Abstract
The origins and dispersal of farming and pastoral nomadism in southwestern Asia are complex, and there is controversy about whether they were associated with cultural transmission or demic diffusion. In addition, the spread of these technological innovations has been associated with the dispersal of Dravidian and Indo-Iranian languages in southwestern Asia. Here we present genetic evidence for the occurrence of two major population movements, supporting a model of demic diffusion of early farmers from southwestern Iran—and of pastoral nomads from western and central Asia—into India, associated with Dravidian and Indo-European–language dispersals, respectively.
Farming and animal domestication are recent phenomena in human history, occurring from 10,000 years before present (YBP) onward. Farming arose independently in several parts of the world, including in a region in the Middle East known as the “Fertile Crescent,” which extends from Israel through northern Syria to western Iran. From this region, agriculture expanded in both western and eastern directions. The expansion toward Europe is the most thoroughly studied (Ammerman and Cavalli-Sforza 1984) and began ∼9,000 YBP. The spread of the farming economy toward the east, into the area from Iran to India, started a little later, between the 6th and the 5th millennia b.c. The Neolithic revolution in the Iranian region and in the Indus valley reached its zenith by 6,000 YBP and involved strong urban civilizations such as the Sumerian, the Elamite, and the Harappan. Another major innovation, probably beginning later than agriculture, was the domestication of animals, which is thought to have led to dramatic population expansions in Eurasia. Pastoral nomadism developed in the grasslands of central Asia east of the Volga-Don region, as well as in southeastern Europe, opening up the possibility of rapid movements of large population groups (Zvelebil 1980). The spread of these new technologies has been associated with the dispersal of Dravidian and Indo-Iranian languages in southern Asia (Renfrew 1987; Cavalli-Sforza 1988). Specifically, Elamo-Dravidian languages (Ruhlen 1991), which may have originated in the Elam province (Zagros Mountains, southwestern Iran), are now confined to southeastern India and to some isolated groups in Pakistan and northern India. It is hypothesized that the proto–Elamo-Dravidian language, spoken by the Elamites in southwestern Iran, spread eastward with the movement of farmers from this region (Cavalli-Sforza et al. 1994; Renfrew 1996). A later episode, the arrival of pastoral nomads from the central Asian steppes to the Iranian plateau, ∼4,000 YBP, brought with it the Indo-Iranian branch of the Indo-European language family, which eventually replaced Dravidian languages in Iran and most of Pakistan and northern India, perhaps by an elite-dominance process (Renfrew 1987, 1996; Cavalli-Sforza 1988). The incursion of these “Aryan” peoples coincided with the decadence of important Neolithic cultures, such as the Harappan civilization, by ∼3,000–4,000 YBP.
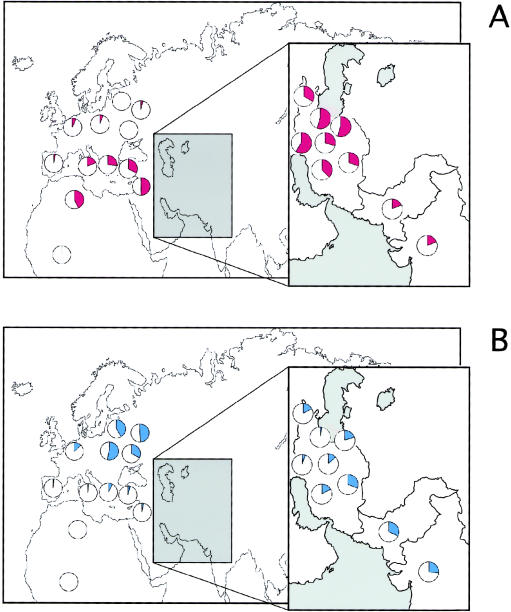
