Abstract
Dopamine-β-hydroxylase (DβH) catalyzes the conversion of dopamine to norepinephrine and is released from sympathetic neurons into the circulation. Plasma-DβH activity varies widely between individuals, and a subgroup of the population has very low activity levels. Mounting evidence suggests that the DBH structural gene is itself the major quantitative-trait locus (QTL) for plasma-DβH activity, and a single unidentified polymorphism may account for a majority of the variation in activity levels. Through use of both sequencing-based mutational analysis of extreme phenotypes and genotype/phenotype correlations in samples from African American, European American (EA), and Japanese populations, we have identified a novel polymorphism (−1021C→T), in the 5′ flanking region of the DBH gene, that accounts for 35%–52% of the variation in plasma-DβH activity in these populations. In EAs, homozygosity at the T allele predicted the very low DβH–activity trait, and activity values in heterozygotes formed an intermediate distribution, indicating codominant inheritance. Our findings demonstrate that −1021C→T is a major genetic marker for plasma-DβH activity and provide new tools for investigation of the role of both DβH and the DBH gene in human disease.
Dopamine-β-hydroxylase (DβH [MIM 223360]) catalyzes the conversion of dopamine to norepinephrine. It is localized in catecholamine-containing vesicles of adrenergic and noradrenergic cells (Oka et al. 1967; Kemper et al. 1987). DβH protein is released in response to stimulation (Viveros et al. 1968), and DβH activity, derived largely from sympathetic nerves, can be measured in human plasma (Weinshilboum and Axelrod 1971a; Weinshilboum 1978). Plasma (or serum) activity levels vary widely among individuals (Weinshilboum et al. 1973) but are stable within individuals over time (Fahndrich et al. 1982). Family and twin studies have indicated that the majority of interindividual variability in plasma-DβH activity is genetically determined (Ross et al. 1973; Weinshilboum et al. 1973; Oxenstierna et al. 1986).
In a large study of European Americans (EAs), Weinshilboum et al. (1975) identified a subgroup, consisting of 3%–4% of the population, who have very low levels of plasma-DβH activity. Parents of these individuals have an average enzyme activity that is intermediate between the very low DβH–activity subgroup and a population selected at random. Pedigree segregation analyses of these families suggested that the very low DβH–activity trait is monogenically inherited by an autosomal codominant mechanism (Weinshilboum 1983). Weinshilboum (1978) postulated the existence, at an unspecified locus, of a functional low-activity allele, DBHL, with a frequency of ∼20% in the EA population. A single locus was estimated to account for 46%–75% of the plasma DβH–activity variance in the population (Elston et al. 1979; Goldin et al. 1982; Wilson et al. 1990). Studies using protein-phenotype markers showed strong evidence for linkage between a major locus controlling plasma-DβH activity and the ABO blood-group locus (Goldin et al. 1982; Wilson et al. 1988). The DBH gene was subsequently cloned, mapped to chromosome 9q34, and shown, by molecular studies, to be linked to ABO (Lamouroux et al. 1987; Craig et al. 1988; Gelernter et al. 1991; Perry et al. 1991). Molecular markers at the DBH locus were then shown to associate with variation in both plasma-DβH activity (Wei et al. 1997; Cubells et al. 1998, 2000) and cerebrospinal-fluid (CSF) levels of immunoreactive DβH protein (Cubells et al. 1998). Taken together, these findings strongly suggest that DBH itself is the major quantitative-trait locus (QTL) for plasma-DβH activity—and that it contains the DBHL allele. The molecular identity of the DBHL allele remains to be determined.
Altered plasma-DβH activity has been reported in a variety of psychiatric and neurological disorders (Weinshilboum and Axelrod 1971b; Lieberman et al. 1972; Fujita et al. 1978; Matuzas et al. 1982; Nagatsu et al. 1982). Given the evidence summarized above, DBH may therefore be an important disease-causing or disease-modifying candidate gene. Disease-related variation in DβH activity could be interpreted more readily if the molecular basis of heritable variation in plasma DβH were known. Comprehensive studies examining the relationship between polymorphic variation at DBH and plasma-DβH activity have not been published. Here we report a sequencing-based mutational analysis of the DBH gene from subjects with extreme plasma-DβH phenotypes. We then examine genotype/phenotype correlations in samples from African American (AA), Japanese (Jp), and EA populations.
Plasma and DNA samples from 58 unrelated AA and 174 unrelated EA subjects (self-identified population groups; those of mixed or other heritage were excluded) were collected during the course of several ongoing genetic studies. The groups included both healthy individuals and those with psychiatric and substance-abuse disorders. As discussed elsewhere (Cubells et al. 1998), sampling from a variety of diagnostic groups is unlikely to obscure fundamental genetic influences on plasma-DβH activity. Plasma and DNA samples were obtained from 53 healthy, unrelated ethnic-Japanese subjects recruited at the Institute for Comprehensive Medical Sciences, Fujita Health University, Toyoake, Japan. All subjects gave informed consent for participation in molecular-genetic studies.
