Heart disease is a major cause of morbidity and premature mortality in the United States. Cardiovascular disease affects >59 million Americans, ranks first among all diagnostic categories necessitating hospitalizations, and causes 1 in every 2.4 deaths (American Heart Association Web site). Although heart disease in developed countries often arises through complex interactions between genetics, lifestyle choices, and environment, a considerable fraction of cardiovascular disease—including cardiomyopathy, arrhythmias, and congenital heart malformations—are due to heritable, monogenic traits. Application of human molecular genetic strategies to elucidate the molecular etiologies of some of these conditions has provided mechanistic insights into the cellular events and signals that affect heart structure and function (Milewicz and Seidman 2000).
Cardiomyopathy represents a diverse group of heart-muscle disorders, which are further subdivided on the basis of their anatomic and hemodynamic findings. More than 80% of cardiomyopathies are classified as dilated or congestive. These disorders increase both myocardial mass and volume, such that, despite moderate myocyte enlargement, or hypertrophy, the heart appears thin walled and distended. Diminished contractile function is the critical hemodynamic feature of dilated cardiomyopathy, an abnormality that triggers complex neurohumoral responses, which increase circulatory volume so as to maintain cardiac output. Although such events are initially compensatory, these responses ultimately become maladaptive and contribute to clinical deterioration and onset of heart failure. Only 50% of patients with dilated cardiomyopathy survive >5 years after diagnosis (Grogan et al. 1995); premature death occurs from unmitigated pump failure and from comorbidities such as thromboembolic events and arrhythmias. Despite current strategies to aggressively manage dilated cardiomyopathy, the disorder remains a common cause of heart failure and a prevalent diagnosis in individuals referred for cardiac transplantation.
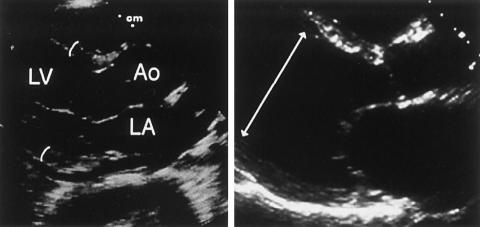
Diagnosis of dilated cardiomyopathy is often made through noninvasive cardiac imaging, particularly through two-dimensional echocardiography (fig. 1). Measurements obtained throughout the cardiac cycle provide systolic and diastolic dimensions that both index the amount of chamber enlargement and provide an estimate of contractile function, through calculations of the fractional shortening of heart muscle and the fraction of blood ejected with each beat. Echocardiographic findings of dilated cardiomyopathy usually prompt a search for underlying causes of cardiac enlargement and diminished contractile function. Coronary artery disease, viral myocarditis, thyroid disease, immunologic processes, toxins (such as alcohol and the antineoplastic agent danorubycin), and infiltrative processes (hemochromatosis, sarcoidosis, or storage diseases) are well-known causes of dilated cardiomyopathy. In many instances, however, an underlying pathology is not discovered, and the diagnosis of “idiopathic” dilated cardiomyopathy ensues.
Figure 1.
Echocardiographic view of the normal and a dilated heart. Left panel, Parasternal long-axis view of a normal heart in end-diastole, illustrating normal left-ventricular (LV) wall thickness (1.2 cm; curved lines) and cavity dimensions. A normal-sized left atrium (LA) and aortic root (Ao) are shown on this view. Right panel, Dilated cardiomyopathy causing marked increases in the LV diastolic diameter (7.7 cm, arrowed line) and LA enlargement.
The histopathology of idiopathic dilated cardiomyopathy is usually nonspecific: degenerating myocytes that exhibit mild-to-moderate hypertrophy without disarray (i.e., without misalignment of contiguous myocytes), and an absence of inflammatory cells. Increased interstitial fibrosis is often evident, but amounts can vary quite considerably. Unique microscopic findings can help to refine the more general diagnosis of dilated cardiomyopathy into a pathological subtype; for example, myocyte loss and marked fibrofatty replacement with preferential right-ventricular involvement indicates arrhythmogenic right-ventricular dysplasia (Thiene et al. 1988).
Over the past two decades, there has been increased recognition that many “idiopathic” dilated cardiomyopathies are familial. Although medical reports of familial cardiomegaly were published as early as 1948 (Evans 1948), a clinical estimate of prevalence came only in 1992, when Michels and colleagues reported that >20% of individuals with dilated cardiomyopathy had an affected first-degree relative (Michels et al. 1992). Several large population-based studies have confirmed these data and have delineated that dilated cardiomyopathy can be transmitted as autosomal (dominant and recessive), X-linked, or mitochondrial traits (Keeling et al. 1995; Grünig et al. 1998; Mestroni et al. 1999). Not only have these observations fostered molecular genetic analyses aimed at the definition of disease-causing mutations, but these studies have also altered clinical practice. Screening evaluations for clinically silent dilated cardiomyopathy in family members at risk for inheriting dilated cardiomyopathy has fostered early interventions that both reduce symptoms and decrease the morbidity and mortality associated with arrhythmias or thromboembolic events. Although early initiation of treatments may also retard progression and prolong the pretransplantation phase of the disease, more-definitive therapies for dilated cardiomyopathy await better mechanistic understandings of the molecular basis for this disease.
Clinical and Genetic Heterogeneity
There is considerable heterogeneity in the clinical features of dilated cardiomyopathy resulting from a single gene mutation. Age at disease onset ranges from early childhood to late in senescence, although most cases of genetic or acquired dilated cardiomyopathy become apparent during the fourth or fifth decade of life. The natural history of disease can vary considerably with regard to severity of symptoms and survival, even in affected members of the same family. Family studies of dilated cardiomyopathy sometimes demonstrate additional phenotypes that cosegregate with the disease (fig. 2); these can be subtle and clinically unimportant or can profoundly disturb another organ system. Additional cardiac phenotypes (i.e., conduction-system disease and mitral valve prolapse) and extracardiac findings (i.e., skeletal muscle abnormalities and sensorineural hearing loss) have been described. Recognition of adjunct findings in dilated cardiomyopathy was particularly important for early genetic-mapping studies, since middle-age onset of disease often constrained pedigree sizes and limited the number of families that were informative for linkage analyses. Since adjunct phenotypes are sometimes expressed in advance of dilated cardiomyopathy, these have been used as surrogate markers of cardiac status, which have fostered the definition of multiple genetic loci for familial dilated cardiomyopathy.
Figure 2.
Diversity of phenotypes accompanying heritable dilated cardiomyopathy. A, Electrocardiographic tracings, revealing conduction-system abnormalities in patients with dilated cardiomyopathy. Top, First-degree atrioventricular (AV) block with prolonged interval (horizonal bar = .4 s) between atrial P wave (↓) and subsequent ventricular QRS wave. Middle, Second-degree AV block; note that a QRS complex does not follow the third or sixth P wave. Bottom, Presence of atrial fibrillation, with absence of P waves and irregular rhythm of ventricular QRS complexes. B, Plantar (and palmar) keratosis, which accompanies dilated cardiomyopathy in Naxos syndrome. C, Audiogram showing normal hearing (triangles) in comparison to sensorineural hearing loss in patients with dilated cardiomyopathy due to a mutation at chromosome 6q23-24. White circles (○) and crosses (×) denote sound transmitted through air into the right and left ear, respectively. Note that hearing loss is symmetric and affects air and bony (black circles [●]) transmission of sound.
