Abstract
Classic tetrahydrobiopterin (BH4) deficiencies are characterized by hyperphenylalaninemia and deficiency of monoamine neurotransmitters. In this article, we report two patients with progressive psychomotor retardation, dystonia, severe dopamine and serotonin deficiencies (low levels of 5-hydroxyindoleacetic and homovanillic acids), and abnormal pterin pattern (high levels of biopterin and dihydrobiopterin) in cerebrospinal fluid. Furthermore, they presented with normal urinary pterins and without hyperphenylalaninemia. Investigation of skin fibroblasts revealed inactive sepiapterin reductase (SR), the enzyme catalyzing the final two-step reaction in the biosynthesis of BH4. Mutations in the SPR gene were detected in both patients and their family members. One patient was homozygous for a TC→CT dinucleotide exchange, predicting a truncated SR (Q119X). The other patient was a compound heterozygote for a genomic 5-bp deletion (1397–1401delAGAAC) resulting in abolished SPR-gene expression and an A→G transition leading to an R150G amino acid substitution and to inactive SR as confirmed by recombinant expression. The absence of hyperphenylalaninemia and the presence of normal urinary pterin metabolites and of normal SR-like activity in red blood cells may be explained by alternative pathways for the final two-step reaction of BH4 biosynthesis in peripheral and neuronal tissues. We propose that, for the biosynthesis of BH4 in peripheral tissues, SR activity may be substituted by aldose reductase (AR), carbonyl reductase (CR), and dihydrofolate reductase, whereas, in the brain, only AR and CR are fully present. Thus, autosomal recessive SR deficiency leads to BH4 and to neurotransmitter deficiencies without hyperphenylalaninemia and may not be detected by neonatal screening for phenylketonuria.
Introduction
Tetrahydrobiopterin (BH4) deficiencies are severe neurological disorders characterized by hyperphenylalaninemia and by monoamine-neurotransmitter deficiency and caused by mutations in the genes encoding the enzymes responsible for BH4 biosynthesis and regeneration (Blau et al. 2001). BH4 is an essential cofactor for enzymes such as nitric oxide synthase, phenylalanine-3-hydroxylase, and the rate-limiting catecholamine- and serotonin-biosynthesis enzymes tyrosine-4-hydroxylase and tryptophan-5-hydroxylase. The biosynthesis of BH4 starts from GTP via reactions catalyzed by the enzymes GTP cyclohydrolase I (GTPCH [E.C.3.5.4.16]), 6-pyruvoyltetrahydropterin (PTP) synthase (PTPS [E.C.4.6.1.10]), and sepiapterin reductase (SR [E.C.1.1.1.153]). Alternatively, the final two-step reduction of the intermediate PTP to BH4 can be effected by aldose reductase (AR) and carbonyl reductase (CR) (fig. 1). Two additional enzymes, pterin-4a-carbinolamine dehydratase (E.C.4.2.1.96) and dihydropteridine reductase (DHPR [E.C.1.6.99.7]), are involved in the regeneration of BH4 from intermediates formed during the hydroxylation of aromatic amino acids (Thöny et al. 2000).
Figure 1.
Biosynthesis of tetrahydrobiopterin and proposed alternative routes in SR deficiency (for details, see text)
Most patients with autosomal recessive BH4 deficiencies present with progressive neurological deterioration, regardless of their different enzyme defects. The clinical manifestation is variable, but common symptoms are mental retardation, convulsions, disturbance of tone and posture, abnormal movements, hypersalivation, swallowing difficulties, and temperature instability and oculogyric crises (Blau et al. 1996). These patients can be detected by neonatal screening for phenylketonuria (PKU [MIM 261600]), owing to abnormally high levels of phenylalanine in blood. The autosomal dominant form of GTPCH deficiency, dopa-responsive dystonia (DRD [MIM 600225]), however, presents without hyperphenylalaninemia, and patients with DRD can be diagnosed only by the typical clinical signs and symptoms, by responsiveness to l-dopa (Nygaard and Wooten 1998), and by investigation of cerebrospinal fluid (CSF).
Recently, we investigated production of neopterin and biopterin in cytokine-stimulated fibroblasts from patients with different forms of BH4 deficiencies. Furthermore, we measured the activity of all enzymes involved in BH4 metabolism in normal and stimulated cells. By this method, we showed that both the classic forms of BH4 deficiency and DRD can be differentiated in fibroblasts (Bonafé et al. 2001). In this study, we investigated fibroblasts from two patients with severe neurotransmitter depletion without hyperphenylalaninemia who were initially diagnosed with a “central” form of DHPR deficiency (MIM 261630) (Blau et al. 1998). Pterin metabolites and enzymatic activities revealed SR deficiency, which was confirmed by DNA mutation analysis. We thus describe a new autosomal recessive form of BH4 deficiency with monoamine-neurotransmitter depletion and without hyperphenylalaninemia, and we propose alternative routes, in different tissues, for the final step in the BH4-biosynthesis pathway.