DβH activity was assayed in plasma samples from all subjects, as described elsewhere (Cubells et al. 1998). The DBH gene is ∼23 kb in length and contains 12 exons (Kobayashi et al. 1989). The genomic sequence is publicly available (Genbank accession numbers AC000404 and AC001227). We searched for potential functional polymorphisms by sequencing a total of 6,443 bp of DBH, including the proximal 1,468 bp of the 5′ upstream area, all exons (2,744 bp), and 2,182 bp of intronic sequence, spanning ⩾49 bp flanking each exon. A minimum of eight individuals were selected to represent extreme values of DβH activity. This group included at least four individuals (two EA and two Jp) with very low activity (defined here as <2.5 nmol octopamine/min/ml plasma [hereafter expressed as “nmol/min/ml”]), two EAs with near-average activity, and two EAs with activity >2 SD above the mean. Twelve subjects, eight (six EA and two Jp) with very low activity, were included for the sequencing of the proximal 1.0 kb of the promoter region and of exon 1. Sequencing was performed, with minor modifications, as described elsewhere (Zabetian et al. 2000). PCR primer sequences and reaction conditions are available on request.
All newly detected polymorphisms that appeared to aggregate according to DβH activity in the sequenced subset of samples, as well as all previously described nonsynonymous polymorphisms, were selected as functional candidates and were genotyped in the entire AA, EA, and Jp samples. The average rate of PCR nonamplification for each population-specific genotype group was 1.6%, and the maximum was 4.0%. Genotypes were determined by RFLPs, by digestion of PCR products with the appropriate restriction enzymes, as shown in table 1. RFLP genotyping was confirmed by the sequencing of representative samples of each polymorphism.
Table 1.
Allele Frequencies of DBH Polymorphisms in Three Population Samples
|
Mean ± Binomial SE, for Allele Frequency in |
||||||
| Location | Polymorphisma | Amino Acid Change | Restriction Site for Genotyping | AA (2n=116) | EA (2n=348) | Jp (2n=106) |
| 5′ Flanking | −1021C→T | … | MwoIb | .198 ± .037 | .224 ± .022 | .160 ± .036 |
| Exon 3 | 499G→Cc | E167Q | BsrI | NDd | ND | ND |
| Exon 3 | 589G→Ac | A197T | BstUI | .129 ± .031 | .083 ± .015 | .144 ± .035 |
| Exon 3 | 675G→Cc | K225N | ApaIb | ND | ND | ND |
| Exon 4 | 706G→Cc | E236Q | BanII | ND | ND | ND |
| Exon 4 | 826G→Ac | D276N | BsiEI | .052 ± .021 | .003 ± .003 | ND |
| Intron 4 | IVS4+601T→C | … | BsmBIb | .348 ± .045 | .524 ± .034 | .837 ± .036 |
| Exon 5 | 908T→Cc | L303P | BslI | ND | ND | ND |
| Exon 5 | 910G→Te | A304S | MwoI | .147 ± .033 | .062 ± .013 | .010 ± .010 |
| Exon 11 | 1603C→Te | R535C | BstUI | .035 ± .017 | .043 ± .011 | ND |
Nucleotide positions are numbered according to either cDNA sequence, for exons, or genomic sequence, for the 5′ flanking region, beginning at the A of the ATG initiator Met codon; positions for introns are numbered according to the genomic sequence, starting from the G of the donor-site invariant GT.
Artificial restriction site.
From Halushka et al. (1999).
ND=not detected in sample.
From Kobayashi et al. (1989).
An initial plot of genotypes at −1021C→T and of population-specific variance against mean DβH activity produced three approximately straight lines passing through the origin (data not shown), suggesting that a square-root transformation would be appropriate for stabilization of the variance. Maximum likelihood was therefore used to fit a Box-Cox power transformation (Box and Cox 1964) and to test equality of the power parameter to 0.5, on the assumption of normality and a common variance. On the basis of the result of this test, an analysis of variance of square-root DβH activity was used to test for genotype effects, population effects, and genotype × population interaction. In addition, deviation from additive gene action was tested and was found to be nonsignificant. Because heritability can be estimated more accurately if Hardy-Weinberg equilibrium (HWE) can be assumed, deviations from HWE were assessed using the HWSIM program (Cubells et al. 1997; also see the HWSIM Web site). Since some of the analyses contained small numbers of observations in some cells, P values for all analyses were estimated empirically, with Monte Carlo simulations (10,000 iterations in each case) based on observed allele frequencies. Significance levels were estimated as the proportion of times that the simulated distribution reached or exceeded the observed deviation from HWE. Heritability was estimated, under the assumption of HWE, as follows: h2=[2p(1-p)δ2]/[2p(1-p)δ2+s2], where the additive gene effect is δ, the gene frequency is p, s2 is the estimate of the common variance about the genotype mean, and its standard error is obtained by application of the large-sample “delta” method (DeGroot 1989), by double differentiation of the log likelihood to obtain the variance-covariance matrix of δ and s2. A pooled estimate of heritability was also calculated under the assumption of HWE, as a weighted average of the individual population heritabilities, with weights inversely proportional to their variances and summing to 1.