The most common mode of inheritance of familial dilated cardiomyopathy is autosomal dominant (table 1). Incomplete penetrance is sometimes evident and presumably reflects the influence of modifying genes and environment. Dominant loci that cause isolated dilated cardiomyopathy have been found on chromosomes 1q32 (MIM 601494, Durand et al. 1995; MIM 191045, Kamisago et al. 2000), 2q31 (MIM 604145, Siu et al. 1999), 5q33-34 (MIM 601411, Tsubata et al. 2000), 6q12-16 (MIM 605582, Sylvius et al. 2001), 9q13-22 (MIM 600884, Krajinovic et al. 1995), 14q11 (MIM 160760, Kamisago et al. 2000), 15q14 (MIM 102540, Olson et al. 1998), and 15q22 (MIM 191010, Olson et al. 2001). Dominant loci that cause dilated cardiomyopathy in association with an additional phenotype have been mapped to chromosome 10q21-23 (mitral valve prolapse [MIM 601494, Bowles et al. 1996]), chromosomes 1p1-q21, 2q14-22, and 3p22-25 (conduction-system disease [MIM 150330, Fatkin et al. 1999; MIM 604288, Jung et al. 1999; MIM 601154, Olson et al. 1996]), chromosomes 2q35 and 6q23 (muscular dystrophies [MIM 126560, Li et al. 1999; MIM 602067, Messina et al. 1997]), and chromosome 6q23-24 (sensorineural hearing loss [MIM 605362, Schönberger et al. 2000]). Dominant loci for arrhythmogenic right-ventricular dysplasia reside on chromosomes 1q42 (MIM 600996, Rampazzo et al. 1995), 2q32.1-32.2 (MIM 602087, Rampazzo et al. 1997), 3p23 (MIM 604400, Ahmad et al. 1998), 10p12-14 (MIM 604401, Li et al. 2000), 14q12-22 (MIM 602086, Severini et al. 1996), and 14q23-24 (MIM 107970, Rampazzo et al. 1994) Little is known about recessive mutations that cause dilated cardiomyopathy, with the exception of the mutation that causes Naxos syndrome (arrhythmogenic right-ventricular dysplasia, wooly hair, and keratinosis), which maps to chromosome 17q21 (MIM 601214, Coonar et al. 1998), and a clinically related disorder encoded on chromosome 6p24 (MIM 605676, Norgett et al. 2000). Chromosome Xq28 mutations (Barth syndrome [MIM 302060, Bolhuis et al. 1991]) cause dilated cardiomyopathy in the context of neutropenia and short stature. Specific dystrophin defects (Xp21) can also lead to dilated cardiomyopathy with more modest skeletal muscle pathology than typically occurs in Duchenne muscular dystrophy (MIM 302045, Towbin et al. 1993). In addition to disease loci in which cardiac phenotype predominates, many multisystem diseases and neuromuscular disorders exhibit dilated cardiomyopathy as one component of the phenotype (Kaplan and Fontaine 2001).
Table 1.
Loci with Dilated Cardiomyopathy as the Predominant Phenotype
| Locus | Trait | Additional Phenotype | Disease Gene |
| 1q32 | AD | None | Cardiac troponin T |
| 1q32 | AD | None | ? |
| 2q31 | AD | None | ? |
| 5q33-34 | AD | None | δ-sarcoglycan |
| 6q12-16 | AD | None | ? |
| 9q13-22 | AD | None | ? |
| 14q11 | AD | None | Cardiac β myosin heavy chain |
| 15q14 | AD | None | Cardiac actin |
| 15q22.1 | AD | None | α-tropomyosin |
| 10q21-23 | AD | Mitral valve prolapse | ? |
| 1p1-q21 | AD | Conduction disease | Lamin A/C |
| 2q14-q22 | AD | Conduction disease | ? |
| 3q22-25 | AD | Conduction disease | ? |
| 6q23 | AD | Conduction disease and skeletal myopathy | ? |
| 2q35 | AD | Skeletal myopathy | Desmin |
| 6q23-24 | AD | Sensorineural hearing loss | ? |
| 1q42-43 | AD | ARVD and exercise-induced ventricular tachycardia | Cardiac ryanodine receptor |
| 2q32.1-32.3 | AD | ARVD | ? |
| 3p23 | AD | ARVD | ? |
| 10p12-14 | AD | ARVD | ? |
| 14q12-22 | AD | ARVD | ? |
| 14q23-24 | AD | ARVD | ? |
| 6p24 | AR | Wooly hair and keratoderma | Desmoplakin |
| 17q21 | AR | ARVD, wooly hair, and keratoderma | Plakoglobin |
| Xp21 | X | Skeletal myopathy | Dystrophin |
| Xq28 | X | Short stature and neutropenia | Tafazzin |
Mechanisms of Human Dilated Cardiomyopathy
Definition of the genetic causes for some cardiac conditions has provided mechanistic insights into heretofore poorly understood disease processes. For example, the discovery of the genetic basis for long-QT syndrome showed that this enigmatic sudden-death syndrome is a heritable disorder of cardiac ion channels (Keating and Sanguinetti 2001); elucidation of the molecular basis for “idiopathic” hypertrophic cardiomyopathy has also redefined this pathology as a disorder of the sarcomere (Seidman and Seidman 2001). These paradigms initially heightened expectations that genetic studies would also provide a unified pathogenetic mechanism for “idiopathic” dilated cardiomyopathy. Instead, these studies indicate a great diversity of genetic causes for dilated cardiomyopathy. Although the number of disease genes identified to date remains limited, and many more will undoubtedly be defined, the current repertoire of mutations that cause dilated cardiomyopathy suggest a multiplicity of pathways (fig. 3) by which the heart can fail.
Figure 3.
Schematic of the multiple mechanisms leading to dilated cardiomyopathy. Insufficiencies of force production and force transmission, deficits of myocyte energy, and apoptosis or necrosis can each result in ventricular dilation and deficits in myocardial contraction. Note that single gene mutations (inside circle) and some acquired causes (outside circle) of dilated cardiomyopathy perturb the same pathways. The functional consequences of some mutations remain unknown.