Patients and Methods
Patients
The two patients described are registered in the international database BIODEF (Blau et al. 1996). Patient 360 (BIODEF) is a 14-year-old adolescent male born of consanguineous parents of Turkish origin. He presented with psychomotor retardation, spasticity, dystonia, microcephaly, and growth retardation (Blau et al. 1998). Patient 229 (BIODEF) is a 9-year-old boy born of unrelated parents of Turkish origin. He presented with progressive psychomotor retardation, spasticity, tremor, ataxia, dystonic posturing and falls (initially misinterpreted as epileptic seizures), depressive and aggressive behavior, and oculogyric crises (Blau et al. 1999); there were also marked diurnal fluctuations. Both patients were initially classified as having a “central” form of DHPR deficiency, because of a CSF-pterin pattern typical of this disorder. Table 1 summarizes the most-important biochemical findings. Plasma phenylalanine, urinary and plasma total neopterin and total biopterin, and red blood cell DHPR activity were all normal. After BH4 administration, both patients responded to the oral loading test with phenylalanine, suggesting reduced phenylalanine hydroxylation in the liver (Blau et al. 1999). DNA analysis of all coding exons of the QDPR gene failed to detect any mutation (I. Dianzani and A. Romstad, personal communication). Neurotransmitter-precursor therapy was started, in patients 229 and 360, at the ages of 5 years and 10 years, respectively. In both subjects, administration of 5-hydroxytryptophan resulted in severe vomiting and/or leucopenia, whereas administration of l-dopa resulted in clinical improvement. Selegiline (l-deprenyl) was introduced only in patient 360.
Table 1.
Summary of Diagnostic Biochemical and Molecular Data
|
Patient |
|||
| Biochemical finding(s) | 360 | 229 | Controls |
| Age at diagnosis (years) | 10 | 5 | … |
| Plasma phenylalanine (mmol/liter) | <90 | <90 | 26–98 |
| CSFa(nmol/liter): | |||
| Total neopterin | 28 | 24 | 9–30 |
| Total biopterin | 83 | 77 | 10–44 |
| Dihydrobiopterin | 57 | 42 | <14 |
| 5HIAA | 14 | 4 | 88–178 |
| HVA | 111 | 49 | 144–801 |
| Urine (mmol/mol creatinine): | |||
| Total neopterin | .65 | .78 | .2–1.7 |
| Total biopterin | .58 | .70 | .5–2.7 |
| Fibroblasts: | |||
| Total neopterin (pmol/mg) | 169 | 238 | 18–98 |
| Total biopterin (pmol/mg) | 29 | 57 | 154–303 |
| SR activity (mU/mg) | <10 | <10 | 99–185 |
| Red blood cells: | |||
| SR activityb (mU/mg) | .34 | .36 | .33–1.86 |
| Genomic DNA | 1303–1304TC→CT (homozygous) | 1397A→G, 1397–1401delAGAAC | Wild type |
| cDNA | 354–355TC→CT | 448A→G | Wild type |
| Effect on protein | Q119X | R150G | Wild type |
Methods
Study of pterin metabolism in fibroblasts
Cell culturing, production of neopterin and of biopterin in fibroblasts after stimulation with cytokines for 24 h, and SR-activity measurements in nonstimulated fibroblasts were performed as described elsewhere (Bonafé et al. 2001). For fibroblast cultures, the appropriate informed consent was obtained from all subjects.
SR activity in red blood cells
SR activity in red blood cells was assayed according to the method described by Ferre and Naylor (1988), with sepiapterin as a substrate and under assay conditions essentially the same as those for the fibroblasts (see the preceding paragraph). The assay monitors the conversion of sepiapterin to dihydrobiopterin, which is then measured as the oxidized product, biopterin. Because the same reaction can be catalyzed by CR (Park et al. 1991), this assay measures activities of both SR and CR. Also, substrates used in the CR assay can be reduced by SR (Katoh and Sueoka 1984).
Mutation analysis
All oligonucleotide primers were custom synthesized by Microsynth. For SR cDNA analysis, total RNA was extracted from skin fibroblasts of the two patients (RNeasy Mini Kit; Qiagen). On the basis of the published SR cDNA sequence (GenBank accession number M76231; Ichinose et al. 1991), a 756-bp fragment harboring 96% of the coding sequence was amplified by one-step reverse-transcription PCR (RT-PCR) (OneStep RT-PCR Kit; Qiagen), by the cDNA-specific primers SR12 (5′-[8]GCGGGCTGGGGCGTGCTGTG-3′) and SR13 (5′-[763]GGGCTCCAGACTTGAACTCG-3′) (the nucleotide numbers in square brackets refer to the published cDNA sequence, and numbering starts with 1 at adenosine in the ATG-start codon). PCR amplification was performed under standard conditions, at an annealing temperature of 56°C and in 35 cycles. To analyze the corresponding SPR gene, genomic DNA from the two patients was extracted from fibroblasts, and DNA from their parents was extracted from whole blood cells by standard procedures (QIAmp DNA Mini Kit; Qiagen). The three coding exons of SPR were each amplified at an annealing temperature of 54°C and in 35 cycles, by the following primer pairs: SR14 (5′-[187]CAGCAACCAAGGGAACCAGA-3′) and SR15 (5′-[933]GCAAGGGGCTCGGGAAAGTT-3′) to yield a 747-bp fragment for exon 1; SR16 (5′-[810]GCAAGTGGAGGCGAGGTGTA-3′) and SR17 (5′-[1858]GAGCGTCTTCCCCATTTCAC-3′) to yield a 1,049-bp fragment for exon 2; and SR18 (5′-[141]AATAGAAATGGGAATGTCAG-3′) and SR19 (5′-[880]GGGATAGAGACACCAATACC-3′) to yield a 740-bp fragment for exon 3 (the nucleotide numbers in square brackets refer to the deposited genomic DNA sequences) (Ohye et al. 1998; GenBank accession numbers AB017547, for exons 1 and 2, and AB017548, for exon 3).
