Abstract
Mutations in the extracellular domain of the 55-kD tumor-necrosis factor (TNF) receptor (TNFRSF1A), a key regulator of inflammation, define a periodic-fever syndrome, TRAPS (TNF receptor–associated periodic syndrome [MIM 142680]), which is characterized by attacks of fever, sterile peritonitis, arthralgia, myalgia, skin rash, and/or conjunctivitis; some patients also develop systemic amyloidosis. Elsewhere we have described six disease-associated TNFRSF1A mutations, five of which disrupt extracellular cysteines involved in disulfide bonds; four other mutations have subsequently been reported. Among 150 additional patients with unexplained periodic fevers, we have identified four novel TNFRSF1A mutations (H22Y, C33G, S86P, and c.193−14 G→A), one mutation (C30S) described by another group, and two substitutions (P46L and R92Q) present in ∼1% of control chromosomes. The increased frequency of P46L and R92Q among patients with periodic fever, as well as functional studies of TNFRSF1A, argue that these are low-penetrance mutations rather than benign polymorphisms. The c.193−14 G→A mutation creates a splice-acceptor site upstream of exon 3, resulting in a transcript encoding four additional extracellular amino acids. T50M and c.193−14 G→A occur at CpG hotspots, and haplotype analysis is consistent with recurrent mutations at these sites. In contrast, although R92Q also arises at a CpG motif, we identified a common founder chromosome in unrelated individuals with this substitution. Genotype-phenotype studies identified, as carriers of cysteine mutations, 13 of 14 patients with TRAPS and amyloidosis and indicated a lower penetrance of TRAPS symptoms in individuals with noncysteine mutations. In two families with dominantly inherited disease and in 90 sporadic cases that presented with a compatible clinical history, we have not identified any TNFRSF1A mutation, despite comprehensive genomic sequencing of all of the exons, therefore suggesting further genetic heterogeneity of the periodic-fever syndromes.
Introduction
Until recently, the dominantly inherited periodic-fever syndromes represented a poorly understood group of inflammatory diseases. Clinically, these disorders are characterized by recurrent prolonged (>1 wk) attacks of fever associated with abdominal pain, myalgia, migratory erythematous skin rash, conjuctivitis, and/or periorbital edema (McDermott et al. 1997). In contrast to patients with familial Mediterranean fever (FMF [MIM 249100]), which is recessively inherited, patients with dominantly inherited periodic fever usually have a poor response to colchicine but a favorable response to corticosteroids. Dominantly inherited periodic fevers have been described mostly in families of northern-European descent (Williamson et al. 1982; Gertz et al. 1987; McDermott et al. 1997; Zaks and Kastner 1997).
In 1998, linkage studies identified, in the same region of chromosome 12p13, a candidate interval for two clinical variants—benign autosomal dominant familial periodic fever (FPF) (Mulley et al. 1998) and familial Hibernian fever (FHF [MIM 142680]) (McDermott et al. 1998). One of the several candidate genes in this interval is TNFRSF1A, the gene encoding the 55-kD receptor for tumor-necrosis factor (TNF) α (TNF-α). Subsequently, in 1999 we identified six mutations in the TNFRSF1A sequence encoding the extracellular domain of the p55 receptor in seven affected families, including those previously denoted as having FPF and FHF (McDermott et al. 1999). This was the first report of TNFRSF1A mutations in human disease.
In our initial report, all of the mutations were single-nucleotide missense substitutions, with five of the six mutations disrupting highly conserved cysteine residues involved in extracellular disulfide bonds. Although five of the first seven families were of either Irish or Scottish descent, one was Finnish and another was French Canadian. In light of the more diverse ethnic distribution, we have proposed the term “TNF receptor–associated periodic syndrome” (TRAPS) for those patients with TNFRSF1A mutations.
In vitro studies of leukocytes from patients bearing the C52F mutation indicated an impairment in the normal activation-induced cleavage of cell-surface p55 receptors. Receptor clearance is thought to have a negative homeostatic effect, both by preventing repeated stimulation through membrane p55 and by creating a pool of antagonistic soluble receptors, and thus a cleavage defect could at least partially account for the hyperinflammatory phenotype in TRAPS.
