Abstract
The terminal 22q13.3 deletion syndrome is characterized by severe expressive-language delay, mild mental retardation, hypotonia, joint laxity, dolichocephaly, and minor facial dysmorphisms. We identified a child with all the features of 22q13.3 deletion syndrome. The patient's karyotype showed a de novo balanced translocation between chromosomes 12 and 22, with the breakpoint in the 22q13.3 critical region of the 22q distal deletion syndrome [46, XY, t(12;22)(q24.1;q13.3)]. FISH investigations revealed that the translocation was reciprocal, with the chromosome 22 breakpoint within the 22q subtelomeric cosmid 106G1220 and the chromosome 12q breakpoint near STS D12S317. Using Southern blot analysis and inverse PCR, we located the chromosome 12 breakpoint in an intron of the FLJ10659 gene and located the chromosome 22 breakpoint within exon 21 of the human homologue of the ProSAP2 gene. Short homologous sequences (5-bp, CTG[C/A]C) were found at the breakpoint on both derivative chromosomes. The translocation does not lead to the loss of any portion of DNA. Northern blot analysis of human tissues, using the rat ProSAP2 cDNA, showed that full-length transcripts were found only in the cerebral cortex and the cerebellum. The FLJ10659 gene is expressed in various tissues and does not show tissue-specific isoforms. The finding that ProSAP2 is included in the critical region of the 22q deletion syndrome and that our proband displays all signs and symptoms of the syndrome suggests that ProSAP2 haploinsufficiency is the cause of the 22q13.3 deletion syndrome. ProSAP2 is a good candidate for this syndrome, because it is preferentially expressed in the cerebral cortex and the cerebellum and encodes a scaffold protein involved in the postsynaptic density of excitatory synapses.
Introduction
Recent approaches to the analysis of chromosome terminal regions led to the detection of several patients who had chromosome 22q distal deletions (Nesslinger et al. 1994; Flint et al. 1995; Precht et al. 1998; Prasad et al. 2000). The terminal 22q13.3 deletion syndrome is characterized by severe expressive-language delay and mild mental retardation; minor dysmorphisms have been reported in some subjects (Wong et al. 1997; Precht et al. 1998; Prasad et al. 2000). On the basis of clinical and molecular studies of patients with either macroscopic or submicroscopic cytogenetic rearrangements, Wong and colleagues (1997) narrowed (to 70 kb) the shortest region of deletion overlap (SRO) in which the gene(s) involved in 22q13.3 deletion syndrome should be localized.
We describe a child who has a de novo balanced translocation between chromosomes 12 and 22, with the breakpoint in the 22q13.3 critical region. The child has all the features of terminal deletion 22q syndrome. Molecular investigations have demonstrated that the translocation disrupts the human homologue of the proline-rich synapse-associated protein 2 (ProSAP2) (Boeckers et al. 1999a) gene on chromosome 22 and the FLJ10659 gene on chromosome 12. Two facts make ProSAP2 a good candidate for the 22q13.3 deletion syndrome: (1) it is expressed preferentially in the cerebral cortex and the cerebellum, and (2) it encodes a scaffold protein involved in the postsynaptic density (PSD) of excitatory synapses. Recent breakthroughs have, in fact, shown that mutations of genes regulating synaptic vesicle transport result in mental retardation (Chelly 2000; Toniolo and D’Adamo 2000).
Subject and Methods
Subject
The proband is a 4.5-year-old boy. Family history is negative for mental retardation and other neurological or psychiatric disorders. Pregnancy, delivery, and neonatal life were unremarkable. Early motor milestones were slightly delayed—the infant sat at age 7 mo and walked at age 19 mo. However, language development was severely compromised, in that the boy was unable to utter single words until he was 2 years old. At age 4.5 years (last evaluation), his verbal expression was limited to a few words; he had mild mental retardation (overall IQ 54) and sharply limited verbal abilities (verbal IQ 32; performance IQ 70). Neurological examination revealed mild hypotonia and minor dysmorphic features (dolichocephaly, epicanthic folds, and saddle nose with bulbous tip) are evident. Results of audiometric testing, needle electromyography, and cranial magnetic resonance imaging are normal.