The amplification products from genomic DNA and cDNA were separated on 1% agarose gels, purified (Concert Gel Extraction Systems; Life Technologies; Gibco BRL), and directly sequenced using fluorescence-labeled terminator reagents and an automated sequencer (ABI Prism 310, Applied Biosystems). The primers used for PCR amplification were also used for sequence analysis, with the addition of the primer pair SR22 (5′-[1190]GGGAGGGCTGGGGAAGAAGAA-3′) and SR23 (5′-[1574]AGGACAGGGACGGCAGACTT-3′), which were used, with the primer pair composed of SR16 and SR17, to sequence the exon 2–specific fragment.
Recombinant expression of human SR in Escherichia coli
To express wild-type and mutant R150G SR proteins in E. coli cells, corresponding cDNA fragments were cloned into the expression vector pGEMEX-2-Nde, which contained an isopropyl-2-thio-β-d-galactopyranoside (IPTG)–inducible promoter (Thöny et al. 1994). As described above, RT-PCR was performed with RNA from wild-type or mutant fibroblasts as template, with the primer pair SR20 (5′-CGGAATTCATATGGAGGGCGGGCTGGGGCGTGCTGTG-3′) and SR21 (5′-CGGGATCCTATTTGTCATAGAAGTCCACGTGGGCTCCAGACTTGAACTCG-3′); SR20 contains a recognition site for NdeI, and SR21 contains a recognition site for BamHI (both underlined). The 0.8-kb PCR product was gel purified and ligated into the NdeI and BamHI sites of pGEMEX-2-Nde, to generate plasmids pHSR9 and pHSR10, which harbor wild-type and mutant R150G SR proteins, respectively. Expression of SR proteins was achieved, in E. coli BL-21 cells (Promega), by standard induction with IPTG, according to a protocol recommended by New England Biolabs.
Escherichia coli harboring pHSR9 with wild-type SR, harboring pHSR10 with R150G SR, or transformed with the vector alone (pGEMEX control) were used for expression studies. Cells from 500-ml cultures were harvested by centrifugation, resuspended in 2 ml of ice-cold lysis buffer containing 50 mM potassium phosphate (pH 6.4) and 0.4 mg of lysozyme/ml, frozen in liquid nitrogen, lysed by thawing at 8°C, and centrifuged at 4°C for 20 min at 2,000 g in a SS34 rotor (Sorvall). To obtain “clear lysates,” supernatants were filtered at 4°C through a Sephadex G50 Nick spin column (Pharmacia) and were used for protein determination (Bio-Rad), enzyme-activity measurement, and western blot analysis.
To measure enzyme activity, 1 μg, 5 μg, and 10 μg of each clear lysate was incubated in a reaction mixture containing 0.1 M potassium phosphate, pH 6.4, 0.25 mM reduced nicotinamide adenine dinucleotide phosphate (NADPH), and 0.125 mM sepiapterin, according to assay conditions described elsewhere (Bonafé et al. 2001). After 30 min of incubation, the product, 7,8-dihydrobiopterin, was oxidized and measured as biopterin metabolite, in a high-performance liquid chromatography (HPLC) system with fluorescence detection (Curtius et al. 1991). One unit of SR activity is defined as the amount that catalyzes the production, from the sepiapterin substrate, of 1 μmol of biopterin/min.
For western blot analysis, electrophoresis through 12.5%-SDS–polyacrylamide gel was performed according to the method of Laemmli (1970), with a prestained SDS-PAGE low-range protein standard from Bio-Rad. Proteins were blotted onto nitrocellulose sheets (Bio-Rad). As a first antibody, the rabbit anti–mouse SR antibody (provided by Y. S. Park) was applied in a 1:25,000 dilution. As a second antibody, a commercially available goat anti–rabbit IgG alkaline phosphatase conjugate from Bio-Rad was employed. Immunostaining was performed by the colorimetric method for alkaline phosphatase, by use of NBT/BCIP solution (Roche).
Results
SR Deficiency Revealed by Metabolic and Enzymatic Studies of Primary Skin Fibroblasts
On the basis of our recent observation that the molecular basis of the various BH4 disorders can be differentiated by analysis of primary patients’ fibroblasts (Bonafé et al. 2001), we examined cytokine-stimulated cells for pterin metabolites and/or nonstimulated cells for BH4-enzymatic activities. Cultured skin fibroblasts from the two patients presented here were stimulated with cytokines for 24 h and revealed elevated neopterin production and low biopterin production, in both cases (table 1). These findings suggested an enzymatic block in the biosynthesis of BH4. Moreover, the pattern was similar to that found in patients with PTPS deficiency (MIM 261640); however, the PTPS activity was essentially normal when measured in fibroblasts from both patients (0.3 and 0.6 μU/mg; control value 0.4–1.6 μU/mg). The GTPCH and DHPR activities in these fibroblasts were also in the normal range (data not shown). In contrast, when we assayed SR under standard conditions with sepiapterin as substrate, activity was below detection level in fibroblasts from either patient (<10 μU/mg; control value 99–188 μU/mg), and, when we measured SR in red blood cells, activity was in the low-normal range (table 1). This suggested a primary defect in the BH4-biosynthetic enzyme SR.