Four additional TRAPS mutations in the extracellular domain of TNFRSF1A have subsequently been reported by other groups: C30S (Dodé et al. 2000), R92P (Aganna et al. 2001), C55S (Jadoul et al. 2001), and C70Y (Simon et al. 2001). Families were of French, Dutch, Belgian, and Dutch-Indonesian descent, respectively. Individuals affected with each mutation manifested typical TRAPS inflammatory attacks, and amyloidosis was documented in patients with either the C70Y mutation or the C55S mutation.
Here we report a new group of patients with TRAPS, including both familial and sporadic cases. We identified four mutations, two of which were cysteine substitutions, that were not present among a panel of ethnically matched control chromosomes. Two other substitutions, neither of which involved cysteines, were found in ∼1% of control chromosomes. On the basis of their clinical presentation, their higher frequency among periodic fever patients, and functional studies of TNFRSF1A in vitro and in vivo, these latter two substitutions are likely to be low-penetrance mutations rather than polymorphisms. We have also investigated the carrier haplotypes associated with mutations observed in unrelated patients, the penetrance of the 16 known TRAPS mutations, and possible genotype-phenotype relationships. Our data strongly suggest that cysteine substitutions lead to a higher risk of amyloidosis—and, probably, to a higher penetrance—than do the noncysteine mutations. Finally, we report evidence for further genetic heterogeneity of periodic-fever syndromes, with one clinically consistent family not having linkage to chromosome 12p13 and with 90 sporadic cases without demonstrable mutations in the coding region of TNFRSF1A.
Subjects, Material, and Methods
Patients and DNA Specimens
The present study included patients referred to the National Institutes of Health for clinical and/or molecular diagnosis of unexplained periodic fevers. The study was approved by the human-experimentation committee at the National Institutes of Health, and consent was obtained from all participants. Genomic DNA was extracted from whole blood by a commercially available kit (Puregene; Gentra Systems).
TNFRSF1A Mutation Detection
Genomic DNAs from 150 patients (137 unrelated individuals) with symptoms compatible with TRAPS were sequenced for exons 2–5 of TNFRSF1A, encoding the extracellular domain, as described elsewhere (McDermott et al. 1999). Samples of genomic DNA from 90 patients were analyzed by the combination of denaturing high-performance liquid chromatography (DHPLC) and sequence analysis, for all 10 exons of TNFRSF1A. Patients from two families initially negative for a mutation in the extracellular domain of the gene were also sequenced through the complete coding region and promoter (450 bp). The forward and reverse oligonucleotide primers used to amplify regulatory sequences of the TNFRSF1A gene were 5′-TCACTGCTGTGATCCATCTGGG-3′ and 5′-TTCTCACCAGTGGCAGCAGCAG-3′. Other exons were sequenced as described elsewhere (McDermott et al. 1999), except that exons 8 and 9 were analyzed separately, with the following primers: exon 8, forward primer 5′-TCGAGCTGCATCAGTAGCTCT-3′ and reverse primer 5′-TTCTACGTGGGGGTTGGGAC-3′; exon 9, forward primer 5′-TAAGTCCCAACCCCCACGTAG-3′ and reverse primer 5′-CAGCCTCCTCGTCTCCAG-3′.
Mutation detection was performed by fluorescent sequencing with dye-primer chemistry (Amersham), on an ABI 377 automated sequencer, and by DHPLC (Transgenomics). PCR was performed with AmpliTaq Gold (PE Biosystems) in 50-μl reactions with 10–50 ng of genomic DNA and with the same primers, described above, that were used for exon sequencing. DHPLC analysis of amplicons was performed at temperatures modified for each of the exons, according to WAVE-MAKER 4.0 software (Transgenomics). Amplicons showing a DHPLC elution shift were confirmed in a repeat sequencing analysis. The sequencing data were analyzed by SEQUENCHER 3.0 (Gene Codes).