To date, there is little genetic evidence to support or contradict these linguistic and archeological observations, and the genetic impact of these events has not been evaluated. In the present study, a set of 459 Y chromosomes from several populations located in a key geographical position between the Fertile Crescent, central Asia, the Indus valley, and northern India (table 1) were analyzed, and the results were compared with data from neighboring Pakistani populations. Y-chromosome haplogroups (HGs) were defined by the analysis of 11 biallelic markers (SRY-1532, YAP, SRY-8299, sY81, 12f2, M9, 92R7, SRY-2627, LLY22g, Tat, and RPS4Y) (Bergen et al. 1999; Rosser et al. 2000, and references therein). Two Y-chromosome lineages, HG 9 and HG 3, show frequency clines that may reflect population movements in southwestern Asia (fig. 1A and B). The frequency distribution of these two HGs in the study populations is reported in table 1, together with relevant data from the literature. HG 9, defined by the 12f2 deletion, is largely confined to caucasoid populations, with its highest frequencies being found in Middle Eastern populations (fig. 1A). This HG has been proposed as an indicator of the demic diffusion of farming in Europe (Semino et al. 1996). In Iranian populations, HG 9 shows very high frequencies (∼30%–60%). Populations from the southeastern Caspian region and the Zagros Mountains exhibit the highest frequencies so far observed (∼60%) (fig. 1A). High frequencies of HG 9 have been found throughout the Fertile Crescent region (Hammer et al. 2000): Palestinians, 51%; Lebanese, 46%; and Syrians, 57%. The incidences of HG 9 in Pakistan (18%) and northern India (19%) indicate a decreasing-frequency cline from Iran toward India.
Table 1.
Frequency Distribution of HG 9 and HG 3 in Human Populations from Different Regions
|
Frequencyb(%) |
||||
| Regiona | N | HG 9 | HG 3 | Source |
| Iran:c | ||||
| Azarbaijan | 83 | 34 | 17 | Present study |
| Zagros Mountains | 34 | 59 | 6 | Present study |
| Western Caspian | 32 | 53 | 3 | Present study |
| Eastern Caspian | 25 | 56 | 20 | Present study |
| Tehran region | 50 | 30 | 14 | Present study |
| Central-north | 79 | 39 | 9 | Present study |
| Central-south | 72 | 38 | 17 | Present study |
| Eastern provinces | 26 | 35 | 31 | Present study |
| Pakistan | 708 | 18 | 32 | Present study |
| India: | ||||
| Gujurat | 58 | 19 | 26 | Present study |
| Jaunpur | 152 | NT | 20 | Zerjal et al. (1999) |
| Indians mixed | 72 | NT | 15 | Hammer et al. (1998) |
| Uttar Pradesh | 62 | 7 | NT | Semino et al. (1996) |
| Sri Lanka | 83 | NT | 15 | Hammer et al. (1998) |
| Middle East: | ||||
| Lebanon | 24 | 46 | 4 | Hammer et al. (2000) |
| Syria | 91 | 57 | 9 | Hammer et al. (2000) |
| Palestine | 73 | 51 | 0 | Hammer et al. (2000) |
| Europe: | ||||
| Turkey | 167 | 33 | 5 | Rosser et al. (2000) |
| Russia | 122 | 4 | 47 | Rosser et al. (2000) |
| Ukraine | 27 | 0 | 30 | Rosser et al. (2000) |
| Latvia | 34 | 0 | 41 | Rosser et al. (2000) |
| Poland | 112 | 4 | 54 | Rosser et al. (2000) |
| Greece | 36 | 28 | 8 | Rosser et al. (2000) |
| Italy | 99 | 20 | 2 | Rosser et al. (2000) |
| Spain | 126 | 3 | 2 | Rosser et al. (2000) |
| Africa: | ||||
| Algeria | 27 | 41 | 0 | Rosser et al. (2000) |
| Sub-Saharan Africa | 199 | 1 | 0 | Hammer et al. (2000) |
Data from the present study are underlined.