Cytogenetic Investigations
Chromosome analysis was performed on the proband's and parents’ blood, using standard high-resolution techniques (Dutrillaux 1981). Whole-chromosome painting (with a chromosome 22–specific library [Cytocell]) and FISH (with 22q subtelomere–specific cosmid 106G1220 [Vysis]), were performed on the proband's metaphase spreads. Other FISH experiments were performed with YACs that were selected according to their position on the Genome Database. YAC DNA was labeled with biotin, by nick translation. The labeled probes were visualized with fluorescein isothiocyanate–avidin (Vector), and the chromosomes were counterstained with 4′,6-diamino-2-phenyl-indole (Sigma). Hybridizations were analyzed with a Zeiss Axioplan epifluorescence microscope, and images were captured with the Power Gene FISH System (PSI).
DNA Analysis
For Southern blot analysis, 20-μg aliquots of genomic DNA from the proband and his parents were digested using EcoRI, EcoRV, and HindIII and were separated by size on a 0.7% agarose/0.5× TBE gel. The DNA was then transferred to a nylon membrane (Boehringer Mannheim).
Two micrograms of DNA from clone n85a3 (Genome Database accession number AC000036) was cut with EcoRV, isolated on a 0.4% agarose/1× TAE gel, and purified with Qiaex II (Qiagen). Approximately 200 ng of each fragment was labeled with [32P] dCTP by random priming. Labeled probes were preannealed with COT-1 DNA (Gibco) for 30 min at 65°C, then hybridized overnight at 65°C in Church's buffer (Church and Gilbert 1984), washed, and photographed using Kodak X-Omat AR film.
Inverse PCR was performed on MscI-digested ligated (in 1-ml volumes to facilitate self-ligation of individual fragments) genomic DNA, using nested sets of primers at different positions. Amplified fragments were isolated on a 1% agarose/Tris-acetate-EDTA gel, purified with Qiaex II, and sequenced with a Big Dye terminator cycle sequencing kit (Applied Biosystems). Sequence analysis was performed on an ABI Prism 310 genetic analyzer. All primer sequences and amplification protocols are available, on request, from the authors.
Northern analysis was performed on human multiple-tissue northern blots (Clontech Laboratories), according to the manufacturer's protocols. [32P]-labeled rat ProSAP2 cDNA (Boeckers et al. 1999a) and FLJ10659 cDNA (IMAGE: 3450646) were used as probes.
Results
Cytogenetic and Molecular Studies
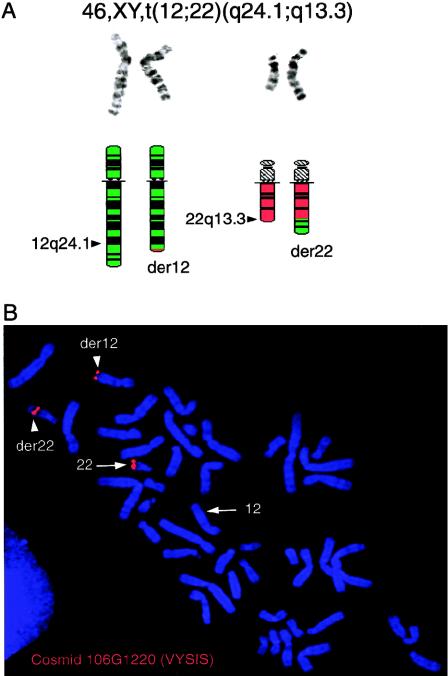
Standard cytogenetic investigations in the proband showed a chromosome translocation: 46, XY, t(12;22)(q24.1;q13.3) (fig. 1). No 22q bands were visible on der(12). The translocation’s reciprocity was demonstrated by FISH, using the 22q subtelomere–specific cosmid 106G1220 (Ning et al. 1996). No signals were seen on der(12) after FISH with a chromosome 22 painting library, whereas two signals, one on der(12) and one on der(22), were visible after FISH with 106G1220 (fig. 1). Thus, the translocation appeared to be reciprocal, with the chromosome 22 breakpoint within probe 106G1220. Fine localization of the breakpoint on chromosome 12q was performed using YAC clones localized in 12q24 (contig WC-841). Signals from the overlapping YACs (948-E-9 and 901-A-5) were found on both derivative chromosomes, whereas YAC 764-F-2 gave a signal only on der(12). Thus, the breakpoint on chromosome 12q lies near STS D12S317 (115 cM from the 12p telomere).
Figure 1.