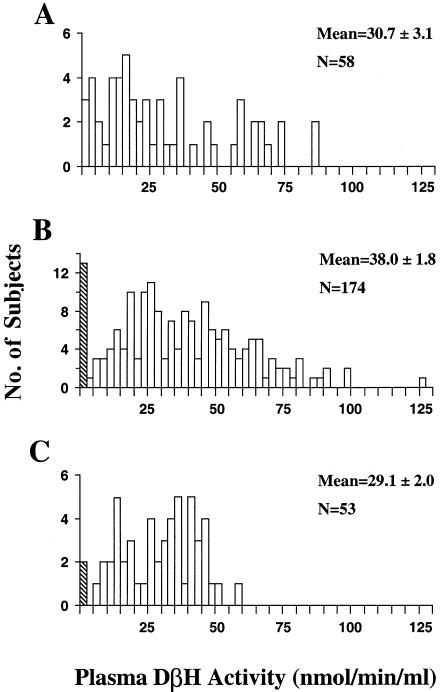
The plasma DβH–activity data are summarized in figure 1. There was a small but statistically significant difference, in mean activity values, between EAs and Jps (P=.003), but there were no differences between AAs and either Jps or EAs (P>.1). In the EA group, a cluster of individuals with activity values <2.5 nmol/min/ml was identified by visual inspection of frequency histograms (fig. 1), and was defined as the “very low DβH–activity” subgroup. This corresponds closely with the threshold of <2.6 nmol/min/ml defined by Weinshilboum et al. (1975) for the very low DβH–activity subgroup (corrected, for differences in methodology, by a factor of 18.9, per Weinshilboum [1978]). This subgroup constituted 7.5% of EAs and 3.8% of Jps in the present study. In the AA group, however, no discrete cluster of very low levels was readily discernible, with 5.2% of the activity values <2.5 nmol/min/ml.
Figure 1.
Distribution of plasma-DβH activity in AA (A), EA (B), and Jp (C). Subjects are grouped in successive 2.5-nmol/min/ml increments. The hatched bar indicates the results for the very low DβH-activity group. The EA and Jp mean activity values differed significantly (P<.01, one-way ANOVA; followed by Dunnett C post-hoc test).
The sequencing of the eight individuals representing phenotypic extremes revealed 18 single-nucleotide polymorphisms (SNPs) not previously reported (ID numbers obtained from the Database of Single Nucleotide Polymorphisms). Fourteen of these were located in introns, four in the 5′ flanking region, and none in the coding sequence. The most proximal SNP in the 5′ flanking region occurred at position −496. Two of these SNPs—one located in the 5′ flanking region (−1021C→T), the other in intron 4 (IVS4+601T→C)—associated with the very low DβH–activity subgroup in the sequenced samples and were included in further genotyping studies of the complete sample populations.
Four of the eight previously described nonsynonymous SNPs were not detected in any of the three population samples, and only two (589G→A and 910G→T) occurred in all three samples (table 1). There were no deviations from HWE for genotypes at any of the polymorphisms in either the AA group or the Jp group. In the EA sample, a small but statistically significant (P=.034) deviation from HWE was seen in genotypes at −1021C→T, but at none of the other polymorphisms. However, this observed P value does not reach significance after application of the Bonferroni correction for three tests.
The −1021C→T polymorphism was strongly associated with untransformed DβH-activity values in all three population samples (P<.001). Because of significant heterogeneity of variance between genotype groups, an appropriate transformation function was estimated, as described above. The value of the power parameter was .5083, whether separate genotypic means or additive gene effects were fitted, and the nonadditivity (dominance variance) was not significant (P=.72). This estimate was not significantly different (P=.9) from .5, the square-root function. Table 2 gives the population-/genotype-specific means (and the standard errors of the mean) for square-root DβH activity. An analysis of variance of square-root DβH activity indicated significant effects of genotype at −1021C→T (P<.001) and of population (P<.001), but not of genotype × population interaction (P=.51). The proportion of variability (R2) due to genotype at −1021C→T was .35, .51, and .52 for the AAs, EAs, and Jps, respectively. Addition of 1603C→T to the model resulted in a small but significant (P=.0024) increase of .02 in R2. The added contribution of the remaining four polymorphisms detected in our population samples was not significant (P>.05).
