For this reason formula (1) tends to underestimate Ne. However, because the number of loci used is still small, our estimate of Ne = 52,000 to 96,000 should be taken with caution.
This caution notwithstanding, as our estimate of the effective population size of the common ancestor of human and chimpanzee is about 5 to 9 times higher than the estimate (∼10,000) of the effective population size of humans from various genetic polymorphism data (e.g., Nei and Graur 1984; Takahata 1993; Zhao et al. 2000), the human lineage apparently has undergone a significant reduction in effective population size since its separation from the chimpanzee lineage. It is not clear why this reduction has occurred, but one possibility might be that the human lineage has gone through many local extinction and recolonization events; such events can greatly reduce the effective size of a species (Maruyama and Kimura 1980).
Acknowledgments
This study was supported by NIH grants GM55759 and GM30998. We thank Ning Yu for help, and two anonymous reviewers, J. Hey, M. Jensen-Seaman, D. Pilbeam, M. Ruvolo, and N. Takahata for suggestions.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST server, http://www.ncbi.nlm.nih.gov/BLAST/
- Genome Channel, http://genome.ornl.gov/GCat/species.shtml (for 53 DNA segments studied)
- RepeatMasker, http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker
- Silver Project Home Page, http://sayer.lab.nig.ac.jp/~silver/homoNuc.html (for accession numbers of coding regions)
References
- Bailey WJ, Fitch DH, Tangle DA, Czelusniak J, Slightom JL, Goodman M (1991) Molecular evolution of the ψη-globin gene locus: gibbon phylogeny and the hominoid slowdown. Mol Biol Evol 8:155–184 [DOI] [PubMed] [Google Scholar]
- Bohossian HB, Skaletsky H, Page DC (2000) Unexpectedly similar rates of nucleotide substitution found in male and female hominids. Nature 406:622–625 [DOI] [PubMed] [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS (1991) “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 19:4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorit RL, Akashi H, Gilbert W (1995) Absence of polymorphism at the ZFY locus on the human Y chromosome. Science 268:1183–1185 [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D, Gouy M (1994) HOVERGEN: a database of homologous vertebrate genes. Nucleic Acids Res 22:2360–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G, Sosinsky A, Ben-Asher E, Avidan N, Sonkin D, Bahar A, Rosenthal A, Clifton S, Roe B, Ferraz C, Demaille J, Lancet D (2000) Sequence, structure, and evolution of a complete human olfactory receptor gene cluster. Genomics 63:227–245 [DOI] [PubMed] [Google Scholar]
- Goodman M (1961) The role of immunochemical differences in the phyletic development of human behavior. Hum Biol 33:131–162 [PubMed] [Google Scholar]
- Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP (1998) Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol 9:585–598 [DOI] [PubMed] [Google Scholar]
- Goodman M, Tagle DA, Fitch DH, Bailey W, Czelusniak J, Koop BF, Benson P, Slightom JL (1990) Primate evolution at the DNA level and a classification of hominoids. J Mol Evol 30:260–266 [DOI] [PubMed] [Google Scholar]
- Graur D, Li WH (2000) Fundamentals of molecular evolution. 2nd ed. Sinauer Associates, Sunderland, MA [Google Scholar]
- Horai S, Satta Y, Hayasaka K, Kondo R, Inoue T, Ishida T, Hayashi S, Takahaka N (1992) Man’s place in Hominoidea revealed by mitochondrial DNA genealogy. J Mol Evol 35:32–43 [DOI] [PubMed] [Google Scholar]
- Hudson RR (1983) Testing the constant-rate neutral allele model with protein sequence data. Evolution 37:203–217 [DOI] [PubMed] [Google Scholar]
- Jaruzelska J, Zietkiewicz E, Labuda D (1999) Is selection responsible for the low level of variation in the last intron of the ZFY locus? Mol Biol Evol 16:1633–1640 [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Heißig F, von Haeseler A, Pääbo S (1999) DNA sequence variation in a non-coding region of low recombination on the human X chromosome. Nat Genet 22:78–81 [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC (1975) Evolution at two levels in humans and chimpanzees. Science 188:107–116 [DOI] [PubMed] [Google Scholar]
- Labuda D, Striker G (1989) Sequence conservation in Alu evolution. Nucleic Acid Res 17:2477–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH (1993) Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol 36:96–99 [DOI] [PubMed] [Google Scholar]