A (top), Cut-out of normal chromosomes 12 and 22, aligned with their homologue translocation derivatives der(12) and der(22) in G-banding at a resolution of ⩾550 bands. A (bottom), Corresponding high-resolution G-banding ideograms of chromosomes 12 (green) and 22 (red) and their derivatives der(12) (green with a small red portion) and der(22) (red with a green portion). Arrowheads indicate the breakpoints in subbands 12q24.1 and 22q13.3. The reciprocal exchange of chromosome material involves a larger region from chromosome 12 than from chromosome 22 (see ideogram); no 22q material is visible on der(12), when examined by cytogenetics or FISH with a 22-painting library. B, Results of FISH with the 22q subtelomeric probe 106G1220 (VYSIS). The normal chromosome 22 (arrow) and both der(12) and der(22) (arrowheads) show a signal demonstrating the translocation's reciprocity, with the chromosome 22 breakpoint within probe 106G1220.
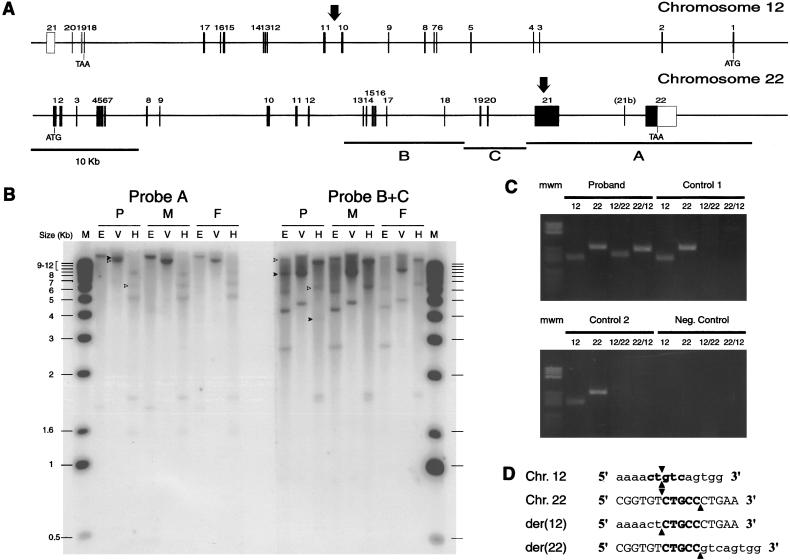
After Southern blot hybridization with fragments corresponding to bases 22039–41000 (fragment A), 4238–15981 (fragment B), and 15981–22039 (fragment C) from cosmid n85a3 (fig. 2), several bands (fig. 2) showed a decrease in relative signal strength in the proband, whereas additional bands (fig. 2) appeared. The breakpoint's location could be narrowed to the region shared by the fragments with reduced signal, from the EcoRV site at position 295109 to the HindIII site at position 299969 (all nucleotide positions refer to the chromosome 22 sequence, locus NT_024379 [Dunham et al. 1999]).
Figure 2.
A, Genomic structure of human ProSAP2 and FLJ10659 genes. The left-to-right orientation of both maps is from centromere to telomere. Sizes of exons (boxes) and introns are drawn to scale. Vertical arrows indicate the breakpoint’s location on both chromosomes. Horizontal lines (A, B, and C) indicate the positions and sizes of the restriction fragments isolated from cosmid n85a3 and used as Southern hybridization probes. B, Southern blot analysis. Genomic DNA (10-μg) from the proband (P), his mother (M), and father (F), digested with EcoRV (V), EcoRI (E), or HindIII (H), was loaded in each lane. C, Selective amplification of der(12) and der(22) in the proband. Genomic DNA from the proband and two normal subjects, as well as a negative control sample that contained no DNA, was amplified with primer pairs spanning the breakpoints on chromosome 12, 22, der(12) (12/22), and der(22) (22/12). D, Alignment of chromosome 12, 22, der(12) and der(22) sequences at the breakpoint. Chromosome 12–derived sequence is shown in lowercase letters, and chromosome 22–derived sequence is shown in uppercase letters. Breakpoints are indicated by arrowheads. The CTG(C/T)C sequence at the breakpoint on both chromosome 12 and 22 is shown in bold. The orientation of sequences is as in panel A.
The breakpoint was cloned by inverse PCR and was confirmed by specific amplification of both der(12) and der(22) breakpoints (fig. 3). Sequencing of the breakpoints showed that the chromosome 12 sequence is exactly split between the derivative chromosomes, whereas 5 bases around the chromosome 22 breakpoint are present on both derivative chromosomes (fig. 3). The chromosome 22 breakpoint lies at position 296489/296494, within exon 21 of the human homologue of rat ProSAP-2 (Boeckers et al. 1999a), Shank-3 (Lim et al. 1999), and SPANK-2 (Naisbitt et al. 1999). The chromosome 12 breakpoint is localized at ∼5 kb from the D12S317 locus (contig NT_024379), between exons 10 and 11 of the gene for the hypothetical protein FLJ10659 (XM_006601) (fig. 3).