Microsatellite and Single-Nucleotide Polymorphism (SNP) Analysis
Genotyping in the chromosomal region of the TNFRSF1A locus was performed with the following markers: D12S99, D12S356, TNFRP55, CD4, D12S1695, D12S77, and D12S364, as described elsewhere (McDermott et al. 1998; Mulley et al. 1998; Eskadele et al. 2000). The two intragenic SNPs utilized in the present study were recently reported (Bazzoni et al. 2000) and have been submitted to HGSBASE, with accession numbers SNP000005194 and SNP000005195. Allele frequencies of G (.47) and of A (.53), for SNP000005194 (exon 1 SNP), and of C (.32) and of T (.68), for SNP000005195 (intron 5 SNP), were determined in 71 healthy controls. TNFRSF1A intragenic SNPs of our patients were determined by DNA sequencing.
Genotyping for the Muckle-Wells syndrome (MWS [MIM 191900])/familial cold urticaria (FCU [MIM 120100]) locus on distal chromosome 1q44 was performed with the D1S2682 and D1S423 microsatellites, which define the boundaries of the MWS/FCU critical interval (Cuisset et al. 1999; Hoffman et al. 2000; McDermott et al. 2000). Since the marker D1S423 was not informative, we also used two other microsatellites close to the centromeric boundary of the candidate interval, D1S1609 and D1S547. Microsatellite haplotypes were constructed so as to minimize the number of intrafamilial recombinations.
RNA Extraction
Total RNA was extracted from leukocytes by use of Trizol Reagent (GIBCO-BRL), as described elsewhere (Centola et al. 2000). First-strand cDNA synthesis was performed with the GIBCO Superscript Preamplification System, according to the manufacturer’s recommendations. The cDNA template was amplified by PCR with the following primers: forward primer 5′-ATTGGACTGGTCCCTCACCTAGGG-3′ and reverse primer 5′-TCTCACACTCCCTGCAGTAC-3′.
Construction of a Minigene to Analyze Aberrant Splicing
DNA fragments comprising exon 2, intron 3, and exon 3 were obtained, by PCR amplification, from the proband and control DNA samples, by use of the following oligonucleotides: forward primer 5′-CGAGAATTCATGCGTGCTCCTGGAGCTGTTGGTG-3′ and reverse primer 5′-TCGCTCGAGTCACCTTTCGGCATTTGGAGCAGCTG-3′. The forward primer contained an EcoRI restriction site and four nucleotides (ATGC), to create a methionine codon in-frame with the open reading frame of exon 2; the reverse primer contained an XhoI site followed by a stop codon. PCR amplification was performed with AmpliTaq Gold (PE Applied Biosystems), and cycling conditions were 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, for 30 cycles, followed by extension at 72°C for 7 min. The amplicons were ligated into pCR2.1 vector by use of the TOPO TA cloning kit (Invitrogen), and the resulting colonies were analyzed, by restriction digestion and dye-primer sequencing, for expected mutation-containing inserts. Verified inserts were cloned into pcDNA3 expression vector (Invitrogen) by ligation of the XhoI/EcoRI insert fragments from TA clones with XhoI/EcoRI-digested vector. Expression plasmids were transfected into Cos-7 cells by use of Lipofectamine reagent (Gibco/BRL), as described by the manufacturer. After 24 h of expression, RNA was extracted from transfected cells by use of Trizol reagent (Gibco/BRL), and bulk cDNA was prepared by the GIBCO Superscript Preamplification System, according to the manufacturer’s recommendations.
Cell Activation and Flow-Cytometric Analysis
TNFRSF1A shedding assays were performed as described elsewhere (McDermott et al. 1999). In brief, blood cells were washed and incubated at 37°C for 10 min in Hanks’ balanced salt solution containing 1% fetal calf serum (FCS), with or without 4-β-phorbol 12-myristate 13-acetate (PMA; Sigma). Cells were washed with HBSS/1% FCS at 4°C, and erythrocytes were lysed by use of ammonium chloride potassium lysing buffer (BioWhittaker). Cells were incubated with phycoerythrin (PE)-conjugated anti-TNFRSF1A monoclonal antibodies (Caltag) and isotype controls (Becton Dickinson), for 30 min at 4°C, and were washed three times with phosphate-buffered saline containing 0.5% bovine serum albumin. Expression of membrane TNFRSF1A on monocytes was analyzed by flow cytometry, by a FACS Calibur flow cytometer (Becton Dickinson). Soluble levels of TNFRSF1A were detected by solid-phase enzyme-linked immunosorbent assay, as described elsewhere (McDermott et al. 1999).