For HG 9 and HG 3, the allelic-state combinations of the 11 biallelic markers analyzed—SRY-1532, YAP, SRY-8299, sY81, 12f2, M9, 92R7, SRY-2627, LLY22g, Tat, and RPS4Y—are G, Alu-, G, A, 8 kb, C, C, C, C, T, and C; and A, Alu-, G, A, 10 kb, G, T, C, C, T, and C, respectively. NT = not tested.
Divided on the basis of geographical origin (ascertained until the grandfather’s generation): Zagros Mountains (Kordestan, Lorestan, Elam and Khuzestan), western Caspian (Gilan), eastern Caspian (Mazandaran), central-north (Zanjan, Markazi, Hamadan, and Semnan), central-south (Fars, Esfahan, and Hormozgan), and eastern provinces (Khorasan, Baluchestan, and Kerman).
Figure 1.
Frequency distribution of HG-9 (A) and HG-3 (B) Y chromosomes in southwestern Asia. The large map represents published data, and the inset represents new data. The comparative data are from Semino et al. (1996), Hammer et al. (1998), Zerjal et al. (1999), Hammer et al. (2000), and Rosser et al. (2000).
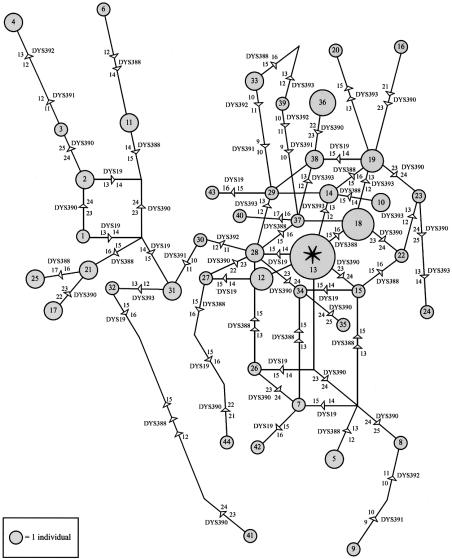
The most likely region of origin of a given HG can be recognized on the basis of two characteristics: it has the highest frequency and the highest diversity. Founder effects and drift in small populations can also lead to high HG frequencies, but this will usually affect neighboring populations differently and be accompanied by low diversity. Genetic diversity within HG 9 was therefore examined by the typing of HG-9 chromosomes from populations in Iran, Pakistan and India, at six microsatellite loci (DYS19, DYS388, DYS390, DYS391, DYS392, and DYS393). If the number of mutations has been low, the haplotype (Ht) that underwent expansion is likely to be the one with the most common allele for each short tandem repeat (STR) (in this case, Ht 13: 14-15-23-10-11-12). This Ht is present in our sample and is most frequent in the Iranian populations examined, as illustrated in the median-joining network (Bandelt et al. 1999) (fig. 2). Both the high incidence and the global haplotypic diversity of Iranian HG-9 chromosomes (D=.97), which are scattered throughout the median-joining network, suggest that this region was the geographical origin of HG 9. Consistently, high haplotypic-diversity values of HG-9 chromosomes are also observed in the Zagros Mountains (D=.97) and southeastern Caspian regions (D=.98), where HG 9 exhibits its highest frequencies. These STR diversity values argue against drift being responsible for the increased HG-9 frequencies in these regions. Altogether, the clinal variation and haplotypic diversity of this Y-chromosomal lineage support a model in which farming dispersal was accompanied by major population movements, probably originating in what was historically defined as Elam, towards the Indus valley, and this movement was associated with the dispersal of Dravidian languages (Renfrew 1996).
Figure 2.