Table 2.
DβH Activity Values Specific to Genotypes at −1021C→T in Three Population Samples
|
Mean ± SEM |
|||
| Sample andGenotype (n) | DβH Activity(nmol/min/ml) | Square-RootDβH Activity(square-root nmol/min/ml) | h2a |
| AA: | |||
| TT (4) | 6.0 ± 4.2 | 2.07 ± .76 | |
| CT (15) | 15.8 ± 3.7 | 3.62 ± .44 | |
| CC (39) | 39.0 ± 3.7 | 5.96 ± .30 | |
| Overall | .31 ± .12 | ||
| EA: | |||
| TT (16) | 4.1 ± 2.1 | 1.47 ± .36 | |
| CT (46) | 25.2 ± 2.0 | 4.80 ± .22 | |
| CC (112) | 48.1 ± 2.1 | 6.74 ± .15 | |
| Overall | .44 ± .09 | ||
| Jp: | |||
| TT (1) | .4 | .66 | |
| CT (15) | 15.0 ± 1.7 | 3.79 ± .21 | |
| CC (37) | 35.6 ± 1.8 | 5.85 ± .20 | |
| Overall | .52 ± .28 | ||
Under the assumption of HWE.
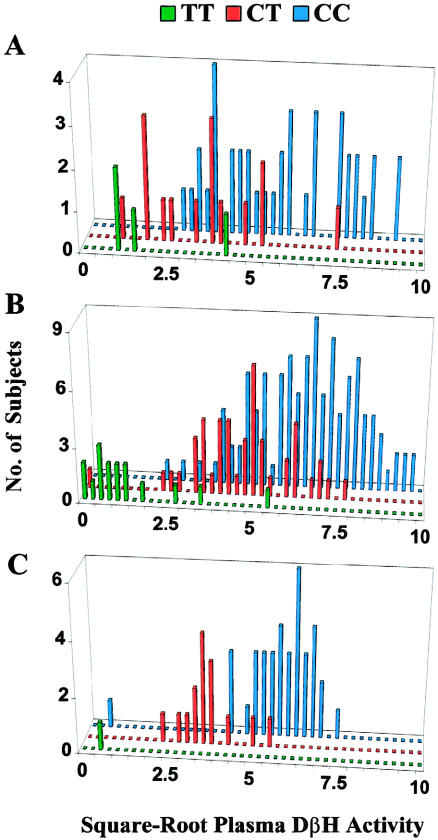
The heritabilities ± SE of square-root DβH activity under the assumption of HWE and additive gene action were h2=.31±.12, for AAs; .44±.09, for EAs; and .52±.28, for Jps (h2=.50 for EAs, when there is no assumption of HWE). The overall (mean ± SEM) heritability was .40 ± .07 when data from all three populations were pooled. Histograms of square-root DβH activity versus genotype at −1021C→T are presented in figure 2. In EAs, the TT genotype was strongly associated (P<.0001) with the very low DβH–activity trait, since 12 of the 13 subjects with the trait were TT homozygotes. The remaining individual was heterozygous at −1021C→T but was the only subject in any of the three populations who was homozygous at the nonsynonymous exon 11 polymorphism 1603C→T. Four TT homozygotes had transformed activity values that exceeded the very low DβH–activity threshold (<1.58 square-root nmol/min/ml), but all were below the population mean (5.74 square-root nmol/min/ml). Transformed activity values in subjects who were heterozygous at −1021C→T formed a distribution intermediate between the two homozygous groups in the EA and Jp samples, in a way consistent with autosomal codominant inheritance of the very low DβH–activity trait (fig. 2).
Figure 2 .
Distribution of square-root plasma-DβH activity, by genotype, at DBH polymorphism −1021C→T in AA (A), EA (B), and Jp (C). Green bars denote T/T homozygotes, red bars denote C/T heterozygotes, and blue bars denote C/C homozygotes. A single C/C homozygote with an extreme activity value of 11.2 square-root nmol/min/ml was omitted from the EA distribution plot, for the purpose of graphical clarity.