Detection of DNA Mutations in Human SPR
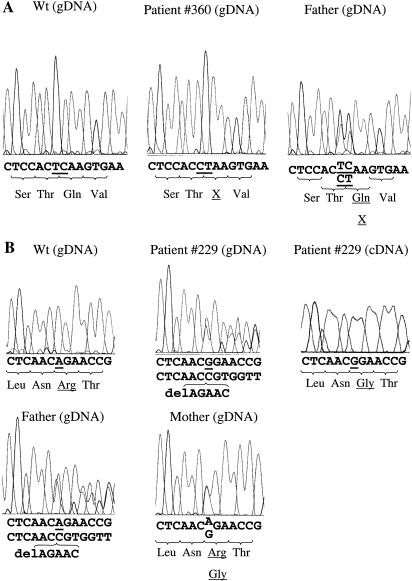
Next, we examined both the SR cDNA and all three exons of the corresponding SPR gene, by PCR amplification and direct DNA sequencing. Analysis of patient 360 revealed, in exon 2, a homozygous TC→CT dinucleotide transition at cDNA position 354–355 and at genomic DNA position 1303–1304 (fig. 2A). This exchange predicted the mutant allele Q119X with a premature stop codon at amino position 119. Heterozygosity could be confirmed in at least the father’s genomic DNA, whereas the mother’s DNA was not available.
Figure 2.
A, Mutations in SPR, in patient 360 and in his father. B, Mutations in SPR and SR cDNA, in patient 229 and in his parents.
In patient 229, cDNA sequencing of the RT-PCR product exhibited a homozygous A→G missense mutation at position 448, predicting the amino acid exchange R150G (fig. 2B). Genomic DNA analysis revealed compound heterozygosity in the proband, with one allele carrying the corresponding A→G nucleotide exchange at position 1397 in exon 2, as found in the cDNA. In addition, exon 2 contained a second alteration, a 5-bp deletion spanning nucleotide position 1397–1401 (allele 1397–1401delAGAAC). This deletion predicted a frameshift after triplet Asn149, leading to a subsequent stop codon at position 152 and, thus, because the cDNA of this allele was not amplifiable from the patient’s fibroblasts, to mRNA degradation. Analysis of genomic DNA of the parents showed that the mother was heterozygous for the 1397A→G transition, leading to the mutant allele R150G, and that the father was heterozygous for the mutant allele 1397–1401delAGAAC.
Recombinant Expression of Mutant R150G and Wild-Type SR Alleles in E. coli
To verify the effect, at the protein level, of the identified mutant allele R150G, we assayed wild-type and R150G SR proteins as recombinant proteins in E. coli. The mutant allele Q119X was not tested, because ∼55% of the predicted SR was deleted. SR activities in bacterial lysate of the clones harboring wild-type and R150G SR proteins are represented in figure 3A. The specific activity of wild-type SR was 6.2 U/mg, whereas the negative control (i.e., bacteria harboring the pGEMEX-expression vector without SR cDNA) showed background SR activity (0.14 U/mg) that probably corresponds to the CR activity. Lysate containing the mutant R150G SR showed a specific activity similar to that of the negative control (0.13 U/mg). Expression of SR proteins in bacterial cells was verified by western blot analysis, showing a single immunoreactive band for the SR monomer in lysates from both wild-type and mutant R150G SR proteins but not for that in lysate from the pGEMEX control (fig. 3B).
Figure 3.
Recombinant expression of wild-type and mutant R150G SR proteins in bacterial cells. A, SR activity assayed by determination of biopterin production with sepiapterin as substrate and with different amounts of soluble-protein extracts from E. coli expressing either the wild type, R150G, or no SR (vector control) from the expression vector pGEMEX. B, Western blot analysis using the same extracts as in panel A. Each lane contains 0.05 mg of total protein, and cross-reactive material was detected by use of a rabbit anti–mouse SR antibody (arrow). Note that, although the human recombinant SR is a 28-kD monomer, it runs at ∼34 kD in the 12.5% SDS-PAGE with the molecular-weight standard here used.