Figure 3.
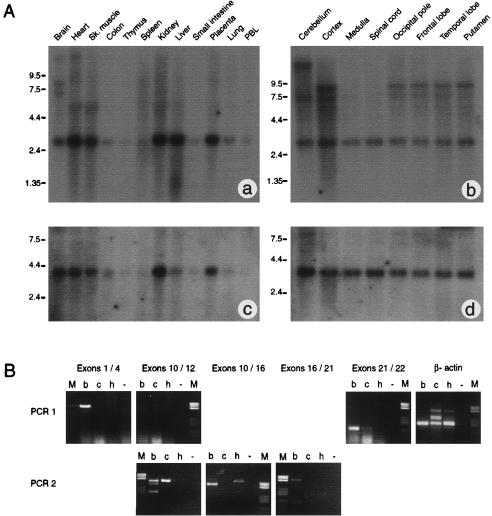
A, Expression of ProSAP2 and FLJ10659. Commercial northern blot analysis was performed using 2 μg per lane of poly-A+ RNA from human tissues (panels a and c), and human brain tissues (panels b and d). The filters were hybridized to rat ProSAP2 cDNA (panels a and b) or IMAGE clone 3450646 (panels c and d). Filters a and b were exposed for 2 w, and filters c and d were exposed for 40 h. B, PCR analysis of human ProSAP2 expression. Groups of exons spanning most of the gene’s coding region (exons 1/4, 10/12, 10/16, 16/21, and 21/22) were amplified from human brain (b), cerebellum (c), and heart (h) cDNAs using a nested PCR protocol. Negative control amplifications (−) were always included. Human β-actin cDNA was used as a control, and amplified at similar levels in all tissues. Some products (exons 1/4, 10/12, and 21/22) were visible after the primary amplification (PCR 1), but others (exons 10/16 and 16/21) required a secondary amplification (PCR 2). All amplified fragments were isolated and sequenced. The smallest fragment (200 bp) from the secondary amplification of exons 10/12 from brain tissue does not contain exon 11. The 540-bp fragment from the secondary amplification of exon 10/16 from brain tissue does not contain exons 11 and 12, but the 600-bp fragment from heart in the same panel was amplified from genomic DNA. The 128-bp fragment from the primary amplification of exons 21/22 from brain tissue does not contain exon 21b. All other fragments have the expected sequence.
Structure of the Human ProSAP2 and FLJ10659 Genes
The structure of the human ProSAP2 gene was deduced by comparing the chromosome 22 sequence (NT_001128) with the rat ProSAP2 cDNA (NP_067707), using BLAST2. The structure of the FLJ10659 gene was already known and can be browsed using LocusLink. The ProSAP2 gene spans a 60-kb region on chromosome 22, between arylsulfatase A (ARSA) and acrosin, and is coded by 22 exons (fig. 2). An additional 27-bp exon (exon 21b) was found by comparing the human sequence to rat Shank-3A cDNA (NP_067708); however, it may not be functional, because it does not appear to have a correct 3′-splice sequence. In fact, PCR analysis shows that exon 21b is not present in the ProSAP2 transcript from human brain tissue (fig. 3).
Exon 1 contains the ATG start codon, if we assume that the human ProSAP2 translation start codon is the same as that in rat and mouse cDNAs. Exon 22 contains the stop codon and, presumably, the whole 3′ nontranslated portion of the cDNA. This portion has not been sequenced in rats or mice. BLAST analysis of the human genomic sequence against human expressed-sequence tags (data not shown) suggests that AATAAA motifs at positions 308209 and 308619 are the most-frequently used polyadenylation signals. Nucleotide-sequence identity is 86% overall and is conserved throughout the gene (data not shown). The gene codes for a 1731aa protein. Intron-exon boundaries, except for exon 21b, conform to the canonical sequence (data not shown).
The FLJ10659 gene spans a 63-kb region of chromosome 12 and is composed of 21 exons (fig. 2). The ATG start codon is located on exon 1, and the stop codon is located on exon 18. The gene codes for a 541aa protein.