Several previous population-based studies have found associations between polymorphisms in the DBH gene and plasma-DβH activity, in individuals of European heritage (Wei et al. 1997; Cubells et al. 1998, 2000). These polymorphisms have included one in the 5′ flanking region (−4784–(−)4803del) and a synonymous SNP in exon 2 (444G→A). However, the allele frequencies and distributions of genotype-specific mean activities for each of these polymorphisms are not compatible with those predicted for the DBHL allele, and neither of these previous allelic associations holds in any sample except the EA sample (J.F.C., C.P.Z., G.M.A., and J.G., unpublished observations). Therefore, these previous associations appear to arise from linkage disequilibrium (LD) between the examined alleles and nearby functional variants. In contrast, −1021C→T accounts for one third or more of the variance in plasma-DβH activity, in all three of the ethnically diverse populations examined in the present study. Such cross-population consistency in genotype-phenotype relationships supports a causal association between a polymorphism and trait outcome, rather than an association based solely on LD (Risch 2000). Moreover, the frequency (.224±.022) of the T allele in the EA sample examined here is very close to the predicted frequency (.20) for DBHL (Weinshilboum 1978). The mean ± SEM and range (38.0±1.8 and 0–125 nmol/min/ml) of activity values for EAs in this study are similar to those obtained in Weinshilboum et al.'s (1975) study of 227 unrelated healthy adult EAs (36.1±1.5 and 0–129.6 nmol/min/ml, corrected as described above). Thus, the present study provides the strongest evidence yet for the molecular identity of the DBHL allele, predicted some 25 years ago to account for a large proportion of the genetic variation in human plasma-DβH activity.
Several additional considerations suggest that −1021C→T may itself be the major functional polymorphism at the DBH locus, accounting for variation in plasma-DβH activity:
-
1.
The DBHL allele appears to lower plasma-DβH activity by diminishing the levels of circulating DβH protein, rather than by decreasing the activity of homospecific enzymes (Dunnette and Weinshilboum 1976). A 5′-upstream polymorphism such as −1021C→T, differentially regulating DBH transcription, would readily account for this type of biochemical phenotypic variation.
-
2.
Reporter-gene experiments in transgenic mice suggest that the region between base pair −600 and base pair −1100 contains elements critical for human DBH-gene expression (Hoyle et al. 1994). To date, specific cis-acting motifs, including CREB- and Phox2a/2b-binding sites, have only been identified in more-proximal 5′ flanking regions (Kim et al. 1998).
-
3.
−1021C→T is located in a noradrenergic cell-type–specific DNase I hypersensitive site of the DBH gene (Ishiguro et al. 1993).
These observations do not directly demonstrate a functional role for −1021C→T, nor do they rule out the possibility that LD with an as-yet-unidentified functional variant accounts for the strong association with DβH activity. However, these converging lines of evidence suggesting the testable hypothesis that −1021C→T alters transcriptional regulation of DBH expression warrant further study.
In the EA sample, genotypes at −1021C→T deviated slightly from HWE, with a greater number of TT homozygotes being observed than were expected, possibly because of a sampling bias due to the inclusion of subjects with psychiatric illnesses. Regardless of the source of the deviation, if the TT genotype is associated with the very low DβH–activity trait, then one would expect to see that trait in a larger proportion of individuals within the sample; and this is precisely what was observed: the very low DβH–activity subgroup constituted a larger proportion of our EA sample (7.5%) than it did in Weinshilboum et al.'s (1975) study of adult EAs (3.1%).
The clinical significance of plasma-DβH activity levels has been the subject of controversy for some time. Initially, some 3 decades ago, it was hoped that plasma-DβH activity might both provide a direct measure of noradrenergic and adrenergic synaptic activity and be a sensitive indicator of sympathetic function. This idea proved overly simplistic, since it later became apparent that the wide individual variation in activity levels is primarily genetically determined, with estimates of heritability being .61–.98 (Elston et al. 1979; Oxenstierna et al. 1986; Wilson et al. 1990). Also, even extreme manipulations of sympathetic function, such as cold-pressor stimulation and strenuous exercise, had minimal effects on plasma-DβH levels, despite producing large increases in circulating catecholamines (Winer and Carter 1977; Peronnet et al. 1985). Yet, these findings do not exclude the role of plasma DβH as an indicator of long-term trends in—or of functional reserve of—the sympathetic nervous system. Our findings in the present study suggest that low plasma-DβH levels result from diminished expression of the DBH gene associated with the −1021T allele. Homozygosity at this allele, resulting in very low plasma-DβH activity, might likewise limit the maximum potential production of DβH enzyme within noradrenergic neurons. Normally, this may have no significant physiological consequences. However, in disease states in which degeneration of the sympathetic nervous system occurs, such as in idiopathic Parkinson Disease (IPD) (Wakabayashi and Takahashi 1997), this may be an important issue; for example, a growing body of evidence suggests that mild autonomic dysfunction in IPD is quite common but that severe symptoms are rare (Turkka et al. 1986; Magalhaes et al. 1995; Druschky et al. 2000). Individuals with IPD who are homozygous for the DBHL allele may have a decreased sympathetic functional reserve and a diminished capacity to compensate for the loss of adrenergic neurons. These patients might thus have an earlier onset of—and more severe symptoms of—autonomic dysfunction. Our results make this hypothesis directly testable.