Statistical Analysis
Statistical analyses were performed by the SAS 6.12 and Epi Info 6 statistical software packages (Centers for Disease Control and Prevention [CDC]). Comparison of cysteine mutations, in patients versus a control group of healthy individuals and in patients with amyloidosis versus patients without amyloidosis, were performed by χ2 statistics. Results were reported as odds ratios with 95% confidence intervals. All P values were Yates corrected.
Results
TNFRSF1A Mutational Survey
At the National Institutes of Health Clinical Center, a referral center for periodic-fever syndromes, we collected DNA samples from >800 patients suffering from periodic fever and accompanying symptoms. The 150 patients who underwent screening for mutations in TNFRSF1A were selected on the basis of a clinical history suggestive of TRAPS (i.e., attacks lasting >1 wk, characteristic skin rash, and/or nonresponsiveness to colchicine). The patients were screened for mutations in exons 2–5 of TNFRSF1A, where all of the initially reported mutations clustered (McDermott et al. 1999). We used automated sequencing of PCR-amplified genomic DNAs, since, in our initial report, all but one of the mutations was private. Ninety patients underwent further comprehensive analysis of the entire TNFRSF1A coding region (i.e., 10 exons). This new cohort of patients with TRAPS with demonstrable TNFRSF1A mutations included several multiplex families and 10 sporadic cases of various ethnic backgrounds. Representative pedigrees are shown in figure 1.
Figure 1.
TRAPS pedigrees with novel TNFRSF1A mutations. Affected individuals are indicated by blackened symbols; individuals who are mutation positive but asymptomatic are represented by gray-shaded symbols. For the H22Y, R92Q, and c.193−14 G→A mutations, only representative families are shown.
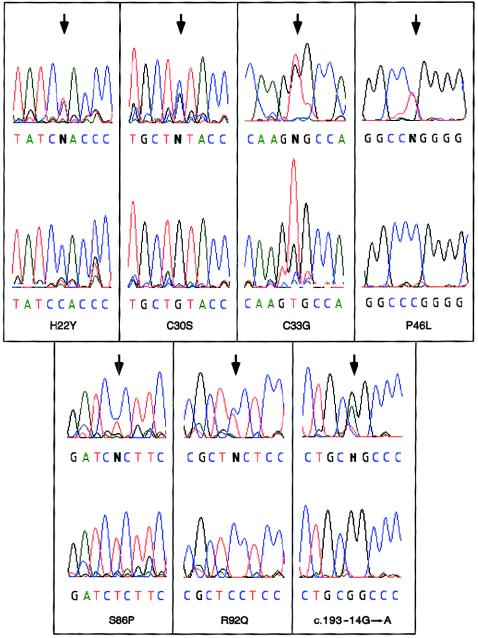
Among these patients, we identified seven new TNFRSF1A mutations, one of which was recently reported by another group (Dodé et al. 2000). The mutations are listed in table 1, and corresponding DNA-sequence electropherograms are shown in figure 2. Amino acid positions are defined relative to the signal peptide–cleavage site. H22Y is a novel mutation detected in the father and three affected children in a family of Scottish/German background and in one case, of mixed northern-European background, that initially appeared to be sporadic, with lifelong unexplained periodic fevers leading to 170 hospitalizations and 13 exploratory surgeries. Subsequently, we found that the latter patient’s father, who is also of mixed northern-European background, also suffered from mild symptoms of TRAPS and bore the H22Y mutation. This substitution was not observed either in the only available healthy member (i.e., the mother) of each H22Y-positive family or in any of >100 northern-European control chromosomes screened by genomic sequencing.
Table 1.