- ——— (1997) Molecular evolution. Sinauer Associates, Sunderland, MA [Google Scholar]
- Maruyama T, Kimura, M (1980) Genetic variability and effective population size when local extinction and recolonization of subpopulations are frequent. Proc Natl Acad Sci USA 77:6710–6714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL (2000a) Estimate of the mutation rate per nucleotide in humans. Genetics 156:297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2000b) Contrasting evolutionary histories of two introns of the Ducheme muscular dystrophy gene, Dmd, in humans. Genetics 156:297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M (1986) Stochastic errors in DNA evolution and molecular phylogeny. In: Gershowitz H, Rucknagel DL, Tashian RE (eds) Evolutionary perspectives and the new genetics. Alan R. Liss, New York, pp 133–147 [PubMed] [Google Scholar]
- Nei M, Graur D (1984) Extent of protein polymorphism and the neutral mutation theory. Evol Biol 17:73–118 [Google Scholar]
- Nowak RM, Paradiso JL (1983) Walker’s mammals of the world. Vol. 1. The Johns Hopkins University Press, Baltimore and London [Google Scholar]
- Reynolds V, Reynolds F (1965) Chimpanzees of the Budongo forest. In: DeVore I (ed) Primate behavior: field studies of monkeys and apes. Holt, Rinehart and Winston, New York, pp 368–424 [Google Scholar]
- Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569 [DOI] [PubMed] [Google Scholar]
- Ruvolo M (1997) Molecular phylogeny of the hominoids: inferences from multiple independent DNA sequence data sets. Mol Biol Evol 14:248–265 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425 [DOI] [PubMed] [Google Scholar]
- Sarich VM, Wilson AC (1967) Immunological time scale for hominid evolution. Science 158:1200–1203 [DOI] [PubMed] [Google Scholar]
- Satta Y, Klein J, Takahata N (2000) DNA archives and our nearest relative: the trichotomy problem revisited. Mol Phylogenet Evol 14:259–275 [DOI] [PubMed] [Google Scholar]
- Shen P, Wang F, Underhill PA, Franco C, Yang WH, Roxas A, Sung R, Lin AA, Hyman RW, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ (2000) Population genetic implications from sequence variation in four Y chromosome genes. Proc Natl Acad Sci USA 97:7354–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CG, Ahlquist JE (1987) DNA hybridization evidence of hominoid phylogeny: results from an expanded data set. J Mol Evol 26:99–121 [DOI] [PubMed] [Google Scholar]
- Sigurdardóttir S, Helgason A, Gulcher JR, Stefansson K, Donnelly P (2000) The mutation rate in the human mtDNA control region. Am J Hum Genet 66:1599–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N (1990) A simple genealogical structure of strongly balanced allelic lines and trans-species evolution of polymorphism. Proc Natl Acad Sci USA 87:2419–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1993) Allelic genealogy and human evolution. Mol Biol Evol 10:2–22 [DOI] [PubMed] [Google Scholar]
- Takahata N, Satta Y, Klein J (1995) Divergence time and population size in the lineage leading to modern humans. Theor Popul Biol 48:198–221 [DOI] [PubMed] [Google Scholar]
- Tremblay M, Vézina H (2000) New estimates of intergeneration time intervals for the calculation of age and origins mutations. Am J Hum Genet 66:651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI (1991) Inferences of species phylogeny in relation to segregation of ancient polymorphisms. Genetics 127:429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Li WH (1985) Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci USA 82:1741–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X (2000) Data analysis in molecular biology and evolution. Kluwer Academic Publishers, Boston [Google Scholar]
- Yu N, Zhao Z, Fu YX, Ramsay M, Jenkins T, Leskinen E, Patty L, Jorde LB, Sambuughin N, Li W-H. Global patterns of human DNA sequence variation in a 10-Kb region on chromosome 1. Mol Biol Evol (in press) [DOI] [PubMed] [Google Scholar]
- Zhao, Z, Jin L, Fu YX, Ramsay M, Jenkins T, Leskinen E, Pamilo P, Trexler M, Patthy L, Jorde LB, Sambuughin N,Yu N, Li W-H (2000) Worldwide DNA sequence variation in a 10 kilobase noncoding region on chromosome 22. Proc Natl Acad Sci USA 97:11354–11358 [DOI] [PMC free article] [PubMed] [Google Scholar]