Deficits in Force Generation
Cardiac function is dependent on force production by the sarcomere, the fundamental unit of contraction in all muscle cells. Although many human mutations in the protein constituents of the sarcomere have been identified, only recently have these deficits been recognized as being associated with dilated cardiomyopathy. Missense mutations in β cardiac myosin heavy chain and an in-frame deletion of cardiac troponin T (Kamisago et al. 2000) cause autosomal dominant dilated cardiomyopathy. Many previously reported defects in each of these genes cause hypertrophic cardiomyopathy (a primary myocardial disorder characterized by increased heart mass and preserved contractile function). Recently, however, specific defects have been defined that produce early-onset heart failure that occurs without associated findings. Dilated cardiomyopathy due to a deletion in cardiac troponin T is thought to disrupt calcium-sensitive troponin C interactions that are critical for actin-myosin ATPase activity. A decrease in this enzymatic activity could reduce the energy needed to power sarcomere contraction. The location of β cardiac myosin missense mutations that cause dilated cardiomyopathy suggests that these may also reduce contractile force. Structural data (Rayment et al. 1993) indicate that one mutation leads to a substitution of an amino acid involved in tight binding of actin by myosin, an interaction that is essential for initiation of the power stroke of contraction. Another dilated-cardiomyopathy defect alters a residue in one of several flexible joints of myosin that undergo conformational changes during the contractile cycle and that transmit movement and directionality throughout the thick filament.
Deficits in Force Transmission
Physiologic heart function requires that contractile force generated by the sarcomere be effectively transmitted to the extracellular matrix. Although little is known about the precise biophysical mechanism by which this occurs, multiple filamentous proteins found in muscle cells that link the contractile apparatus to the sarcolemma are thought to function in the propagation of force to the extracellular matrix. Cardiac actin is centrally located within the sarcomere, where it participates in force production and is linked to cytoskeletal components that transmit force. Two mutations in cardiac actin cause early onset of autosomal dominant dilated cardiomyopathy without additional clinical phenotypes (Olson et al. 1998). Since the location of actin missense mutations alters amino acid residues involved in actin-cytoskeletal—rather than actin-myosin—interactions, the consequence of actin mutations that cause dilated cardiomyopathy is thought to be diminution of force transmission from the sarcomere to the cytoskeleton.
Two missense mutations in α-tropomyosin were recently reported (Olson et al 2001) that also cause dilated cardiomyopathy. Each is located on the surface of the molecule, in positions that may be involved in electrostatic interactions between α-tropomyosin and actin or other thin-filament proteins. Diminished interactions could disrupt the structural and/or functional integrity of the thin filament and could impede force production by the sarcomere or force transmission to the myocyte cytoskeleton.
Intermediate-filament proteins, such as desmin, join actin to the dystrophin-sarcoglycan complex, located just beneath the plasma membrane of all muscle cells. Components of the sarcoglycan complex also extend outside of myocytes into the extracellular matrix. Mutations in several of these molecules are widely recognized as causes of skeletal muscular dystrophies—degenerative muscle disorders that often exhibit some myocardial involvement. Surprisingly, mutations in dystrophin, desmin, and δ-sarcoglycan have also been identified in patients with dilated cardiomyopathy, in whom there is either subclinical or no skeletal muscle involvement (Muntoni et al. 1993; Li et al. 1999; Tsubata et al. 2000). The probable mechanism by which these defects cause the heart to fail is by ineffectual transmission of force. However, why allelic mutations cause predominant clinical disease in skeletal—versus cardiac—muscle remains an unanswered question. The location of a dystrophin mutation may influence the pattern of muscle involvement; defects in promoter elements, 5′ gene deletions, and mid-rod mutations may appear to more selectively affect heart muscle (Muntoni et al. 1993, 1997; Milasin et al. 1996).
Given the many proteins that participate in force transmission, deficits in this pathway might be expected to be a prevalent cause of dilated cardiomyopathy. So far, only three genes coding for these proteins have been analyzed, in small families and in sporadic cases. Analyses of actin and desmin sequences (Tesson et al. 2000), in 41 probands with familial disease and 22 sporadic cases of dilated cardiomyopathy, identified no mutations. A study of adult-onset dilated cardiomyopathy (Arbustini et al 2000) identified only 13 dystrophin mutations in 201 samples; in four individuals, familial transmission of disease was evident. Another study, of 27 patients with dilated cardiomyopathy, reported no dystrophin mutations (Michels et al. 1993).
Mutations in plakoglobin (also called “γ-catenin”) have been demonstrated to cause arrhythmogenic right-ventricular cardiomyopathy with palmoplantar keratoderma (fig. 2B) and wooly hair (Naxos syndrome [McKoy et al. 2000]). Mutations in desmoplakin lead to a very similar phenotype (Norgett et al. 2000), although with more-pronounced left-ventricular involvement. These rare recessive syndromes share many histopathological features with autosomal dominant arrhythmogenic right-ventricular dysplasia, including myocyte death and prominent right-ventricular fibrofatty replacement. Both desmoplakin and plakoglobin have critical functions in desmosomes and adherens junctions, where intermediate filaments are anchored to the plasma membrane to create tight junctions with contiguous cells. Since these junctions are presumed to be important for overall organ function of the heart and skin, a likely consequence of defective cell-cell junctions in the heart would be impaired force propagation.
Understanding of the mechanism by which plakoglobin defects cause disease has also been fostered by analyses of plakoglobin-null mice (Bierkamp et al. 1996; Ruiz et al. 1996); myocardium from these mutants has decreased compliance, because of reduced cell-cell adhesions. When the heart is subjected to mechanical stress, the animals die of ventricular rupture. Whether plakoglobin mutations are sufficient to promote myocyte death or whether this occurs as a function of constant mechanical stress is unknown.
Deficits in Energy Production
Recessive mutations in genes that encode transport proteins or enzymes involved in cardiac fatty acid β-oxidation can cause dilated cardiomyopathy (Roe and Ding 2001). Mitochondrial β-oxidation provides an important source of energy during periods of fasting; defects in this pathway damage the myocardium indirectly, through an inadequate supply of energy, or directly, owing to toxic effects of intermediary metabolites. Intracellular accumulation of intermediary metabolites (such as long-chain acylcarnitines) may be arrhythmogenic; these could account for the increased incidence of sudden death associated with these disorders, relative to other cardiac disorders.
Carnitine is required for entry of long-chain fatty acids into mitochondria, and carnitine deficiencies prevent metabolism of long-chain fatty acids. Mutations in proteins that participate in carnitine transport and metabolism are heritable causes of dilated cardiomyopathy that are transmitted as recessive traits. Dilated cardiomyopathy is also a feature of mutations in the organic cation transporter protein, which transports carnitine into cells; in translocase, which shuttles carnitine and acylcarnitine into the mitochondria; and in carnitine palmitoyltransferase II (Roe and Ding 2001), which catalyzes carnitine derivatives into acyl-CoA. Cardiomyopathy can be the presenting manifestation of these carnitine-deficiency states, but skeletal muscle disease and metabolic defects (hypoketotic hypoglycemia) are usually evident. Cardiac function is normal in heterozygous carriers.
Many of the reported human mutations in proteins involved in β-oxidation are associated with hypertrophic—rather than dilated—cardiomyopathy. Notable exceptions are defects in the mitochondrial trifunctional protein that affect long-chain l-3-hydroxylacyl-CoA protein (Brackett et al. 1995). These defects cause skeletal myopathy, dilated cardiomyopathy, and hepatic steatosis.