Summary of New TRAPS Mutations
|
No. of |
||||
| Mutation(Nucleotide Change) | Location | Patients | Families | Ethnicity |
| H22Y (151 C→T) | Exon 2 | 6 | 2 | Scottish/German, mixed northern European |
| C30S (176 G→C) | Exon 2 | 3 | 1 | Irish |
| C33G (184 T→G) | Exon 2 | 2 | 1 | Puerto Rican |
| P46L (254 C→T) | Exon 2 | 1 | 1 | African American/northern European |
| S86P (343 T→C) | Exon 4 | 4 | 1 | Scottish/Irish |
| R92Q362 G→A | Exon 4 | 9 | 9 | Mexican (1), Italian (2), Irish (1), English (1), Portuguese (1), Ashkenazi (1), mixed northern European (2) |
| Splice (c.193−14 G→A) | Intron 3 | 5 | 2 | Scottish/Arab |
Figure 2.
DNA sequence electropherograms for the seven TNFRSF1A mutations identified in the present study. For each sequence, the upper tracing is from a patient, and the lower tracing is from a normal control.
C30S and C33G mutations were found at the same highly conserved cysteine residues where we previously had reported the C30R and C33Y mutations, and neither C30S nor C33G was found in any of >100 northern-European control chromosomes. The C30S mutation was observed in an Irish American family with three affected members, and has also recently been described in a French family (Dodé et al. 2000). The C33G mutation was found in a father and daughter originally from Puerto Rico, with histories of recurrent fever, abdominal pain, and arthralgia since birth. They had been treated with corticosteroids for many years. The father had developed progressive hepatic amyloidosis, eventually necessitating liver transplantation. The mother and their other three children were healthy and negative for this mutation.
S86P is a new private mutation found in a three-generation family, of Scottish/Irish descent, with four affected members (fig. 1) who experienced episodes of severe abdominal pain, high fever, and arthralgia lasting 6–8 wk. This substitution was observed in none of 100 northern-European control chromosomes. One of the affected sibs in the third generation is symptomatic but declined genetic testing.
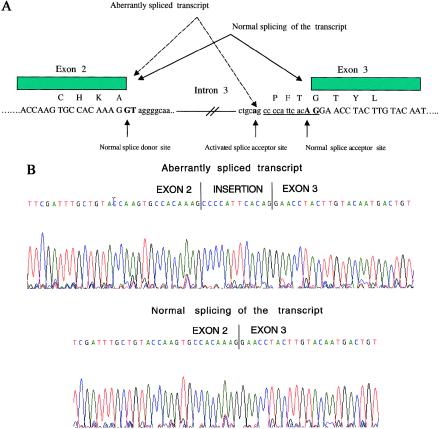
Screening of intronic sequences flanking the exons identified one splicing mutation. In a family of Scottish descent and in a family of Arab descent (fig. 1; Scottish family is not shown), we observed a G→A change in intron 3 at position −14 relative to codon 193 (fig. 2). This substitution was not detected in any of 100 control chromosomes and potentially could create a new consensus splice-acceptor sequence (i.e., AG) in the presence of a preceding polypyrimidine tract (fig. 3A). We obtained mRNA from one of the patients carrying this substitution, performed reverse transcriptase–PCR, and detected a band slightly larger than the normal-size transcript. Sequencing of the cloned larger band revealed a transcript encoding an additional four amino acids, as would be expected with activation of this new splice-acceptor site in intron 3.
Figure 3.
A, Diagram of normal and aberrantly spliced TNFRSF1A transcript. The conventional splice-donor site, splice-acceptor site, and mutated nucleotide in intron 3 are in boldface. The c.193−14 G→A mutation creates a new splice-acceptor site, as shown. Single-letter designations for amino acids are shown immediately above the first nucleotide in each codon. Four additional amino acids that are present in the alternatively spliced transcript are encoded by the nucleotides that are underlined. B, DNA-sequence electropherograms showing in vitro splicing of c.193−14 G→A transcripts (top) and normal transcripts (bottom) in Cos-7 cells.