Discussion
The first two cases of SR deficiency described in this article were distinct from other cases of autosomal recessive forms of BH4 deficiency in that they presented without hyperphenylalaninemia and thus cannot be detected by the neonatal screening programs for PKU. SR deficiency becomes the fourth form of BH4 deficiency to be associated with disturbance in biogenic amine metabolism. The two defects in the biosynthesis of BH4—GTPCH and PTPS deficiencies—as well as a defect in its regeneration—DHPR deficiency—are characterized by deficiency of catecholamines and of serotonin (Blau et al. 2001). They are all autosomal recessively inherited and present with hyperphenylalaninemia, in contrast to the autosomal dominant form of GTPCH deficiency (DRD), which is characterized by normal plasma phenylalanine levels. SR deficiency and DRD can be diagnosed only by investigation of CSF metabolites, in conjunction with the study of biopterin metabolism and enzymatic measurements in cultured skin fibroblasts. When BH4 biosynthesis in fibroblasts from patients with BH4 deficiency is induced, for 24 h, with tumor-necrosis factor–α and interferon-γ, pterin patterns characteristic of the different BH4 disorders are generated (Bonafé et al. 2001). Fibroblasts from patients with autosomal recessive GTPCH deficiency and DRD produce low amounts of neopterin and biopterin. On the other hand, those from patients with PTPS deficiency produce high amounts of neopterin and almost no biopterin. This pattern of high neopterin and low biopterin was also found in our two patients with SR deficiency, indicating a defect in the biosynthesis of BH4 (table 1). The absence of SR activity in unstimulated fibroblasts, in conjunction with the normal PTPS activity, confirmed the diagnosis. Furthermore, the homozygous mutant allele detected in patient 360, Q119X, although expressed at the mRNA level, is certainly a null allele, because it predicts an early stop codon at ∼45% of the normal polypeptide. The deletion detected in one allele of SPR in patient 229, 1397–1401delAGAAC, leads to aberrant gene expression and mRNA degradation. The second mutant allele, R150G, which is at the same site as the other mutant allele, leads to a completely inactive mutant protein, as confirmed by recombinant expression in bacterial cells.
For many years, there has been speculation as to why no patients with SR deficiency have been detected. It has been proposed that such a deficiency may be either fatal in utero or possibly compensated by the activity of other reductase(s). As shown in this article, neither proposition obtains, and alternative reductases replacing absent SR activity, at least in peripheral tissues, may be responsible for the phenotype (see below). SR is an enzyme catalyzing the NADPH-dependent two-step reduction of PTP to BH4 (fig. 1). On the basis of kinetic, crystallographic, and nuclear magnetic-resonance studies, the initial step is the reduction at the C-1’-oxo side-chain of PTP, resulting in the formation of 1’-hydroxy-2’-oxopropyl-tetrahydropterin (2’-oxo-TP). Internal rearrangement of the oxo group via side-chain isomerization leads to 1’-oxo-2’-hydroxypropyl-tetrahydropterin (1’-oxo-TP [6-lactoyl-tetrahydropterin]) (Auerbach et al. 1997). Subsequently, 1’-oxo-TP is reduced to BH4 (fig. 1, pathway A). In contrast, Katoh and Sueoka (1987) have suggested that SR catalyzes the isomerization of 1’-oxo-TP to 2’-oxo-TP and that isomerase function can be further stimulated by NADPH or the oxidized form of nicotinamide adenine dinucleotide phosphate (Katoh and Sueoka 1988). However, Smith (1987) has demonstrated that rat erythrocyte SR catalyzes tetrahydropterin substrates at the relative velocity of sepiapterin→1’-oxo-TP→PTP→2’-oxo-TP and that the 1’-oxo position of PTP is reduced first. Thus, pathway A in figure 1 seems to be involved in de novo biosynthesis of BH4.
SR is a homodimeric protein belonging to the family of short-chain dehydrogenases/reductases and is inhibited by N-acetyl derivatives of both serotonin and dopamine (Smith et al. 1992). Although SR was thought to be specific for the 1’-oxo group of sepiapterin and PTP, it was well documented that for that group it has a broad substrate specificity to carbonyl compounds and thus can reduce many “typical” aldo-keto reductase substrates, such as p-nitrobenzaldehyde and 9,10-phenanthrenequinone (Katoh and Sueoka 1984, 1989; Sueoka and Katoh 1985). Both mouse and human SPR genes span a region of 4–5 kb, and the reading frames are split into three exons (Ota et al. 1995; Ohye et al. 1998; Lee et al. 1999). No alternative splice variants have been observed, and, except for the horse liver enzyme (Katoh 1971), not much is known about the expression and regulation of mammalian SR. The enzyme has been purified from and characterized in rat erythrocytes (Sueoka and Katoh 1982; Oyama et al. 1990), rat liver (Smith 1987; Citron et al. 1990), silkworm (Iino et al. 1992), Chlorobium tepidum (Cho et al. 1999), Dictyostelium discoideum (Kim et al. 2000), and Drosophila melanogaster (Primus and Brown 1994; Ruiz-Vazquez et al. 1996; Seong et al. 2000).
In conjunction with two other NADPH-dependent reductases, SR can produce BH4 from PTP, without isomerase reaction (Milstien and Kaufman 1986; Levine et al. 1990). Although it has been shown that SR reduces 1’-oxo-TP to BH4, neither SR inhibitors nor SR antibodies were able to inhibit BH4 production, indicating that SR alone is not responsible for the synthesis of 1’-oxo-TP. The enzyme catalyzing this step was later identified as AR (Milstien and Kaufman 1989; Steinerstauch et al. 1989). By use of antibodies specific to AR, BH4 biosynthesis was inhibited to ∼60% in crude rat-brain extracts, suggesting both that the reaction sequence catalyzed by AR accounts for approximately half of BH4 biosynthesis flux in rat brain and that reduction of 1’- and 2’-oxo groups proceeds at approximately equal rates (Milstien and Kaufman 1989). AR, in addition to converting PTP to 1’-oxo-TP, also catalyzes the reduction of 2’-oxo-TP to BH4 (Park et al. 1991). Thus, in the presence of CR (Wermuth 1982), an alternative pathway converting PTP to BH4 in the absence of SR could be functional in vivo. CR can reduce PTP both to 1’-oxo-TP and to 2’-oxo-TP; however, the rate of production is much more preferable for the 1’-oxo intermediate (Park et al. 1991).