Median-joining network (Bandelt et al. 1999) showing phylogenetic relationships of Iranian Y-chromosomes Hts within HG 9. The network contains 44 Y-microsatellite Hts represented in a sample of 80 Y chromosomes. Microsatellite Hts, defined by states at six microsatellite loci—DYS19, DYS388, DYS390, DYS391, DYS392 and DYS393—are represented by circles with area proportional to their frequencies in the sample. Branch lengths are proportional to the number of mutational steps and parallel links in a reticulation represent the same mutational changes. The putative ancestral Ht, Ht 13 (14-15-23-10-11-12), is labeled with an asterisk (*).
HG 3, defined by a back mutation at SRY-1532, is virtually absent from African, eastern Asian, and Native American populations and is found at its highest frequency in central Asia (Hammer et al. 1998; Karafet et al. 1999; Zerjal et al. 1999)—Russia, 50% and the Altai, 52%—with a decreasing-frequency cline westward into Europe (Zerjal et al. 1999; Rosser et al. 2000); this evidence suggests central Asia as the source region of this marker. The distribution of HG 3 in Iran shows marked differences between western and eastern provinces (southwestern Caspian [3%] vs. eastern provinces [31%]) (fig. 1B), with a decreasing-frequency cline towards India (Pakistan [32%], northern India [26%]). When the very low frequencies of HG 3 in the Middle East (Hammer et al. 2000) are taken into account, the frequency pattern of HG 3 in southwestern Asia (table 1) supports the idea that Indo-European speakers spread from Central Asia into modern Iran via an eastern-Caspian route, as well as into India. The relatively high frequency and haplotypic diversity (D=.90) of HG 3 in our Indian sample suggests that the number of individuals entering from the west was large. This view is supported by the presence of HG 3 throughout most of the Indian subcontinent (table 1), showing that this lineage spread over a vast area.
As a result of coalescence analysis, the mutations defining HG 9 and HG 3 have been dated to ∼14,800 and ∼7,500 YBP, respectively (Karafet et al. 1999; Hammer et al. 2000). We have used microsatellite-diversity analysis to estimate the most recent common ancestor of our set of Iranian, Pakistani, and Indian HG-9 and HG-3 chromosomes, through use of the mean variance of microsatellite repeats, averaged across the six loci (Slatkin 1995; Kittles et al. 1998). We have estimated a Y-chromosomal microsatellite mutation rate by pooling the data available in the literature (Heyer et al. 1997; Bianchi et al. 1998; Foster et al. 1998; Kayser et al. 2000), for the six microsatellite loci used in our study. Thus, 10 mutational events in a total of 5,431 meioses were observed, giving an average mutation-rate (μ) estimate of 1.8×10-3 (95% confidence interval [CI] 9.8×10-4–3.1×10-3). The coalescence times obtained (table 2) provide an upper-limit estimate for the time when the populations carrying these HGs started to expand in size in the areas considered here. However, coalescence-time estimates must be interpreted with caution. Several factors—such as the distinctive demographic histories of the populations sampled and the diverse mutation rates of microsatellite loci among different Y-chromosome backgrounds (or HGs)—may distort estimates of age. Consequently, the history of this single locus is not necessarily the history of the population, because of drift, selection, or male-specific behavior.
Table 2.
Estimated Ages for HG 9 and HG 3 in Southwestern Asia
|
Ageb (95% CIc) at Generation Time = |
||
| HG and Region (Mean Variancea) | 20 Years | 30 Years |
| HG 9: | ||
| Iran (.57) | 6,300 (3,700–11,600) | 9,500 (5,500–17,400) |
| Pakistan (.47) | 5,200 (3,000–9,600) | 7,800 (4,500–14,400) |
| India (.36) | 4,000 (2,300–7,300) | 6,000 (3,500–11,000) |
| HG 3: | ||
| Iran (.38) | 4,200 (2,500–7,800) | 6,300 (3,700–11,600) |
| Pakistan (.37) | 4,100 (2,400–7,600) | 6,200 (3,600–11,300) |
| India (.33) | 3,700 (2,100–6,700) | 5,500 (3,200–10,100) |
Of the microsatellite repeats, averaged across loci.