Expression Pattern of Human ProSAP2 and FLJ10659
Northern blot analysis of human tissues, using a full-length rat ProSAP2 cDNA probe, has shown multiple transcripts (fig. 3). A 3-kb band is expressed at varying levels in different tissues, more strongly in heart, kidney, liver, and placenta. Two longer transcripts (7 and 8 kb) are seen exclusively in brain tissue.
Analysis of different CNS regions shows ubiquitous expression of the 3-kb band; however, a 7-kb band is not present in the medulla or spinal cord, and an 8-kb band is specific to cortical samples. We have repeated the hybridization, using two probes from different portions of the human ProSAP2 cDNA: a 400-bp fragment containing exon 11 and a portion of exons 10 and 12 (including a 150-bp fragment that codes for the SH3 domain) and a 1.7-kb portion of exon 21 that has no known similarity to other genes. The exon 21 probe only hybridizes to the longer bands (7, 8, and 12–15 kb), whereas the exon 10-12 probe hybridizes to the longer bands in brain and to additional smaller bands (2–2.3 kb) in several tissues (data not shown). Neither probe hybridizes to the 3-kb band seen with the rat probe.
To determine whether the smaller bands represent differentially spliced ProSAP2 transcripts or are the result of cross-hybridization with other members of the ProSAP family or other PDZ/SH3 domain-containing transcripts, we have amplified fragments spanning most of the coding region of the gene from human cortex, cerebellum, and heart cDNAs (fig. 3). ProSAP2 was always amplified in tissue from the cerebral cortex; however, the cerebellum gave somewhat mixed results, and results were always negative in heart tissue.
These results strongly suggest that human ProSAP2 is expressed primarily in the brain, at least in adults, and that the sizes of its transcripts are 7 and 8 kb. The size difference between the larger bands probably results from different polyadenylation sites. The 12–15-kb band that is visible mainly in the cerebellum may represent an incompletely spliced transcript. Differential splicing events, which have been extensively described in rats (Lim et al. 1999), can also occur in human ProSAP2 (fig. 3). The smaller bands detected in many tissues with the rat cDNA and the exon 10–12 probe are almost certainly cross-hybridizations with other transcripts containing PDZ, SH3, or SAM domains.
The FLJ10659 probe hybridizes to a 4-kb transcript expressed in several tissues, including kidney, brain, heart, and skeletal muscle. The transcipt is expressed in all CNS samples (fig. 3).
Discussion
The introduction of new methods aimed at checking the telomere's integrity is leading to the definition of new malformation syndromes (Knight and Flint 2000; De Vries et al. 2001). Among those recently defined is the 22q distal deletion syndrome. Flint and colleagues (1995) detected the first cryptic 22q deletion while screening for the abnormal inheritance of subtelomeric DNA polymorphisms in subjects with mental retardation. Since then, several instances of 22q distal deletion, not detectable by conventional cytogenetic techniques, have been found. Some were detected while screening for cryptic subtelomeric rearrangements in patients with mental retardation (Knight and Flint 1999). Others (with a generic “chromosomal phenotype”) were detected during exclusion of a deletion at the diGeorge/velocardiofacial region (Goizet et al. 2000; Praphanphoj et al. 2000; Prasad et al. 2000). In fact, commercially available diGeorge probes include 22q distal probes as a control. Although most subjects who have cryptic telomeric rearrangements are affected by mental retardation associated with congenital anomalies and facial dysmorphisms (Knight and Flint 2000), most subjects who have 22q distal deletions are ascertained because of mild mental retardation and delayed speech (Flint et al. 1995; Precht et al. 1998). In these subjects, expressive language is either absent or severely delayed. Less frequently described traits include epicanthic folds, bulbous nose, dysplastic ears, dolichocephaly, hypotonia, joint laxity, and autism (Goizet et al. 2000; Praphanphoj et al. 2000; Prasad et al. 2000). Although these symptoms are somewhat unspecific, it has been suggested that their association with delayed speech and mild mental retardation should prompt clinicians to search for a 22q distal deletion. DNA studies of patients with 22q distal deletion led to the identification of an SRO from below locus D22S97 proximally to below ARSA (Nesslinger et al. 1994). The molecular definition of a terminal microdeletion led Wong and colleagues (1997) to narrow the critical region to the distal 130 kb, from D22S163 to the telomere. Because the distal 60 kb are probably rich in subtelomeric repeats, the proximal remaining 70-kb region is expected to contain one or more genes whose haploinsufficiency accounts for the two main symptoms of the 22q distal deletion (i.e., delayed speech and mild mental retardation) (Wong et al. 1997). In this region are localized the human homologue of ProSAP2, acrosin, an SNRPA1 pseudogene, RABL2B (a member of the RAS oncogene family), and a 60S ribososomal protein L23A pseudogene (Dunham et al. 1999). Because RAB mutations have been associated with X-linked mental retardation (D'Adamo et al. 1998), the finding that RABL2B was deleted in the patient with the critical 22q deletion raised the possibility that its haploinsufficiency was responsible for the patient's abnormal phenotype (Wong et al. 1999). However, the existence of RABL2A (which is located at 2q13), given that it too is expressed and is very close in sequence to RABL2B, makes RABL2B unlikely to be dose sensitive (Wong et al. 1999).