Park et al. (1991) have proposed that, in the case of complete SR deficiency, BH4 could be synthesized only by the concerted action of both AR and CR. Our data indicate that this may be true for some peripheral tissues but not for the brain. Expression of these two reductases is shown to be different in different tissues (Wermuth 1985; Wirth and Wermuth 1985; Forrest and Gonzalez 2000), and our results on fibroblasts from patients with SR deficiency suggest that CR is not fully active in human skin fibroblasts. This suggestion is supported by the fact that, in fibroblasts from patients with SR deficiency, we found high neopterin production and no biopterin production, after stimulation with cytokines (table 1). Obviously, PTP either is not reduced to the 1’- and 2’-oxo intermediates or is first reduced to 1’-oxo-TP and subsequently is converted nonenzymatically to sepiapterin. In the absence of SR and CR, sepiapterin cannot be further reduced to dihydrobiopterin (fig. 1, pathway D). Furthermore, one can speculate that sepiapterin or one of its degradation products may inhibit PTPS, resulting in accumulation of neopterin. Because sepiapterin, used as a substrate in the SR assay, has been shown to be a substrate also for CR (Park et al. 1991), we conclude either that CR is not expressed in skin fibroblasts or that its expression therein is very low.
In mammalian peripheral tissue, such as liver, kidney, or small intestine, both reductases are active, and, in the case of SR deficiency, BH4 is synthesized through the salvage pathway (fig. 1, pathway C). Sepiapterin formed nonenzymatically from 1’-oxo-TP is, in peripheral tissue, reduced to dihydrobiopterin by CR. The final reduction to BH4 is catalyzed by the enzyme dihydrofolate reductase (DHFR). Alternatively, a small portion of BH4 can be synthesized from PTP by the concerted action of CR and AR, via 2’-oxo-TP. A similar, alternative pathway involving two CRs (CR I and CR II) has been described in the lemon mutant silkworm (Iino et al. 2000). The normal activity of SR that we measured in the red blood cells of our patients (table 1) is most probably due to CR activity, which also converts sepiapterin to dihydrobiopterin (Park et al. 1991). In our patients, biosynthesis of BH4 was obviously sufficient, under physiologic conditions, for the hepatic hydroxylation of phenalalanine to tyrosine. Urinary neopterin and biopterin (table 1) and plasma neopterin and biopterin (data not shown) were normal, and blood phenylalanine levels were never elevated. However, on challenge with phenylalanine the intracellular BH4 concentrations may be not sufficient for normal function of the phenylalanine-hydroxylating system. This was documented, in both patients, by an abnormal phenylalanine–loading test, which was corrected by preloading with BH4 (Blau et al. 1999).
Although human brain contains large amounts of AR (Wermuth et al. 1982) and CR (Wermuth 1981), the activity of DHFR is ∼10× lower in the brain than in the liver (Ludwig et al. 1987), and it is ∼2,000× lower than that of DHPR (Kaufman 1991). Thus, sepiapterin is reduced to dihydrobiopterin by CR but cannot be further reduced to BH4, owing to low DHFR activity (fig. 1, pathway B). This pathway would explain our finding of high concentrations of total biopterin and dihydrobiopterin in CSF of patients with SR deficiency (table 1). Sepiapterin, another potential metabolite that may also accumulate in patients with SR deficiency, is a yellow-fluorescent compound and cannot be detected in our HPLC system for blue-fluorescent neopterin and biopterin.
In conclusion, we have described the first patients in whom a new inborn error of BH4 metabolism causes severe monoamine-neurotransmitter deficiency without hyperphenylalaninemia. We have also proposed the metabolic routes, in different tissues, for BH4. The exact mechanisms of action of CR and AR need to be further elucidated.
Acknowledgments
The authors thank Drs. A. Romstad (Glostrup) and I. Dianzani (Turin), for DHPR mutation analysis; Dr. Y. S. Park, for kindly providing the rabbit anti–mouse SR antibodies; W. Leimbacher, for western blot analysis; L. Kierat, for HPLC analysis; and M. Killen, for editorial work. This work was supported by Swiss National Science Foundation grant 31-54183.98.