YBP. An average μ of 0.18% per locus per generation was assumed.
The 95% CI of the μ estimate was taken into account in the calculation of the 95% CI (9.8×10-4–3.1×10-3) for the coalescence estimates.
Despite the important stochastic component of Y-chromosome diversity, clinal variation within Europe has been observed (Semino et al. 1996; Zerjal et al. 1997; Malaspina et al. 1998; Casalotti et al. 1999; Quintana-Murci et al. 1999; Hill et al. 2000; Rosser et al. 2000; Semino et al. 2000), and the same trend seems to emerge from our data from Asia. The geographical distribution, observed clines, and estimated ages of HG-9 and HG-3 chromosomes in southwestern Asia all support a model of demic diffusion of early farmers from southwestern Iran—and nomads from western and central Asia—into India, bringing the spread of genes and culture (including language) to southwestern Asia. Although alternative, more complex explanations are possible, the analysis of the modern male-specific gene pools in these populations suggests that major demographic events, involving migration and admixture, accompanied these important historical and linguistic events.
Acknowledgments
We thank Evelyne Heyer, François Jacquesson, and Chris Ottolenghi for fruitful discussions, and we thank two anonymous reviewers for helpful remarks. We are grateful to Christiana Di Rocco for help with the Y-chromosome STR typing. We acknowledge support from Institut National de la Santé et de la Recherche Médicale and Fondation pour la Recherche Médicale (to L.Q.-M., C.K., and K.M.), Telethon Italy, grant 281/b (to C.K.), The Wellcome Trust (to T.Z., S.Q.M., Q.A., R.Q., and A.M.) and the Cancer Research Campaign (to C.T.-S.). M.A.J. is a Wellcome Trust Senior Fellow supported by grant 057559.
References
- Ammerman AJ, Cavalli-Sforza LL (1984) Neolithic transition and the genetics of populations in Europe. Princeton University Press, Princeton, NJ [Google Scholar]
- Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 [DOI] [PubMed] [Google Scholar]
- Bergen AW, Wang CY, Tsai J, Jefferson K, Dey C, Smith KD, Park SC, Tsai SJ, Goldman D (1999) An Asian-Native American paternal lineage identified by RPS4Y resequencing and by microsatellite haplotyping. Ann Hum Genet 63:63–80 [DOI] [PubMed] [Google Scholar]
- Bianchi NO, Catanesi CI, Bailliet G, Martinez-Marignac VL, Bravi CM, Vidal-Rioja LB, Herrera RJ, López-Camelo JS (1998) Characterization of ancestral and derived Y-chromosome haplotypes of New World native populations. Am J Hum Genet 63:1862–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalotti R, Simoni L, Belledi M, Barbujani G (1999) Y-chromosome polymorphisms and the origins of the European gene pool. Proc R Soc Lond B Biol Sci 266:1959–1965 [Google Scholar]
- Cavalli-Sforza LL (1988) The Basque population and ancient migrations in Europe. Munibe 6:129–137 [Google Scholar]
- Cavalli-Sforza LL, Piazza A, Menozzi P (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Foster EA, Jobling MA, Taylor PG, Donnelly P, de Knijff P, Mieremet R, Zerjal T, Tyler-Smith C (1998) Jefferson fathered slave's last child. Nature 396:27–28 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Karafet T, Rasanayagam A, Wood ET, Altheide TK, Jenkins T, Griffiths RC, Templeton AR, Zegura SL (1998) Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol Biol Evol 15:427–441 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Redd AJ, Wood ET, Bonner MR, Jarjanazi H, Karafet T, Santachiara-Benerecetti S, Oppenheim A, Jobling MA, Jenkins T, Ostrer H, Bonne-Tamir B (2000) Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc Natl Acad Sci USA 97:6769–6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer E, Puymirat J, Dieltjes P, Bakker E, de Knijff P (1997) Estimating Y chromosome specific microsatellite mutation frequencies using deep rooting pedigrees. Hum Mol Genet 6:799–803 [DOI] [PubMed] [Google Scholar]