Within in the field of psychiatry, there has been a great deal of interest in DBH as a disease-modifying gene and in plasma-DβH activity as an intermediate trait. Many conflicting reports on the relationship between plasma/CSF DβH levels and psychiatric disorders have been published (Fujita et al. 1978; Meltzer et al. 1980; Mathew et al. 1981; Matuzas et al. 1982; Arató et al. 1983). Since much of the individual variation in plasma-DβH levels is under genetic control, and since researchers previously had been unable to account for the genetic component of the variance in plasma DβH, such conflicting results are not surprising. Identification of the putative functional polymorphism −1021C→T will allow future clinical studies to evaluate whether DβH-activity differences in disease groups represent differences in genotype frequencies at the DBH locus. In addition, controlling for the large proportion of plasma-DβH variance that is accounted for by the DBH genotype may unmask changes in DβH activity that result from illness, medical treatment, or the influences of other genetic loci. Our demonstration of the −1021C→T polymorphism as a major genetic marker for plasma-DβH activity will allow the role of the DBH gene in human disease to be evaluated in a new light.
Acknowledgments
We are grateful to Ann Marie Wantroba-Lacobelle, David M. Ocame, and Harold Landis for technical assistance. This research was supported by grants from the National Alliance for Research on Schizophrenia and Depression (to J.F.C. and R.T.M), the National Institute on Drug Abuse (Substance Abuse Fellowship 5T32-DA-07238 [to C.P.Z.] and Yale Physician Scientist Training Program grants K-12-DA-00167 and R01- DA12422 [both to J.F.C.]), National Institute on Alcohol Abuse and Alcoholism grant R01-AA11330 (to J.G.), National Institute of Mental Health grant MH 30929 (to Yale University) and grant K02-MH01387 (to J.G.), U.S. Department of Veterans Affairs (Psychiatric Research/Neurosciences Fellowship [to C.P.Z.], Presidential Early Career Award for Scientists and Engineers [to J.F.C.], Merit Review [to J.G.], and the Mental Illness Research Education and Clinical Center), the Connecticut Mental Health Center (support to R.T.M.), U.S. Public Health Service National Institute of General Medical Sciences grant GM 28356 (to R.C.E.), and National Center for Research Resources grant RR 03655 (to R.C.E.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Database of Single Nucleotide Polymorphisms (dbSNP), http://www.ncbi.nlm.nih.gov/SNP/ (for NCBI-assay ID numbers for 18 new SNPs detected in this study [accession numbers 2418871–2418888])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for DBH genomic sequences [accession numbers AC000404 and AC001227])
- HWSIM, http://info.med.yale.edu/genetics/kkidd/programs.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DβH [MIM 223360])
References
- Arató M, Bagdy G, Blümel F, Perényi A, Rihmer Z (1983) Reduced serum dopamine-beta-hydroxylase activity in paranoid schizophrenics. Pharmacopsychiatry 16:19–22 [DOI] [PubMed] [Google Scholar]
- Box GEP, Cox DR (1964) An analysis of transformations. J R Stat Soc B 26:211–252 [Google Scholar]
- Craig SP, Buckle VJ, Lamouroux A, Mallet J, Craig IW (1988) Localization of the human dopamine beta hydroxylase (DBH) gene to chromosome 9q34. Cytogenet Cell Genet 48:48–50 [DOI] [PubMed] [Google Scholar]
- Cubells JF, Kobayashi K, Nagatsu T, Kidd KK, Kidd JR, Calafell F, Kranzler HR, Ichinose H, Gelernter J (1997) Population genetics of a functional variant of the dopamine beta-hydroxylase gene (DBH). Am J Med Genet 74:374–379 [DOI] [PubMed] [Google Scholar]
- Cubells JF, Kranzler HR, McCance-Katz E, Anderson GM, Malison RT, Price LH, Gelernter J (2000) A haplotype at the DBH locus, associated with low plasma dopamine beta-hydroxylase activity, also associates with cocaine-induced paranoia. Mol Psychiatry 5:56–63 [DOI] [PubMed] [Google Scholar]
- Cubells JF, van Kammen DP, Kelley ME, Anderson GM, O'Connor DT, Price LH, Malison R, Rao PA, Kobayashi K, Nagatsu T, Gelernter J (1998) Dopamine beta-hydroxylase: two polymorphisms in linkage disequilibrium at the structural gene DBH associate with biochemical phenotypic variation. Hum Genet 102:533–540 [DOI] [PubMed] [Google Scholar]