Electronic-Database Information
- BIODEF: International Database of Tetrahydrobiopterin Deficiencies, http://www.bh4.org/biodef1.html (for patients 229 and 360) [DOI] [PubMed]
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for SR genomic DNA [accession numbers AB017547 and AB017548] and SR cDNA [accession number M76231])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DRD [MIM 600225], PKU [MIM 261600], PTPS deficiency [MIM 261640], and DHPR deficiency [MIM 261630])
References
- Auerbach G, Herrmann A, Gütlich M, Fischer M, Jacob U, Bacher A, Huber R (1997) The 1.25 Å crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters. EMBO J 16:7219–7230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau N, Barnes I, Dhondt JL (1996) International database of tetrahydrobiopterin deficiencies. J Inherit Metab Dis 19:8–14 [DOI] [PubMed] [Google Scholar]
- Blau N, Thöny B, Cotton RGH, Hyland K (2001) Disorders of tetrahydrobiopterin and related biogenic amines. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Vogelstein B (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw-Hill, New York, pp 1725–1776 [Google Scholar]
- Blau N, Thöny B, Renneberg A, Arnold LA, Hyland K (1998) Dihydropteridine reductase deficiency localized to the central nervous system. J Inherit Metab Dis 21:433–434 [DOI] [PubMed] [Google Scholar]
- Blau N, Thöny B, Renneberg A, Penzien JM, Hyland K, Hoffmann G (1999) Variant of dihydropteridine reductase deficiency without hyperphenylalaninemia: effect of oral phenylalanine loading. J Inherit Metab Dis 22:216–220 [DOI] [PubMed] [Google Scholar]
- Bonafé L, Thöny B, Leimbacher W, Kierat L, Blau N (2001) Diagnosis of dopa-responsive dystonia and other tetrahydrobiopterin disorders by the study of biopterin metabolism in fibroblasts. Clin Chem 47:477–485 [PubMed] [Google Scholar]
- Cho SH, Na JU, Youn H, Hwang CS, Lee CH, Kang SO (1999) Sepiapterin reductase producing l-threo-dihydrobiopterin from Chlorobium tepidum. Biochem J 340:497–503 [PMC free article] [PubMed] [Google Scholar]
- Citron BA, Milstien S, Gutierrez JC, Levine RA, Yanak BL, Kaufman S (1990) Isolation and expression of rat liver sepiapterin reductase cDNA. Proc Natl Acad Sci USA 87:6436–6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtius HC, Blau N, Kuster T (1991) Pterins. In: Hommes FA (ed) Techniques in diagnostic human biochemical genetics. Wiley-Liss, New York, pp 377–396 [Google Scholar]
- Ferre J, Naylor EW (1988) Sepiapterin reductase in human amniotic and skin fibroblasts, chorionic villi, and various blood fractions. Clin Chim Acta 174:271–282 [DOI] [PubMed] [Google Scholar]
- Forrest GL, Gonzalez B (2000) Carbonyl reductase. Chem Biol Interact 129:21–40 [DOI] [PubMed] [Google Scholar]
- Ichinose H, Katoh S, Sueoka T, Titani K, Fujita K, Nagatsu T (1991) Cloning and sequencing of cDNA encoding human sepiapterin reductase—an enzyme involved in tetrahydrobiopterin biosynthesis. Biochem Biophys Res Commun 179:183–189 [DOI] [PubMed] [Google Scholar]
- Iino T, Dohke K, Tsusue M (1992) The purification and characterisation of sepiapterin reductase from fat body of the silkworm Bombyx mori. Zoological Sci 9:119–125 [Google Scholar]
- Iino T, Takikawa SI, Yamamoto T, Sawada H (2000) The enzyme that synthesizes tetrahydrobiopterin from 6-pyruvoyl-tetrahydropterin in the lemon mutant silkworm consists of two carbonyl reductases. Arch Biochem Biophys 373:442–446 [DOI] [PubMed] [Google Scholar]
- Katoh S (1971) Sepiapterin reductase from horse liver: purification and properties of the enzyme. Arch Biochem Biophys 146:202–214 [DOI] [PubMed] [Google Scholar]
- Katoh S, Sueoka T (1984) Sepiapterin reductase exhibits a NADPH-dependent dicarbonyl reductase activity. Biochem Biophys Res Commun 118:859–866 [DOI] [PubMed] [Google Scholar]
- ——— (1987) Isomerization of 6-lactoyl tetrahydropterin by sepiapterin reductase. J Biochem 101:275–278 [DOI] [PubMed] [Google Scholar]
- ——— (1988) Coenzyme stimulation of isomerase activity of sepiapterin reductase in the biosynthesis of tetrahydrobiopterin. J Biochem (Tokyo) 103:286–289 [DOI] [PubMed] [Google Scholar]
- ——— (1989) Properties of carbonyl reductase activity of sepiapterin reductase, an enzyme involved in the biosynthesis of tetrahydrobiopterin. Prog Clin Biol Res 290:381–395 [PubMed] [Google Scholar]
- Kaufman S (1991) Some metabolic relationships between biopterin and folate: implications for the “methyl trap hypothesis.” Neurochem Res 16:1031–1036 [DOI] [PubMed] [Google Scholar]
- Kim YA, Chung HJ, Kim YJ, Choi YK, Hwang YK, Lee SW, Park YS (2000) Characterization of recombinant Dictyostelium discoideum sepiapterin reductase expressed in E. coli. Mol Cells 10:405–410 [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- Lee SW, Park IY, Hahn YS, Lee JE, Seong CS, Chung JH, Park YS (1999) Cloning of mouse sepiapterin reductase gene and characterization of its promoter region. Biochim Biophys Acta 1445:165–171 [DOI] [PubMed] [Google Scholar]