- Hill EW, Jobling MA, Bradley DG (2000) Y-chromosome variation and Irish origins. Nature 404:351–352 [DOI] [PubMed] [Google Scholar]
- Karafet TM, Zegura SL, Posukh O, Osipova L, Bergen A, Long J, Goldman D, Klitz W, Harihara S, de Knijff P, Wiebe V, Griffiths RC, Templeton AR, Hammer MF (1999) Ancestral Asian source(s) of New World Y-chromosome founder haplotypes. Am J Hum Genet 64:817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Roewer L, Hedman M, Henke L, Henke J, Brauer S, Kruger C, Krawczak M, Nagy M, Dobosz T, Szibor R, de Knijff P, Stoneking M, Sajantila A (2000) Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet 66:1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittles RA, Perola M, Peltonen L, Bergen AW, Aragon RA, Virkkunen M, Linnoila M, Goldman D, Long JC (1998) Dual origins of Finns revealed by Y chromosome haplotype variation. Am J Hum Genet 62:1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspina P, Cruciani F, Ciminelli BM, Terrenato L, Santolamazza P, Alonso A, Banyko J, Brdicka R, García O, Gaudiano C, Guanti G, Kidd KK, Lavinha J, Avila M, Mandich P, Moral P, Qamar R, Mehdi SQ, Ragusa A, Stefanescu G, Caraghin M, Tyler-Smith C, Scozzari R, Novelletto A (1998) Network analyses of Y-chromosomal types in Europe, northern Africa, and western Asia reveal specific patterns of geographic distribution. Am J Hum Genet 63:847–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Minch E, Passarino G, Brega A, Santachiara-Benerecetti AS (1999) Further characteristics of proto-European Y chromosomes. Eur J Hum Genet 7:603–608 [DOI] [PubMed] [Google Scholar]
- Renfrew C (1987) Archaeology and language: the puzzle of Indo-European origins. Jonathan Cape, London [Google Scholar]
- ——— (1996) Languages families and the spread of farming. In: Harris DR (ed) The origins and spread of agriculture and pastoralism in Eurasia. Smithsonian Institution Press, Washington, DC, pp 70–92 [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W et al. (2000) Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 67:1526–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlen M (1991) A guide to the world’s languages. Edward Arnold, London [Google Scholar]
- Semino O, Passarino G, Brega A, Fellous M, Santachiara-Benerecetti AS (1996) A view of the Neolithic demic diffusion in Europe through two Y chromosome–specific markers. Am J Hum Genet 59:964–968 [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290:1155–1159 [DOI] [PubMed] [Google Scholar]
- Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, Dashnyam B, Pandya A, Kayser M, Roewer L, Santos FR, Schiefenhovel W, Fretwell N, Jobling MA, Harihara S, Shimizu K, Semjidmaa D, Sajantila A, Salo P, Crawford MH, Ginter EK, Evgrafov OV, Tyler-Smith C (1997) Genetic relationships of Asians and northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet 60:1174–1183 [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, Pandya A, Santos FR, Adhikari R, Tarazona E, Kayser M, Evgrafov O, Singh L, Thangaraj K, Destro-Bisol G, Thomas MG, Qamar R, Mehdi SQ, Rosser ZH, Hurles ME, Jobling MA Tyler-Smith C (1999) The use of Y-chromosomal DNA variation to investigate population history: recent male spread in Asia and Europe. In: Papiha SS, Deka R, Chakraborty R (eds) Genomic diversity: applications in human populations genetics. Plenum Press, New York, pp 91–102 [Google Scholar]
- Zvelebil M (1980) The rise of the nomads in Central Asia. In: Sherratt A (ed) The Cambridge encyclopedia of archaeology, Crown, New York, pp 252–256 [Google Scholar]