We have studied a child who had all the symptoms associated with the 22q distal deletion syndrome (severely delayed expressive language, mild mental retardation, hypotonia, dolichocephaly, epicanthic folds, bulbous nose, and lax joints). The finding that he carries a de novo balanced translocation resulting in the disruption of a gene localized in the 70-kb critical region of chromosome 22q suggests that its haploinsufficiency may be responsible for the 22q distal deletion syndrome phenotype.
The product of the ProSAP2 gene localizes to the PSD, a highly specialized submembranous network of proteins at excitatory synapses of the CNS. The PSD includes a variety of adapter proteins that are involved in localizing receptors, adhesion molecules, and molecules of the intracellular signaling cascade within the synapse (Ziff 1997). Boeckers and colleagues (1999a) isolated cDNAs coding for ProSAP1, a PSD-95/disks-large/ZO-1 (PDZ) domain protein that is highly enriched in the PSD. Homology screening identified a related protein (ProSAP2), which shares a highly conserved PDZ domain (80% identity), a proline-rich Src homology 3 (SH3) binding motif domain that mediates the interaction with cortactin, and a C-terminal sterile-alphamotif (SAM) domain. ProSAP2 contains five ankyrin repeats and an SH3 domain. The ProSAP proteins can bind the scaffold protein Homer and the SAPAP proteins (Boeckers et al. 1999b), which, in turn, bind to PSD-95. They can also form homomultimers, ultimately permitting the formation of a three-dimensional network, linking together the NMDA receptor complex on the PSD, mGluRs in the membrane, and the IP3 receptor on intracellular calcium-storage vesicles (Kennedy 2000).
The ProSAP proteins are coexpressed in many regions of the rat brain, but show a distinct expression pattern in the cerebellum. In fact, in most brain regions, including the cerebral cortex and the hippocampus, ProSAP1 and ProSAP2 appear to be codistributed; however, a complementary distribution is observed in the cerebellum. ProSAP1 is primarily expressed in Purkinje cells, whereas ProSAP2 transcripts are only found in the granular cell layer of cerebellum. (Boeckers et al. 1999b). Expression analysis in human tissues confirms the results in rats and highlights the exclusive expression of full-length Prosap2 in the cerebral cortex and cerebellum.
The chromosome 12 gene disrupted by the translocation codes for the FLJ10659 protein, which contains a PH domain and shows similarity to the human APPL protein (MIM 604299), a protein of unknown function involved in binding to the tyrosine-kinase protein AKT2 (Mitsuuchi et al. 1999). Because the function of FLJ10659 is also currently unknown, we cannot hypothesize about the nature of any correlations between FLJ10659 haploinsufficiency and the phenotype of our patient.
The finding that our patient has symptoms that overlap with those of subjects who have distal 22q deletions suggests that ProSAP2 haploinsufficiency is the cause of this syndrome. The involvement of ProSAP2 protein in the highly specialized cytoskeleton at the PSD is in close agreement with this finding. Another syndrome that is usually associated with haploinsufficiency of a cytoskeletal protein is lissencephaly (Chong et al. 1997; Sapir et al. 1999). The finding of ProSAP2 mutation in subjects with the 22q distal deletion phenotype but no 22q deletion would confirm that it is the causative gene. However, this approach might be difficult because of the rather unspecific phenotype of this syndrome.