Mutations in the mitochondrial genome result in multisystem pathologies with prominent neurologic, metabolic, and cardiac phenotypes. The energy requirements of these tissues are presumably more affected by deficits in oxidative phosphorylation that result from mitochondrial mutations than are other organs. The cardiomyopathy that accompanies most mitochondrial mutations can be either hypertrophic or dilated, and conduction-system abnormalities are seen frequently in association with these pathologies. In addition to the pleiotropic manifestations, cardiomyopathy due to mitochondrial mutations (unlike that due to mutations of mitochondrial proteins encoded within the nuclear genome) are distinguished by matrilineal inheritance. However, family studies indicate that heart involvement in affected members varies considerably, presumably because of mitochondrial heteroplasmy. Further, since mitochondrial mutations spontaneously accumulate with physiologic aging (Melov et al. 1995), demonstration of the causal role of these defects in late-onset dilated cardiomyopathy is problematic (Ruppert and Maisch 2000) and should require biochemical evidence of altered oxidative phosphorylation in heart tissues.
Mutations with Unknown Mechanisms
Tafazzin (also called “G4.5”) is a protein of unknown function, encoded at Xq28, that is mutated in Barth syndrome, which is a triad of dilated cardiomyopathy, neutropenia, and increased 3-methylglutaconicaciduria (Bione et al. 1996). Several different transcripts that differ at their amino terminus and within a central domain are produced from the tafazzin gene. Tafazzins belong to a gene family of acyltransferases involved in phospholipid biosynthesis; other structurally related proteins have unknown functions (Neuwald 1997). Onset of Barth syndrome occurs in infancy, and arrhythmias and heart failure frequently accompany this neonatal dilated cardiomyopathy. Cardiac pathology can vary considerably in affected males, and ventricular noncompaction, as well as cardiac fibroelastosis, have been described (D'Adamo et al. 1997). Both heart morphology and function in female carriers of tafazzin mutations is normal.
Dominant mutations in the nuclear envelope proteins lamin A and C cause progressive conduction-system disease and dilated cardiomyopathy (Fatkin et al. 1999). Longitudinal evaluations of affected kindreds demonstrate progressive prolongation of the normal delay in electrical activation of the atrial and ventricular chambers (fig. 2A); after many years of conduction-system disease, the myocardium dilates, contractile function decreases, and heart failure ensues. Although important functions have been ascribed to lamin proteins, neither the location of these molecules nor their known functions provide insights into the mechanism by which conduction-system disease and cardiomyopathy occurs in cases of lamin mutations. Lamin proteins A, C, and B2 are intermediate filament components of the nuclear envelope, a multimeric structure that is associated with the nucleoplasmic surface of the inner nuclear membrane. These ubiquitously expressed proteins are thought to contribute structural integrity to the nuclear envelope and provide mechanical support for the nucleus (Wilson et al. 2001). Lamins A and C undergo phosphorylation that fosters depolymerization and disassembly of the nucleus during mitosis; dephosphorylation of lamins contributes to the reassembly of the nuclear membrane after cell division. Although their functions in nondividing cells such as cardiac myocytes are largely unknown, lamins A and C contain a chromatin-binding site and may participate in ordering of the genome. They are also associated with nuclear pores, suggesting a potential role in nuclear-cytoplasmic trafficking. In muscle cells, lamins may also function to reduce mechanical stresses imposed on the nucleus by cyclical contractions; if mutations interfere with this lamin function, the structural integrity of the nucleus could be compromised, and premature myocyte death could ultimately result.
Distinct mutations in lamin A/C can also lead to autosomal dominant Emery-Dreifuss muscular dystrophy (Bonne et al. 1999), to the related condition limb-girdle muscular dystrophy (Muchir et al. 2000), and to familial partial lipodystrophy (Dunnigan type [Cao and Hegele 2000; Shackleton et al. 2000; Speckman et al. 2000]), a disorder of adipocyte degeneration and profound insulin-resistant diabetes. Despite marked differences in the predominant clinical phenotypes of these allelic disorders, cardiac disease is an important component of each disorder. Mice that have been engineered to carry lamin A/C mutations also show features of each human condition: mutant animals develop muscular dystrophy and dilated cardiomyopathy and are devoid of white fat (Sullivan et al. 1999).
Arrhythmogenic right-ventricular dysplasia (ARVD) is a distinct morphological type of dilated cardiomyopathy that is transmitted as an autosomal dominant trait and is expressed with incomplete penetrance (Thiene et al. 2000). As in Naxos syndrome, the right ventricle is particularly prone to progressive myocardial atrophy in ARVD. Left-ventricular involvement, although more modest, is usually also present. Marked electrical instability is an important feature of ARVD and accounts for the high incidence of sudden death associated with this disorder. Although ARVD is infrequently diagnosed in the United States, estimates from Italy indicate an incidence of 6/10,000 individuals.
Six disease loci for ARVD have been mapped, but only one disease gene has been identified. The cardiac ryanodine receptor is encoded at 1q42; recently, mutations were reported that cause exercise-induced ventricular tachycardia and sudden death in individuals with features of ARVD (Tiso et al. 2001). However, cardiomyopathy has not been reported with other ryanodine-receptor mutations that are associated with exercise-induced ventricular tachycardia (Laitinen et al. 2001; Priori et al. 2001). Whether these data imply that only specific ryanodine-receptor–gene mutations cause ARVD or implicate functionally related molecules in this cardiomyopathy remains an open question. The cardiac ryanodine receptor is the main calcium-release channel in the myocardial sarcoplasmatic reticulum and fosters electrical excitation with mechanical contraction. Mechanisms by which mutations in this receptor might produce the fibrofatty degeneration that typifies ARVD may therefore include calcium-induced myocyte apoptosis and/or necrosis.
Genetics of Acquired Dilated Cardiomyopathy
De novo mutations, incomplete penetrance, and small family size can limit recognition of the contribution that heritable gene mutations may have in the overall incidence of dilated cardiomyopathy. Nevertheless, a substantial proportion of these clinical disorders is likely acquired. Recent evidence suggests that similar mechanisms by which single gene mutations cause dilated cardiomyopathy may function in nonheritable forms of this disease. Genetic defects in the dystrophin-glycoprotein complex appear to cause hereditary dilated cardiomyopathy, through impairment of force transmission. A comparable mechanism may account for the ventricular dilation and dysfunction that can ensue after viral myocarditis, in that enteroviruses encode a protease that can cleave the dystrophin-glycoprotein complex in model systems (Badorff et al. 1999). Since nitric oxide has been demonstrated to inhibit a coxsackieviral protease through S-nitrosylation (Badorff et al. 2000), elucidation of a definitive role for proteases in the pathogenicity of dilated cardiomyopathy may foster the development of novel therapeutics to prevent the cardiac dysfunction associated with some viral infections.