We also analyzed the effect of this substitution on pre-mRNA splicing, by examining Cos-7 cells transfected with a minigene construct harboring either the mutant A nucleotide or the wild-type G. Cytoplasmic RNA isolated from transfected Cos-7 cells showed that the mutant construct produced aberrantly spliced transcript with an extra four amino acids inserted between exons 2 and 3 (fig. 3B). The wild-type construct resulted in normal splicing of the TNFRSF1A transcript. This substitution cosegregated with affection status in both families but with reduced penetrance in two members of the Arab family and in one member of the Scottish family.
Low-Penetrance TNFRSF1A Mutations
Finally, two additional missense transitions, P46L and R92Q, were identified in patients with periodic fever and at low frequencies in the general population. P46L was detected in one sporadic patient of mixed ancestry who suffered from periodic fevers that sometimes lasted >1 mo and that were associated with severe abdominal pain, vomiting, and leg pain. We found that the patient inherited this substitution from his father, who is of mixed African American and northern-European ancestry. P46L was detected in 1 of 170 northern-European control chromosomes and in 3 of 156 African American control chromosomes, at a combined gene frequency of 1.23%. R92Q was the most common TNFRSF1A sequence change identified in our panel, occurring in 9 of 137 unrelated patients. All were sporadic cases, representing a range of ethnicities. Since, in five cases, this substitution was detected in asymptomatic parents, we investigated the frequency of R92Q in the Irish and North American control populations. R92Q was detected in 2 of 132 Irish control chromosomes and in 6 of 634 North American (mostly northern-European) control chromosomes, at a combined gene frequency of 1.04% in both populations, versus 9 (3.3%) of 274 independent chromosomes in the panel with a clinical picture suggestive of TRAPS (P=.02).
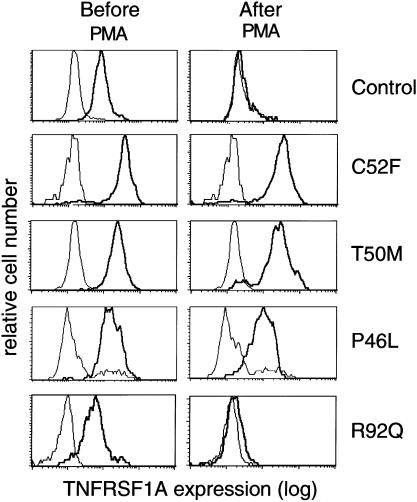
To explore further the significance of P46L and R92Q, we examined cell-surface TNFRSF1A in peripheral blood monocytes before and after in vitro stimulation with PMA. We have previously demonstrated that leukocytes from patients with the C52F mutation show both increased baseline levels of membrane TNFRSF1A and, after stimulation with PMA, a potent inducer of the metalloproteases mediating ectodomain cleavage, reduced receptor cleavage (shedding). Figure 4 shows TNFRSF1A expression on monocytes from patients bearing the C52F, T50M, P46L, or R92Q substitutions and from a control sample. As previously reported, unstimulated C52F monocytes, relative to controls, had more membrane TNFRSF1A and showed minimal receptor clearance, after PMA stimulation. Comparable results, both before and after PMA stimulation, were observed with cells bearing T50M. Consistent with the view that P46L has functional consequences, monocytes from patients bearing this substitution did not completely clear TNFRSF1A after PMA stimulation, although there was more shedding than was seen with either T50M or C52F. Variable degrees of impairment of PMA-induced TNFRSF1A shedding were also observed in monocytes from patients with the H22Y, C30S, or C33G mutations (data not shown).
Figure 4.
Defective clearance of TNFRSF1A in patients with TRAPS. Monocytes were analyzed for TNFRSF1A expression (thicker line in each panel), before (left-hand panels) or after (right-hand panels) PMA activation (100 ng/ml) for 10 min. Single-cell suspensions (106 cells) were incubated with either anti–TNFRSF1A-PE or isotype-matched IgG-PE as control (thinner line in each panel), were washed, and were analyzed by flow cytometry. Fluorescence histograms are shown for controls and for patients with the C52F, T50M, P46L, and R92Q TRAPS mutations.