- DeGroot MH (1989) Probability and statistics, 2d ed. Addison-Wesley, Reading, MA [Google Scholar]
- Druschky A, Hilz MJ, Platsch G, Radespiel-Troger M, Druschky K, Kuwert T, Neundorfer B (2000) Differentiation of Parkinson's disease and multiple system atrophy in early disease stages by means of I-123-MIBG-SPECT. J Neurol Sci 175:3–12 [DOI] [PubMed] [Google Scholar]
- Dunnette J, Weinshilboum R (1976) Human serum dopamine beta-hydroxylase: correlation of enzymatic activity with immunoreactive protein in genetically defined samples. Am J Hum Genet 28:155–166 [PMC free article] [PubMed] [Google Scholar]
- Elston RC, Namboodiri KK, Hames CG (1979) Segregation and linkage analyses of dopamine-beta-hydroxylase activity. Hum Hered 29:284–292 [DOI] [PubMed] [Google Scholar]
- Fahndrich E, Muller-Oerlinghausen B, Coper H (1982) Longitudinal assessment of MAO-, COMT-, and DBH- activity in patients with bipolar depression. Int Pharmacopsychiatry 17:8–17 [DOI] [PubMed] [Google Scholar]
- Fujita K, Ito T, Maruta K, Teradaira R, Beppu H, Nakagami Y, Kato Y, Nagatsu T, Kato T (1978) Serum dopamine-beta-hydroxylase in schizophrenic patients. J Neurochem 30:1569–1572 [DOI] [PubMed] [Google Scholar]
- Gelernter J, Gejman PV, Bisighini S, Kidd KK (1991) Sequence tagged site (STS) TaqI RFLP at dopamine beta-hydroxylase (DBH). Nucleic Acids Res 19:1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin LR, Gershon ES, Lake CR, Murphy DL, McGinniss M, Sparkes RS (1982) Segregation and linkage studies of plasma dopamine-beta-hydroxylase (DBH), erythrocyte catechol-O-methyltransferase (COMT), and platelet monoamine oxidase (MAO): possible linkage between the ABO locus and a gene controlling DBH activity. Am J Hum Genet 34:250–262 [PMC free article] [PubMed] [Google Scholar]
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A (1999) Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet 22:239–247 [DOI] [PubMed] [Google Scholar]
- Hoyle GW, Mercer EH, Palmiter RD, Brinster RL (1994) Cell-specific expression from the human dopamine beta-hydroxylase promoter in transgenic mice is controlled via a combination of positive and negative regulatory elements. J Neurosci 14:2455–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro H, Kim KT, Joh TH, Kim KS (1993) Neuron-specific expression of the human dopamine beta-hydroxylase gene requires both the cAMP-response element and a silencer region. J Biol Chem 268:17987–17994 [PubMed] [Google Scholar]
- Kemper CM, O'Connor DT, Westlund KN (1987) Immunocytochemical localization of dopamine-beta-hydroxylase in neurons of the human brain stem. Neuroscience 23:981–989 [DOI] [PubMed] [Google Scholar]
- Kim HS, Seo H, Yang C, Brunet JF, Kim KS (1998) Noradrenergic-specific transcription of the dopamine beta-hydroxylase gene requires synergy of multiple cis-acting elements including at least two Phox2a-binding sites. J Neurosci 18:8247–8260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kurosawa Y, Fujita K, Nagatsu T (1989) Human dopamine beta-hydroxylase gene: two mRNA types having different 3'-terminal regions are produced through alternative polyadenylation. Nucleic Acids Res 17:1089–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouroux A, Vigny A, Faucon Biguet N, Darmon MC, Franck R, Henry JP, Mallet J (1987) The primary structure of human dopamine-beta-hydroxylase: insights into the relationship between the soluble and the membrane-bound forms of the enzyme. EMBO J 6:3931–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman AN, Freedman LS, Goldstein M (1972) Serum dopamine-beta-hydroxylase activity in patients with Huntington's chorea and Parkinson's disease. Lancet 1:153–154 [DOI] [PubMed] [Google Scholar]
- Magalhaes M, Wenning GK, Daniel SE, Quinn NP (1995) Autonomic dysfunction in pathologically confirmed multiple system atrophy and idiopathic Parkinson's disease: a retrospective comparison. Acta Neurol Scand 91:98–102 [DOI] [PubMed] [Google Scholar]
- Mathew RJ, Ho BT, Davis C, Taylor D, Reck J (1981) Depression, antidepressants, and plasma DBH. Psychiatry Res 5:331–334 [DOI] [PubMed] [Google Scholar]