Chronic ischemia that occurs from coronary-artery atherosclerosis may be the most common cause of dilated cardiomyopathy. Despite this straightforward inciting event, it is unknown whether apoptosis, insufficient energy, or reduced contractile forces are the primary triggers for ventricular dilation.
Animal Models of Dilated Cardiomyopathy
Many of the morphological and hemodynamic features of dilated cardiomyopathy are represented in animal models of this human disease. Characterization of genetic models that arose from spontaneous mutations—or that were produced through the introduction of germline mutations by means of transgenic or recombinatorial strategies—have helped elucidate the consequences of mutations and provide insights into the downstream signals triggered by inciting events. Mouse gene mutations that alter the muscle LIM protein (Arber et al. 1997), desmin (Milner et al. 1996), and δ-sarcoglycan (Nigro et al. 1997) confirm that cytoskeletal dysfunction is one cause of dilated cardiomyopathy. Deficits in myocyte energy and metabolism have been modeled by ablation of a mitochondrial transcription factor (Tfam) in mice (Wang et al. 1999). Animals deficient in lamin A/C (Sullivan et al. 1999) or in the bradykinin receptor B2 (Emanueli et al. 1999) also develop dilated cardiomyopathy, although the signaling pathways triggered by these events remain unclear.
In addition to confirming the multiplicity of pathways by which the heart can fail, studies of animal models for dilated cardiomyopathy have sometimes uncovered unexpected findings. For example, although both human and murine δ-sarcoglycan mutations cause dilated cardiomyopathy, the responses to these gene defects appear to be quite different. Mice lacking δ-sarcoglycan develop coronary-artery spasm, which results in myocardial ischemia, necrosis, and, eventually, dilated cardiomyopathy (Cohn et al. 2001). Administration of the calcium-channel antagonist verapamil reduces coronary-artery ischemia and improves cardiac function. An important role for δ-sarcoglycan in the biology of human coronary arteries has not been recognized. Human mutations in this component of the dystrophin-associated protein complex have been reported (both missense mutations in exon 6 and a 3-bp deletion in exon 9) to cause dilated cardiomyopathy, but studies of the coronary arteries from affected individuals have not indicated ischemia (Tsubata et al. 2000). Rather, human mutations are thought to alter heart function through dominant negative influences on the dystrophin-sarcoglycan complex, analogous to the mechanism by which these cause skeletal muscular dystrophy. Whether these distinct consequences of δ-sarcoglycan mutations reflect different mutations or unique functions of δ-sarcoglycan in human and murine hearts is unknown.
Important insights have also come from strategies aimed at complementation of dilated cardiomyopathy deficits by genetic manipulation of models of disease. Ablation of the muscle LIM protein (MLP [Arber et al. 1997]) in mice causes neonatal dilated cardiomyopathy. However, if these mutant mice are bred with mice deficient in the phospholamban gene (Minamisawa et al. 1999), normal cardiac function is preserved. Phospholamban functions as a physiologic inhibitor of SERCA2a, an ATPase-dependent pump that restores cytosolic calcium to the sarcoplasmic reticulum; ablation of phospholamban would therefore enhance calcium reuptake by the sarcoplasmic reticulum. Functional improvement, by abrogation of phospholamban function, of hearts that are genetically predisposed to dilated cardiomyopathy implicates calcium as a fundamental mediator of some of the pathophysiological components of heart failure. These data raise the possibility that pharmacological inhibition of phospholamban may enhance heart function in dilated cardiomyopathy.
A third insight about the pathogenetic mechanisms of dilated cardiomyopathy comes from a recently described model of ARVD, produced by ablation of the α-actinin–associated LIM domain protein (ALP [Pashmforoush et al. 2001]). This protein colocalizes with α-actinin and plakoglobin at the intercalated disc and seems to play a role in the mechanical stability of the cell-cell contacts in long myocardial fibers. ALP is almost exclusively expressed in the right ventricle, which is concordant with the phenotype of a right-ventricular dysfunction and dilatation, whereas the left-ventricular function is only mildly depressed. Although features of human ARVD are produced by this model, neither fibrofatty infiltrates nor spontaneous arrhythmias have been observed in it. Human ALP maps to 4q35, a genomic location where ARVD loci have not been described. Nevertheless, the intriguing similarities between components of these phenotypes suggest that proteins that are functionally related to ALP may be important in human ARVD.
Conclusions
Dilated cardiomyopathy is often an insidiously progressive disorder that relentlessly progresses toward heart failure. Associated with high morbidity and mortality, this disease produces substantial burdens on patients, families, and health care economic systems. Better understanding of the inciting events that cause dilated cardiomyopathy, as well as elucidation of myocyte responses triggered by these events, have great potential to improve quality of life, lengthen survival of affected individuals, and reduce the need for cardiac transplantation. Although heritable mutations are less common than other causes of dilated cardiomyopathy, insights gleaned from molecular genetic studies provide some immediate and many potential benefits for improving this condition. For families with monogenic causes of dilated cardiomyopathy, gene-based diagnosis has improved presymptomatic recognition of the disease and has enabled early interventions. Further, these discoveries provide new paradigms for considering the many mechanisms by which the heart can fail. Ultimately, these understandings should provide new opportunities for the targeting of therapeutic interventions.