Analysis of the sequences involved in the translocation did not uncover any known features that could be directly responsible for the translocation. There is no homology between the two regions. No repetitive sequences are present at or around the breakpoint on chromosome 12 or 22. There is no unusually high concentration of AT nucleotides or alternating purine/pyrimidine residues that could predispose to rearrangements. The only unique feature in this translocation is a 5-bp sequence, CTG(C/A)C, that is present on chromosomes 12 and 22 at the breakpoint (fig. 2). The presence of short common sequence motifs at the rearrangement sites of other constitutional reciprocal translocations has been reported (Giacalone and Franke 1992; Nothwang et al. 2000). Our findings strengthen the hypothesis that the recombinational machinery recognizes the alignment of two DNA molecules with a small region of sequence homology.
Acknowledgments
The authors wish to thank Dr. Tobias M. Boeckers, for the gift of the rat ProSAP2 cDNA, Drs. Manuela Sironi and Rachele Cagliani, for human RNAs, and the YAC Screening Centre of San Raffaele Biomedical Science Park (Milan) for providing YACs. This work was supported by grant ICS/RC 2000-2169 from the Italian Health Ministry. O.Z. has been supported by grant ICS 190.1/RF00-42 from the Italian Health Ministry.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST 2, http://www.ncbi.nlm.nih.gov/blast/blast.cgi (for BLAST analysis)
- Genome Database, http://www-genome.wi.mit.edu/cgi-bin/contig/phys_map (for locus FLJ10659)
- I.M.A.G.E. Consortium, http://image.llnl.gov/ (for I.M.A.G.E. clone 3450646)
- LocusLink, http://www.ncbi.nlm.nih.gov/LocusLink/ (for FLJ10659 gene structure)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for APPL [MIM 604299])
- YAC Screening Centre, http://w3dibit.hsr.it/YAC/first-yac.html (for all YACs mentioned in this report)
References
- Boeckers TM, Kreutz MR, Winter C, Zuschratter W, Smalla K-H, Sanmarti-Vila L, Wex H, Langnaese K, Bockmann J, Garner CC, Gundelfinger ED (1999a) Proline-rich synapse-associated protein-1/cortactin binding protein 1 (ProSAP1/CortBP1) is a PDZ-domain protein highly enriched in the postsynaptic density. J Neurosci 19:6506–6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckers TM, Winter C, Smalla K-H, Kreutz MR, Bockmann J, Seidenbecher C, Garner CC, Gundelfinger ED (1999b) Proline-rich synapse-associated proteins ProSAP1 and ProSAP2 interact with synaptic proteins of the SAPAP/GKAP family. Biochem Biophys Res Commun 264:247–252 [DOI] [PubMed] [Google Scholar]
- Chelly J (1999) Breakthroughs in molecular and cellular mechanisms underlying X-linked mental retardation. Hum Mol Genet 8:1833–1838 [DOI] [PubMed] [Google Scholar]
- Chong SS, Pack SD, Roschke AV, Tanigami A, Carrozzo R, Smith AC, Dobyns WB, Ledbetter DH (1997 ) A revision of the lissencephaly and Miller-Dieker syndrome critical regions in chromosome 17p13.3. Hum Mol Genet 6:147–155 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81:1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Adamo P, Menegon A, Lo Nigro C, Grasso M, Gulisano M, Tamanini F, Bienvenu T, Gedeon AK, Oostra B, Wu S-K, Tandon A, Valtorta F, Balch WE, Chelly J, Toniolo D (1998) Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nat Genet 19:134–139 [DOI] [PubMed] [Google Scholar]
- De Vries BBA, Knight SJL, Homfray T, Smithson SF, Flint J, Winter RM (2001) Submicroscopic subtelomeric 1q ter deletions: a recognisable phenotype? J Med Genet 38:175–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Hunt AR, Collins IE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al (1999) The DNA sequence of human chromosome 22. Nature 402:489–495 [DOI] [PubMed] [Google Scholar]
- Dutrillaux B, Viegas-Pequignot E (1981) High resolution R- and G-banding on the same preparation. Hum Genet 57:93–95 [DOI] [PubMed] [Google Scholar]
- Flint J, Wilkie AO, Buckle VJ, Winter RM, Holland AJ, McDermid HE (1995) The detection of subtelomeric chromosomal rearrangements in idiopathic mental retardation. Nat Genet 9:132–140 [DOI] [PubMed] [Google Scholar]