In contrast, monocytes from patients bearing the R92Q substitution showed TNFRSF1A membrane staining and receptor shedding comparable to those in controls (fig. 4). Similar results were also observed for monocytes from patients with the c.193−14 G→A mutation (data not shown). Nevertheless, when we examined serum samples from a patient with R92Q that were taken during and between febrile episodes, we found that soluble p55 levels failed to increase with the attack (3,358 pg/ml baseline, 1,997 pg/ml during attack, normal range 749–1,966 pg/ml). Similar findings have recently been reported for the C30S mutation (Dodé et al. 2000). The failure of patients’ soluble TNFRSF1A levels to increase with inflammatory episodes suggests an in vivo functional abnormality that, in the case of R92Q, is not detected by the in vitro shedding assay.
Genotype-Phenotype Studies
When the present study is included, 100 symptomatic patients positive for a total of 16 mutations in TNFRSF1A have been reported in the literature (McDermott et al. 1999; Dodé et al. 2000; Aganna et al. 2001; Jadoul et al. 2001; Simon et al. 2001). Because systemic amyloidosis is the most serious manifestation of TRAPS, sometimes leading to hepatic and/or renal failure and death, we first investigated the possible association of amyloidosis with specific mutations (table 2). Among the 100 patients with TRAPS, this complication has been identified in 14. Despite the fact that only 9 of 16 known mutations in TNFRSF1A involve cysteine residues, 13 of 14 known cases of amyloidosis occurred among patients with cysteine mutations (C30R, C33G, C33Y, C52F, C55S, C70Y, or C88Y); only 1 patient with amyloidosis had T50M. When unaffected carriers of mutations were included, amyloidosis was found in 13 of 59 persons with cysteine substitutions but in only 1 of 58 with noncysteine mutations (P=.002, by Fisher’s exact test; odds ratio 16.11, 95% CI 2.06–341.95).
Table 2.
TNFRSF1A Mutations and Development of Amyloidosis
| Mutation | Frequencyof Amyloidosis |
| H22Y | 0/6 |
| C30R | 2/2 |
| C30S | 0/5 |
| C33G | 1/2 |
| C33Y | 2/17 |
| P46L | 0/1 |
| T50M | 1/18 |
| C52F | 3/5 |
| C55S | 3/3 |
| C70Y | 1/8 |
| S86P | 0/4 |
| C88R | 0/8 |
| C88Y | 1/4 |
| R92P | 0/3 |
| R92Q | 0/9 |
| Splice | 0/5 |
| Total | 14/100a |
Of the 14 cases of amyloidosis, 13 (93%) had cysteine mutations.
We also analyzed family data for the 16 known TRAPS mutations, to compare the penetrance of cysteine and noncysteine mutations. In our study and in data published by other groups (Dodé et al. 2000; Simon et al. 2001), 5 of 59 individuals identified with cysteine TNFRSF1A mutations had no reported symptoms, versus 12 of 58 individuals with noncysteine mutations (P=.11; Aganna et al. 2001; present study). Only 1 of 18 individuals positive for the T50M mutation was asymptomatic. Therefore, the total number of individuals positive for TNFRSF1A mutations was 117, with 100 of them being symptomatic carriers of various mutations and with 17 being asymptomatic carriers of various mutations.
Haplotype Analysis
We next asked whether unrelated carriers of the same mutation share a common founder haplotype. Data relevant to this issue were available for three mutations: T50M, R92Q, and c.193−14 G→A. T50M was initially observed in eight affected members of an Irish family with FHF (including patients 802, 804, and 805 in table 3) and in one available member (patient 806) of a French Canadian family (McDermott et al. 1999). Additionally, we found this mutation in 10 other patients: 3 members (patients 32, 510, and 511) of an Irish American family, 2 patients (patients 466 and 469) from a Scottish American family, 1 additional member (patient 807) of the French Canadian family, 1 patient (patient 594) from the United States who is of unknown descent, and, recently, 3 members of another Scottish American.