- Levine RA, Kapatos G, Kaufman S, Milstien S (1990) Immunological evidence for the requirement of sepiapterin reductase for tetrahydrobiopterin biosynthesis in brain. J Neurochem 54:1218–1224 [DOI] [PubMed] [Google Scholar]
- Ludwig R, Frei E, Kimmig B, Brandeis WE (1987) Dihydrofolate reductase-activity in brain tissue: effect of X-irradiation. Blut 55:483–488 [DOI] [PubMed] [Google Scholar]
- Milstien S, Kaufman S (1986) The biosynthesis of tetrahydrobiopterin in rat brain. In: Cooper BA, Whitehead VM (eds) Chemistry and biology of pteridines. Walter de Gruyter, Berlin, pp 169–181 [Google Scholar]
- ——— (1989) Immunological studies on the participation of 6-pyruvoyl tetrahydropterin (2'-oxo) reductase, an aldose reductase, in tetrahydrobiopterin biosynthesis. Biochem Biophys Res Commun 165:845–850 [DOI] [PubMed] [Google Scholar]
- Nygaard TG, Wooten GF (1998) Dopa-responsive dystonia: some pieces of the puzzle are still missing. Neurology 50:853–855 [DOI] [PubMed] [Google Scholar]
- Ohye T, Hori TA, Katoh S, Nagatsu T, Ichinose H (1998) Genomic organization and chromosomal localization of the human sepiapterin reductase gene. Biochem Biophys Res Commun 251:597–602 [DOI] [PubMed] [Google Scholar]
- Ota A, Ichinose H, Nagatsu T (1995) Mouse sepiapterin reductase: an enzyme involved in the final step of tetrahydrobiopterin biosynthesis. Primary structure deduced from the cDNA sequence. Biochim Biophys Acta 1260:320–322 [DOI] [PubMed] [Google Scholar]
- Oyama R, Katoh S, Sueoka T, Suzuki M, Ichinose H, Nagatsu T, Titani K (1990) The complete amino acid sequence of the mature form of rat sepiapterin reductase. Biochem Biophys Res Commun 173:627–631 [DOI] [PubMed] [Google Scholar]
- Park YS, Heizmann CW, Wermuth B, Levine RA, Steinerstauch P, Guzman J, Blau N (1991) Human carbonyl and aldose reductases: new catalytic functions in tetrahydrobiopterin biosynthesis. Biochem Biophys Res Commun 175:738–744 [DOI] [PubMed] [Google Scholar]
- Primus JP, Brown GM (1994) Sepiapterin reductase and the biosynthesis of tetrahydrobiopterin in Drosophila melanogaster. Insect Biochem Mol Biol 24:907–918 [DOI] [PubMed] [Google Scholar]
- Ruiz-Vazquez P, Silva FJ, Ferre J (1996) Characterization of sepiapterin reductase activity from Drosophila melanogaster. Comp Biochem Physiol B Biochem Mol Biol 113:131–136 [DOI] [PubMed] [Google Scholar]
- Seong C, Baek K, Yoon J (2000) Structure, chromosomal localization, and expression of the Drosophila melanogaster gene encoding sepiapterin reductase. Gene 255:357–361 [DOI] [PubMed] [Google Scholar]
- Smith GK (1987) On the role of sepiapterin reductase in the biosynthesis of tetrahydrobiopterin. Arch Biochem Biophys 255:254–266 [DOI] [PubMed] [Google Scholar]
- Smith GK, Duch DS, Edelstein MP, Bigham EC (1992) New inhibitors of sepiapterin reductase: lack of an effect of intracellular tetrahydrobiopterin depletion upon in vitro proliferation of two human cell lines. J Biol Chem 267:5599–5607 [PubMed] [Google Scholar]
- Steinerstauch P, Wermuth B, Leimbacher W, Curtius HC (1989) Human liver 6-pyruvoyl tetrahydropterin reductase is biochemically and immunologically indistinguishable from aldose reductase. Biochem Biophys Res Commun 164:1130–1136 [DOI] [PubMed] [Google Scholar]
- Sueoka T, Katoh S (1982) Purification and characterization of sepiapterin reductase from rat erythrocytes. Biochim Biophys Acta 717:265–271 [DOI] [PubMed] [Google Scholar]
- ——— (1985) Carbonyl reductase activity of sepiapterin reductase from rat erythrocytes. Biochim Biophys Acta 843:193–198 [DOI] [PubMed] [Google Scholar]
- Thöny B, Auerbach G, Blau N (2000) Tetrahydrobiopterin biosynthesis, regeneration, and functions. Biochem J 347:1–26 [PMC free article] [PubMed] [Google Scholar]
- Thöny B, Leimbacher W, Blau N, Harvie A, Heizmann CW (1994) Hyperphenylalaninemia due to defects in tetrahydrobiopterin metabolism: molecular characterization of mutations in 6-pyruvoyl-tetrahydropterin synthase. Am J Hum Genet 54:782–792 [PMC free article] [PubMed] [Google Scholar]
- Wermuth B (1981) Purification and properties of an NADPH-dependent carbonyl reductase from human brain: relationship to prostaglandin 9-ketoreductase and xenobiotic ketone reductase. J Biol Chem 256:1206–1213 [PubMed] [Google Scholar]
- ——— (1982) Human carbonyl reductases. Prog Clin Biol Res 114:261–274 [PubMed] [Google Scholar]
- ——— (1985) Aldo-keto reductases. Prog Clin Biol Res 174:209–230 [PubMed] [Google Scholar]
- Wermuth B, Burgisser H, Bohren K, von Wartburg JP (1982) Purification and characterization of human-brain aldose reductase. Eur J Biochem 127:279–284 [DOI] [PubMed] [Google Scholar]
- Wirth HP, Wermuth B (1985) Immunohistochemical localisation of aldehyde and aldose reductase in human tissues. Prog Clin Biol Res 174:231–239 [PubMed] [Google Scholar]