- Giacalone JP, Francke U (1992) Common sequence motifs at the rearrangement sites of a constitutional X/autosome translocation and associated deletion. Am J Hum Genet 50:725–741 [PMC free article] [PubMed] [Google Scholar]
- Goizet C, Escoffier E, Taine L, Taupiac E, El Moneim AA, Arveiler B, Bouvard M, Lacombe D (2000) Case with autistic syndrome and chromosome 22q13.3 deletion detected by FISH. Am J Med Genet 96:839–844 [PubMed] [Google Scholar]
- Kennedy MB (2000) Signal-processing machines at the postsynaptic density. Science 290:750–754 [DOI] [PubMed] [Google Scholar]
- Knight SJ, Flint J (2000) Perfect endings: a review of subtelomeric probes and their use in clinical diagnosis. J Med Genet 37:401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Naisbitt S, Yoon J, Hwang J-I, Suh P-G, Sheng M, Kim E (1999) Characterization of the Shank family of synaptic proteins. J Biol Chem 274:29510–29518 [DOI] [PubMed] [Google Scholar]
- Mitsuuchi Y, Johnson SW, Sonoda G, Tanno S, Golemis EA, Testa JR (1999) Identification of a chromosome 3p14.3-21.1 gene, APPL, encoding an adaptor molecule that interacts with the oncoprotein-serine/threonine kinase AKT2. Oncogene 18:4891–4898 [DOI] [PubMed] [Google Scholar]
- Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtscanoff J, Weinberg RJ, Worley PF, Sheng M (1999) Shank, a novel family of postsynaptic density proteins that bind to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron 23:569–582 [DOI] [PubMed] [Google Scholar]
- Nesslinger NJ, Gorski JL, Kurczynski TW, Shapira SK, Siegel-Bartelt J, Dumanski JP, Cullen RF Jr, French BN, McDermid HE (1994) Clinical, cytogenetic, and molecular characterization of seven patients with deletions of chromosome 22q13.3. Am J Hum Genet 54:464–472 [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Rosenberg M, Biesecker LG, Ledbetter DH (1996) Isolation of the human chromosome 22q telomere and its application to detection of cryptic chromosomal abnormalities. Hum Genet 97:765–769 [DOI] [PubMed] [Google Scholar]
- Nothwang HG, Schroer A, van der Maarel S, Kubart S, Schneider S, Riesselmann L, Menzel C, Hinzmann B, Vogt D, Rosenthal A, Fryns J, Tommerup N, Haaf T, Ropers HH, Wirth J (2000) Molecular cloning of Xp11 breakpoints in two unrelated mentally retarded females with X;autosome translocations. Cytogenet Cell Genet 90:126–133 [DOI] [PubMed] [Google Scholar]
- Praphanphoj V, Goodman BK, Thomas GH, Raymond GV (2000) Cryptic subtelomeric translocations in the 22q13 deletion syndrome. J Med Genet 37:58–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad C, Prasad AN, Chodirker BN, Lee C, Dawson AK, Jocelyn LJ, Chudley AE (2000) Genetic evaluation of pervasive developmental disorders: the terminal 22q13 deletion syndrome may represent a recognizable phenotype. Clin Genet 57:103–109 [DOI] [PubMed] [Google Scholar]
- Precht KS, Lese CM, Spiro RP, Huttenlocher PR, Johnston KM, Baker JC, Christian SL, Kittikamron K, Ledbetter DH (1998) Two 22q telomere deletions serendipitously detected by FISH. J Med Genet 35:939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir T, Eisenstein M, Burgess HA, Horesh D, Cahana A, Aoki J, Hattori M, Arai H, Inoue K, Reiner O (1999) Analysis of lissencephaly-causing LIS1 mutations. Eur J Biochem 266:1011–1020 [DOI] [PubMed] [Google Scholar]
- Toniolo D, D'Adamo P (2000) X-linked non-specific mental retardation. Curr Opin Genet Dev 10:280–285 [DOI] [PubMed] [Google Scholar]
- Wong ACC, Ning Y, Flint J, Clark K, Dumanski JP, Ledbetter DH, McDermid HE (1997) Molecular characterization of a 130-kb terminal microdeletion at 22q in a child with mild mental retardation. Am J Hum Genet 60:113–120 [PMC free article] [PubMed] [Google Scholar]
- Wong ACC, Shkolny D, Dorman A, Willingham D, Roe BA, McDermid HE (1999) Two novel human RAB genes with near identical sequence each map to a telomere-associated region: the subtelomeric region of 22q13.3 and the ancestral telomere band 2q13. Genomics 59:326–334 [DOI] [PubMed] [Google Scholar]
- Ziff EB (1997) Enlightening the postsynaptic density. Neuron 19:1163–1174 [DOI] [PubMed] [Google Scholar]