Acknowledgments
This work was supported by grants from the Howard Hughes Medical Institutes and the National Institutes of Health. J.S. has been supported by fellowships from the German Research Council (DFG) (Scho 673/2-1) and from the Bugher Foundation.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- American Heart Association, http://americanheart.org/ (for cardiovascular disease statistics)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for dilated cardiomyopathy [MIM 601494, MIM 191045, MIM 604145, MIM 601411, MIM 605582, MIM 600884, MIM 160760, MIM 102540, MIM 191010], mitral valve prolapse and dilated cardiomyopathy [MIM 601494], conduction-system disease and dilated cardiomyopathy [MIM 150330, MIM 604288, MIM 601154], muscular dystrophies and dilated cardiomyopathy [MIM 126560, MIM 602067], sensorineural hearing loss and dilated cardiomyopathy [MIM 605362], ARVD [MIM 600996, MIM 602087, MIM 604400, MIM 604401, MIM 602086, MIM 107970], Naxos syndrome [MIM 601214], dilated cardiomyopathy with woolly hair and keratoderma [MIM 605676], Barth syndrome [MIM 302060], and X-linked dilated cardiomyopathy [MIM 302045])
References
- Ahmad F, Li D, Karibe A, Gonzalez O, Tapscott T, Hill R, Weilbaecher D, Blackie P, Furey M, Gardner M, Bachinski LL, Roberts R (1998) Localization of a gene responsible for arrhythmogenic right ventricular dysplasia to chromosome 3p23. Circulation 98:2791–2795 [DOI] [PubMed] [Google Scholar]
- Arber S, Hunter JJ, Ross J Jr, Hongo M, Sansig G, Borg J, Perriard JC, Chien KR, Caroni P (1997) MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell 88:393–403 [DOI] [PubMed] [Google Scholar]
- Arbustini E, Diegoli M, Morbini P, Dal Bello B, Banchieri N, Pilotto A, Magani F, Grasso M, Narula J, Gavazzi, Vigano M, Tavazzi L (2000) Prevalence and characteristics of dystrophin defects in adult male patients with dilated cardiomyopathy. J Am Coll Cardiol 35:1760–1768 [DOI] [PubMed] [Google Scholar]
- Badorff C, Fichtlscherer B, Rhoads RE, Zeiher AM, Muelsch A, Dimmeler S, Knowlton KU (2000). Nitric oxide inhibits dystrophin proteolysis by coxsackieviral protease 2A through S-nitrosylation: a protective mechanism against enteroviral cardiomyopathy. Circulation 102:2276–2281 [DOI] [PubMed] [Google Scholar]
- Badorff C, Lee GH, Lamphear BJ, Martone ME, Campbell KP, Rhoads RE, Knowlton KU (1999). Enteroviral protease 2A cleaves dystrophin: evidence of cytoskeletal disruption in an acquired cardiomyopathy. Nat Med 5:320–326 [DOI] [PubMed] [Google Scholar]
- Bierkamp C, Mclaughlin KJ, Schwarz H, Huber O, Kemler R (1996) Embryonic heart and skin defects in mice lacking plakoglobin. Dev Biol 180:780–785 [DOI] [PubMed] [Google Scholar]
- Bione S, D'Adamo P, Maestrini E, Gedeon AK, Bolhuis PA, Toniolo D (1996) A novel X-linked gene, G4.5, is responsible for Barth syndrome. Nat Genet 12:385–389 [DOI] [PubMed] [Google Scholar]
- Bolhuis PA, Hensels GW, Hulsebos TJ, Baas F, Barth PG (1991) Mapping of the locus for X-linked cardioskeletal myopathy with neutropenia and abnormal mitochondria (Barth syndrome) to Xq28. Am J Hum Genet 48:481–485 [PMC free article] [PubMed] [Google Scholar]
- Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, Duboc D, Fardeau M, Toniolo D, Schwartz K (1999) Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet 21:285–288 [DOI] [PubMed] [Google Scholar]
- Bowles KR, Gajarski R, Porter P, Goytia V, Bachinski LL, Roberts R, Pignatelli R, Towbin JA (1996) Gene mapping an autosomal dominant familial dilated cardiomyopathy to chromosome 10q21-23. J Clin Invest 98:1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackett JC, Sims HF, Rinaldo P, Shapiro S, Powell CK, Bennett MJ, Strauss AW (1995) Two alpha subunit donor splice site mutations cause human trifunctional protein deficiency. J Clin Invest 95:2076–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Hegele RA (2000) Nuclear lamin A/C R482Q mutation in Canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum Mol Genet 9:109–112 [DOI] [PubMed] [Google Scholar]
- Cohn RD, Durbeej M, Moore SA, Coral-Vazquez R, Prouty S, Campbell KP (2001) Prevention of cardiomyopathy in mouse models lacking the smooth muscle sarcoglycan-sarcospan complex. J Clin Invest 107:R1–R7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coonar AS, Protonotarios N, Tsatsopoulou A, Needham EW, Houlston RS, Cliff S, Otter MI, Murday VA, Mattu RK, McKenna WJ (1998) Gene for arrhythmogenic right ventricular cardiomyopathy with diffuse nonepidermolytic palmoplantar keratoderma and woolly hair (Naxos disease) maps to 17q21. Circulation 97:2049–2058 [DOI] [PubMed] [Google Scholar]
- D'Adamo P, Fassone L, Gedeon A, Janssen EAM, Bione S, Bolhuis PA, Barth PG, Wilson M, Haan E, Örstavik KH, Patton MA, Green AJ, Zammarchi E, Donati MA, Toniolo D (1997) The X-linked gene G4.5 is responsible for different infantile dilated cardiomyopathies. Am J Hum Genet 61:862–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand JB, Bachinski LL, Bieling LC, Czernuszewicz GZ, Abchee AB, Yu QT, Tapscott T, Hill R, Ifegwu J, Marian AJ, Brugada R, Daiger S, Gregoritch JM, Anderson JL, Quiñones M, Towbin JA, Roberts R (1995) Localization of a gene responsible for familial dilated cardiomyopathy to chromosome 1q32. Circulation 92:3384–3380 [DOI] [PubMed] [Google Scholar]
- Emanueli C, Maestri R, Corradi D, Marchione R, Minasi A, Tozzi MG, Salis MB, Straino S, Capogrossi MC, Olivetti G, Madeddu P (1999) Dilated and failing cardiomyopathy in bradykinin B(2) receptor knockout mice. Circulation 100:2359–2365 [DOI] [PubMed] [Google Scholar]
- Evans W (1948) Familial cardiomegaly. Br Heart J 26:68–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatkin D, MacRae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M, Atherton J, Vidaillet HJ Jr, Spudich S, De Girolami U, Seidman JG, Seidman C (1999) Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med 341:1715–1724 [DOI] [PubMed] [Google Scholar]
- Grogan M, Redfield MM, Bailey KR, Reeder GS, Gersh BJ, Edwards WD, Rodeheffer RJ (1995) Long-term outcome of patients with biopsy-proved myocarditis: comparison with idiopathic dilated cardiomyopathy. J Am Coll Cardiol 26:80–84 [DOI] [PubMed] [Google Scholar]
- Grünig E, Tasman JA, Kücherer H, Franz W, Kübler W, Katus HA (1998) Frequency and phenotypes of familial dilated cardiomyopathy. J Am Coll Cardiol 31:186–194 [DOI] [PubMed] [Google Scholar]