- Matuzas W, Meltzer HY, Uhlenhuth EH, Glass RM, Tong C (1982) Plasma dopamine-beta-hydroxylase in depressed patients. Biol Psychiatry 17:1415–1424 [PubMed] [Google Scholar]
- Meltzer HY, Nasr SJ, Tong C (1980) Serum dopamine-beta-hydroxylase activity in schizophrenia. Biol Psychiatry 15:781–788 [PubMed] [Google Scholar]
- Nagatsu T, Wakui Y, Kato T, Fujita K, Kondo T, Yokochi F, Narabayashi H (1982) Dopamine-beta-hydroxylase activity in cerebrospinal fluid of Parkinsonian patients. Biomed Res 3:95–98 [Google Scholar]
- Oka K, Kijikawa K, Ohuchi T, Yoshida H, Imaizumi R (1967) Distribution of dopamine-β-hydroxylase in subcellular fractions of adrenal medulla. Life Sci 6:461–465 [DOI] [PubMed] [Google Scholar]
- Oxenstierna G, Edman G, Iselius L, Oreland L, Ross SB, Sedvall G (1986) Concentrations of monoamine metabolites in the cerebrospinal fluid of twins and unrelated individuals—a genetic study. J Psychiatr Res 20:19–29 [DOI] [PubMed] [Google Scholar]
- Peronnet F, Cleroux J, Perrault H, Thibault G, Cousineau D, de Champlain J, Guilland JC, Klepping J (1985) Plasma norepinephrine, epinephrine, and dopamine beta-hydroxylase activity during exercise in man. Med Sci Sports Exerc 17:683–688 [DOI] [PubMed] [Google Scholar]
- Perry SE, Summar ML, Phillips JA III, Robertson D (1991) Linkage analysis of the human dopamine beta-hydroxylase gene. Genomics 10:493–495 [DOI] [PubMed] [Google Scholar]
- Risch NJ (2000) Searching for genetic determinants in the new millennium. Nature 405:847–856 [DOI] [PubMed] [Google Scholar]
- Ross SB, Wetterberg L, Myrhead M (1973) Genetic control of plasma dopamine-β-hydroxylase. Life Sci 12:529–532 [DOI] [PubMed] [Google Scholar]
- Turkka JT, Juujarvi KK, Lapinlampi TO, Myllyla VV (1986) Serum noradrenaline response to standing up in patients with Parkinson's disease. Eur Neurol 25:355–361 [DOI] [PubMed] [Google Scholar]
- Viveros OH, Arqueros L, Kirshner N (1968) Release of catecholamines and dopamine-β-hydroxylase from the adrenal medulla. Life Sci 7:609–618 [Google Scholar]
- Wakabayashi K, Takahashi H (1997) Neuropathology of autonomic nervous system in Parkinson's disease. Eur Neurol 38 Suppl 2:2–7 [DOI] [PubMed] [Google Scholar]
- Wei J, Ramchand CN, Hemmings GP (1997) Possible control of dopamine beta-hydroxylase via a codominant mechanism associated with the polymorphic (GT)n repeat at its gene locus in healthy individuals. Hum Genet 99:52–55 [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM (1978) Serum dopamine β-hydroxylase. Pharmacol Rev 30:133–166 [PubMed] [Google Scholar]
- ——— (1983) Biochemical genetics of catecholamines in humans. Mayo Clin Proc 58:319–330 [PubMed] [Google Scholar]
- Weinshilboum R, Axelrod J (1971a) Serum dopamine-beta-hydroxylase activity. Circ Res 28:307–315 [DOI] [PubMed] [Google Scholar]
- ——— (1971b) Reduced plasma dopamine-beta-hydroxylase activity in familial dysautonomia. N Engl J Med 285:938–942 [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM, Raymond FA, Elveback LR, Weidman WH (1973) Serum dopamine-beta-hydroxylase activity: sibling-sibling correlation. Science 181:943–945 [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM, Schorott HG, Raymond FA, Weidman WH, Elveback LR (1975) Inheritance of very low serum dopamine-beta-hydroxylase activity. Am J Hum Genet 27:573–585 [PMC free article] [PubMed] [Google Scholar]
- Wilson AF, Elston RC, Sellers TA, Bailey-Wilson JE, Gersting JM, Deen DK, Sorant AJ, Tran LD, Amos CI, Siervogel RM (1990) Stepwise oligogenic segregation and linkage analysis illustrated with dopamine-β-hydroxylase activity. Am J Med Genet 35:425–432 [DOI] [PubMed] [Google Scholar]
- Wilson AF, Elston RC, Siervogel RM, Tran LD (1988) Linkage of a gene regulating dopamine-β-hydroxylase activity and the ABO blood group locus. Am J Hum Genet 42:160–166 [PMC free article] [PubMed] [Google Scholar]
- Winer N, Carter C (1977) Effect of cold pressor stimulation on plasma norepinephrine, dopamine-β-hydroxylase, and renin activity. Life Sci 20:887–894 [DOI] [PubMed] [Google Scholar]
- Zabetian CP, Gelernter J, Cubells JF (2000) Functional variants at CYP2A6: new genotyping methods, population genetics, and relevance to studies of tobacco dependence. Am J Med Genet 96:638–645 [DOI] [PubMed] [Google Scholar]