- Jung M, Poepping I, Perrot A, Ellmer AE, Wienker TF, Dietz R, Reis A, Osterziel KJ (1999) Investigation of a family with autosomal dominant dilated cardiomyopathy defines a novel locus on chromosome 2q14-q22. Am J Hum Genet 65:1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisago M, Sharma SD, DePalma SR, Solomon S, Sharma P, McDonough B, Smoot L, Mullen MP, Woolf PK, Wigle ED, Seidman JG, Seidman CE (2000) Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med 343:1688–1696 [DOI] [PubMed] [Google Scholar]
- Kaplan JC and Fontaine B (2001) Neuromuscular disorders: gene location. Neuromuscul Disord 11:104–120 [PubMed] [Google Scholar]
- Keating MT, Sanguinetti MC (2001) Molecular and cellular mechanisms of cardiac arrhythmias. Cell 104:569–580 [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Gang Y, Smith G, Seo H, Bent SE, Murday V, Caforio AL, McKenna WJ (1995) Familial dilated cardiomyopathy in the United Kingdom. Br Heart J 73:417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajinovic M, Pinamonti B, Sinagra G, Vatta M, Severini GM, Milasin J, Falaschi A, Camerini F, Giacca M, Mestroni L, Heart Muscle Disease Study Group (1995) Linkage of familial dilated cardiomyopathy to chromosome 9. Am J Hum Genet 57:846–852 [PMC free article] [PubMed] [Google Scholar]
- Laitinen PJ, Brown KM, Piippo K, Swan H, Devaney JM, Brahmbhatt B, Donarum EA, Marino M, Tiso N, Viitasalo M, Toivonen L, Stephan DA, Kontula K (2001) Mutations of the cardiac ryanodine receptor (RyR2) gene in familial polymorphic ventricular tachycardia. Circulation 103:485–490 [DOI] [PubMed] [Google Scholar]
- Li D, Ahmad F, Gardner MJ, Weilbaecher D, Hill R, Karibe A, Gonzalez O, Tapscott T, Sharratt GP, Bachinski LL, Roberts R (2000) The locus of a novel gene responsible for arrhythmogenic right-ventricular dysplasia characterized by early onset and high penetrance maps to chromosome 10p12-p14. Am J Hum Genet 66:148–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Tapscoft T, Gonzalez O, Burch PE, Quinones MA, Zoghbi WA, Hill R, Bachinski LL, Mann DL, Roberts R (1999) Desmin mutation responsible for idiopathic dilated cardiomyopathy. Circulation 100:461–464 [DOI] [PubMed] [Google Scholar]
- McKoy G, Protonotarios N, Crosby A, Tsatsopoulou A, Anastasakis A, Coonar A, Norman, M, Baboonian C, Jeffery S, McKenna WJ (2000) Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease). Lancet 355:2119–2124 [DOI] [PubMed] [Google Scholar]
- Melov S, Shoffner JM, Kaufman A, Wallace DC (1995) Marked increase in the number and variety of mitochondrial DNA rearrangements in aging human skeletal muscle. Nucleic Acids Res 23:4122–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina DN, Speer MC, Pericak-Vance MA, McNally EM (1997) Linkage of familial dilated cardiomyopathy with conduction defect and muscular dystrophy to chromosome 6q23. Am J Hum Genet 61:909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestroni L, Rocco C, Gregori D, Sinagra G, Di Lenarda A, Miocic S, Vatta M, Pinamonti B, Muntoni F, Caforio AL, McKenna WJ, Falaschi A, Giacca M, Camerini F (1999) Familial dilated cardiomyopathy: evidence for genetic and phenotypic heterogeneity: Heart Muscle Disease Study Group. J Am Coll Cardiol 34:181–190 [DOI] [PubMed] [Google Scholar]
- Michels VV, Moll PP, Miller FA, Tajik AJ, Chu JS, Driscoll DJ, Burnett JC, Rodeheffer RJ, Chesebro JH, Tazelaar HD (1992) The frequency of familial dilated cardiomyopathy in a series of patients with idiopathic dilated cardiomyopathy. N Engl J Med 326:77–82 [DOI] [PubMed] [Google Scholar]
- Michels VV, Pastores GM, Moll PP, Driscoll DJ, Miller FA, Burnett JC, Rodeheffer RJ, Tajik JA, Beggs, AH, Kunkel LM, Thibodeau SN (1993) Dystrophin analysis in idiopathic dilated cardiomyopathy. J Med Genet 30:955–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milasin J, Muntoni F, Severini GM, Bartoloni L, Vatta M, Krajinovic M, Mateddu A, Angelini C, Camerini F, Falaschi A, Mestroni L, Giacca M (1996). A point mutation in the 5′ splice site of the dystrophin gene first intron responsible for X-linked dilated cardiomyopathy. Hum Mol Genet 5:73–79 [DOI] [PubMed] [Google Scholar]
- Milewicz DM, Seidman CE (2000) Genetics of cardiovascular disease. Circulation 102 Suppl 4:IV103–IV111 [DOI] [PubMed] [Google Scholar]
- Milner DJ, Weitzer G, Tran D, Bradley A, Capetanaki Y (1996) Disruption of muscle architecture and myocardial degeneration in mice lacking desmin. J Cell Biol 134:1255–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamisawa S, Hoshijima M, Chu G, Ward CA, Frank K, Gu Y, Martone ME, Wang Y, Ross J Jr, Kranias EG, Giles WR, Chien KR (1999) Chronic phospholamban-sarcoplasmic reticulum calcium ATPase interaction is the critical calcium cycling defect in dilated cardiomyopathy. Cell 99:313–322 [DOI] [PubMed] [Google Scholar]
- Muchir A, Bonne G, van der Kooi AJ, van Meegen M, Baas F, Bolhuis PA, de Visser M, Schwartz K (2000) Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B). Hum Mol Genet 9:1453–1459 [DOI] [PubMed] [Google Scholar]
- Muntoni F, Cau M, Ganau A, Congiu R, Arvedi G, Mateddu A, Marrosu MG, Cianchetti C, Realdi G, Cao A, Melis MA (1993) Brief report: deletion of the dystrophin muscle-promoter region associated with X-linked dilated cardiomyopathy. N Engl J Med 329:921–925 [DOI] [PubMed] [Google Scholar]
- Muntoni F, Di Lenarda A, Porcu M, Sinagra G, Mateddu A, Marrosu G, Ferlini A, Cau M, Milasin J, Melis MA, Marrosu MG, Cianchetti C, Sanna A, Falaschi A, Camerini F, Giacca M, Mestroni L (1997) Dystrophin gene abnormalities in two patients with idiopathic dilated cardiomyopathy. Heart 78:608–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF (1997) Barth syndrome may be due to an acyltransferase deficiency. Curr Biol 7:R465–R466 [DOI] [PubMed] [Google Scholar]
- Nigro V, Okazaki Y, Belsito A, Piluso G, Matsuda Y, Politano L, Nigro G, Ventura C, Abbondanza, Molinari AM, Acampora D, Nishimura M, Hayashizaki Y, Puca GA (1997) Identification of the Syrian hamster cardiomyopathy gene. Hum Mol Genet 6:601–607 [DOI] [PubMed] [Google Scholar]
- Norgett EE, Hatsell SJ, Carvajal-Huerta L, Cabezas JC, Common J, Purkis PE, Whittock N, Leigh IM, Stevens HP, Kelsell DP (2000) Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum Mol Genet 9:2761–2766 [DOI] [PubMed] [Google Scholar]
- Olson TM, Keating MT (1996) Mapping a cardiomyopathy locus to chromosome 3p22-25. J Clin Invest 97:528–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson TM, Kishimoto NY, Whitby FG, Michels VV (2001) Mutations that alter the surface charge of alpha-tropomyosin are associated with dilated cardiomyopathy. J Mol Cell Cardiol 33:723–732 [DOI] [PubMed] [Google Scholar]
- Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT (1998) Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science 280:750–752 [DOI] [PubMed] [Google Scholar]
- Pashmforoush M, Pomies P, Peterson KL, Kubalak S, Ross J Jr, Hefti A, Aebi U, Beckerle MC, Chien KR (2001) Adult mice deficient in actinin-associated LIM-domain protein reveal a developmental pathway for right ventricular cardiomyopathy. Nat Med 7:591–597 [DOI] [PubMed] [Google Scholar]
- Priori SG, Napolitano C, Tiso N, Memmi M, Vignati G, Bloise R, Sorrentino VV, Danieli GA (2001) Mutations in the cardiac ryanodine receptor gene (hryr2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation 103:196–200 [DOI] [PubMed] [Google Scholar]
- Rampazzo A, Nava A, Danieli GA, Buja G, Daliento L, Fasoli G, Scognamiglio R, Corrado D, Thiene G (1994) The gene for arrhythmogenic right ventricular cardiomyopathy maps to chromosome 14q23-q24. Hum Mol Genet 3:959–962 [DOI] [PubMed] [Google Scholar]
- Rampazzo A, Nava A, Erne P, Eberhard M, Vian E, Slomp P, Tiso N, Thiene G, Danieli GA (1995) A new locus for arrhythmogenic right ventricular cardiomyopathy (ARVD2) maps to chromosome 1q42-q43. Hum Mol Genet 4:2151–2154 [DOI] [PubMed] [Google Scholar]
- Rampazzo A, Nava A, Miorin M, Fonderico P, Pope B, Tiso N, Livolsi B, Zimbello R, Thiene G, Danieli GA (1997) ARVD4, a new locus for arrhythmogenic right ventricular cardiomyopathy, maps to chromosome 2 long arm. Genomics 45:259–263 [DOI] [PubMed] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM (1993) Three-dimensional structure of myosin subfragment-1; a molecular motor. Science 261:50–58 [DOI] [PubMed] [Google Scholar]
- Roe CR, Ding J (2001). Mitochondrial fatty acid oxidation disorders. In: Scriver C, Beaudet A, Sly WS, Valle D (eds) The metabolic and molecular basis of inherited disease. Vol II. McGraw Hill, Maidenhead, United Kingdom, pp 2297–2326 [Google Scholar]
- Ruiz P, Brinkmann V, Ledermann B, Behrend M, Grund C, Thalhammer C, Vogel F, Birchmeier C, Gunthert U, Franke WW, Birchmeier W (1996) Targeted mutation of plakoglobin in mice reveals essential functions of desmosomes in the embryonic heart. J Cell Biol 35:215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert V, Maisch B (2000) Mitochondrial DNA deletions in cardiomyopathies. Herz 25:161–167 [DOI] [PubMed] [Google Scholar]
- Schönberger J, Levy H, Grünig E, Sangwatanaroj S, Fatkin D, MacRae C, Stacker H, Halpin C, Eavey R, Philbin EF, Katus H, Seidman JG, Seidman CE (2000) Dilated cardiomyopathy and sensorineural hearing loss: a heritable syndrome that maps to 6q23-24. Circulation 101:1812–1818 [DOI] [PubMed] [Google Scholar]
- Seidman JG, Seidman C (2001) The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell 104:557–567 [DOI] [PubMed] [Google Scholar]
- Severini GM, Krajinovic M, Pinamonti B, Sinagra G, Fioretti P, Brunazzi MC, Falaschi A, Camerini F, Giacca M, Mestroni L (1996) A new locus for arrhythmogenic right ventricular dysplasia on the long arm of chromosome 14. Genomics 31:193–200 [DOI] [PubMed] [Google Scholar]
- Shackleton S, Lloyd DJ, Jackson SN, Evans R, Niermeijer MF, Singh BM, Schmidt H, Brabant G, Kumar S, Durrington PN, Gregory S, O'Rahilly S, Trembath RC (2000) LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet 24:153–156 [DOI] [PubMed] [Google Scholar]
- Siu BL, Niimura H, Osborne JA, Fatkin D, MacRae C, Soloman S, Benson W, Seidman JG, Seidman CE (1999) Familial dilated cardiomyopathy locus maps to chromosome 2q31. Circulation 99:1022–1026 [DOI] [PubMed] [Google Scholar]
- Speckman RA, Garg A, Du F, Bennett L, Veile R, Arioglu E, Taylor SI, Lovett M, Bowcock AM (2000) Mutational and haplotype analyses of families with familial partial lipodystrophy (Dunnigan variety) reveal recurrent missense mutations in the globular C-terminal domain of lamin A/C. Am J Hum Genet 66:1192–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B (1999) Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol 147:913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvius N, Tesson F, Gayet C, Charron P, Bénaïche A, Mangin L, Peuchmaurd M, Duboscq-Bidot L, Feingold J, Beckmann JS, Bouchier C, Komajda M (2001) A new locus for autosomal dominant dilated cardiomyopathy identified on chromosome 6q12-q16. Am J Hum Genet 68:241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson F, Sylvius N, Pilotto A, Dubosq-Bidot L, Peuchmaurd M, Bouchier C, Benaiche A, Mangin L, Charron P, Gavazzi A, Tavazzi L, Arbustini E, Komajda M (2000) Epidemiology of desmin and cardiac actin gene mutations in a European population of dilated cardiomyopathy. Eur Heart J 21:1872–1876 [DOI] [PubMed] [Google Scholar]
- Thiene G, Basso C, Calabrese F, Angelini A, Valente M (2000) Pathology and pathogenesis of arrhythmogenic right ventricular cardiomyopathy. Herz 25:210–215 [DOI] [PubMed] [Google Scholar]
- Thiene G, Nava A, Corrado D, Rossi L, Pennelli N (1988) Right ventricular cardiomyopathy and sudden death in young people. N Engl J Med 318:129–133 [DOI] [PubMed] [Google Scholar]
- Tiso N, Stephan DA, Nava A, Bagattin A, Devaney JM, Stanchi F, Larderet G, Brahmbhatt B, Brown K, Bauce B, Muriago M, Basso C, Thiene G, Danieli GA, Rampazzo A (2001) Identification of mutations in the cardiac ryanodine receptor gene in families affected with arrhythmogenic right ventricular cardiomyopathy type 2 (ARVD2). Hum Mol Genet 10:189–194 [DOI] [PubMed] [Google Scholar]
- Towbin JA, Hejtmancik JF, Brink P, Gelb B, Zhu XM, Chamberlain JS, McCabe ER, Swift M (1993) X-linked dilated cardiomyopathy: molecular genetic evidence of linkage to the Duchenne muscular dystrophy (dystrophin) gene at the Xp21 locus. Circulation 87:1854–1865 [DOI] [PubMed] [Google Scholar]
- Tsubata S, Bowles KR, Vatta M, Zintz C, Titus J, Muhonen L, Bowles NE, Towbin JA (2000) Mutations in the human delta-sarcoglycan gene in familial and sporadic dilated cardiomyopathy. J Clin Invest 106:655–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Bruning JC, Kahn CR, Clayton DA, Barsh GS, Thoren P, Larsson NG (1999) Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet 21:133–137 [DOI] [PubMed] [Google Scholar]
- Wilson KL, Zastrow MS, Lee KK (2001) Lamins and disease: insights into nuclear infrastructure. Cell 104:647–650 [PubMed] [Google Scholar]