To test the hypothesis of common ancestry, we constructed haplotypes with both microsatellite and intragenic SNP markers. TNFR1P55 is an intragenic microsatellite marker that resides within intron 2 of TNFRSF1A (based on the genomic structure used by Fuchs et al. 1992), and we tested additional SNPs in exon 1 and in intron 5. D12S99 and D12S356 are telomeric flanking markers, whereas CD4, D12S1695, and D12S77 are centromeric flanking markers. The sequence of these markers and the published genetic distances between them are as follows: D12S99–.02–D12S356–.02–TNFRSF1A–.01–CD4–.03–D12S1695–.03–D12S77. Haplotypes were deduced from the relevant pedigrees. Among five families with T50M (the recently reported Scottish American family was not studied), we identified three intragenic haplotypes: A-150 bp-T, A-152 bp-T, and G-144 bp-C (table 3). In addition, we did not find a common microsatellite founder haplotype associated with the c.193−14 G→A splicing mutation, consistent with the fact that this mutation was detected in two families of very distinct ethnicity (data not shown).
Table 3.
Microsatellite and Intragenic SNP Haplotypes of TRAPS T50M Carrier Chromosomes and of R92Q Carrier Chromosomes[Note]
| Subject (Ethnicity) | Marker D12S99 | Marker D12S356 | Exon 1 SNP | MarkerTNFRp55 | Intron 5 SNP | Marker CD4 | Marker D12S1695 | Marker D12S77 | HaplotypeDesignation |
| T50M |
|||||||||
| Patient 466 (Scottish) | 299/303 | 215/219 | A/G | 144/150 | C/T | 86/86 | 147/155 | 191/183 | 1 |
| Patient 469 (Scottish), child of 466 | 299/297 | 219/219 | A/A | 150/150 | T/T | 86/112 | 147/155 | 191/173 | 1 |
| Patient 594 (unknown) | 303/303 | 215/217 | A/A | 148/150 | T/T | 91/112 | 157/161 | 173/173 | 1 |
| Father of 594, unaffected | ND | ND | A/A | 150/150 | T/T | 91/106 | ND | ND | |
| Patient 32 (Irish) | 293/297 | 219/223 | A/G | 144/152 | C/T | 86/86 | 157/159 | 163/163 | 2 |
| Patient 510 (Irish), child of 32 | 293/297 | 219/223 | A/A | 150/152 | T/T | 86/91 | 155/159 | ND | 2 |
| Patient 511 (Irish), child of 32 | 293/293 | 297/223 | A/A | 150/152 | T/T | 86/91 | 155/159 | ND | 2 |
| Patient 802 (Irish) | ND | 223/225 | A/G | 144/150 | C/T | 106/106 | ND | ND | 3 |
| Patient 804 (Irish) | ND | ?/225 | A/G | 144/152 | C/T | 101/106 | ND | ND | 3 |
| Patient 805 (Irish) | ND | 223/225 | A/G | 144/150 | C/C | 91/106 | ND | ND | 3 |
| Patient 806 (French Canadian) | ND | 219/223 | G/G | 144/144 | C/C | 86/91 | ND | ND | 3 |
| Patient 807 (French Canadian), child of 806 | ND |
219/223 |
G/G |
144/144 |
C/C |
86/91 |
ND |
ND |
3 |
| R92Q |
|||||||||
| Patient 498 (mixed northern European) | 295/297 | 211/211 | G/G | 140/144 | C/C | 91/91 | 147/155 | 177/177 | |
| Mother of 498 | 295/295 | 211/219 | A/G | 140/152 | ND | 91/112 | 147/159 | ND | |
| Patient 522 (Irish) | 293/297 | 219/223 | A/G | 140/150 | C/T | 86/91 | 147/157 | 167/179 | |
| Patient 560 (Mexican) | 297/303 | 217/221 | A/G | 140/150 | C/T | 86/86 | 147/155 | 177/181 | |
| Mother of 560 | 297/301 | 213/217 | G/G | 140/144 | C/C | 86/112 | 155/159 | 177/177 | |
| Patient 618 (English) | 293/301 | 211/213 | A/G | 140/150 | C/T | 91/112 | 159/161 | 163/181 | |
| Patient 644 (Italian) | 297/301 | 217/223 | A/G | 140/150 | C/T | 86/86 | 147/151 | 177/185 | |
| Patient 657 (Portuguese) | 297/? | 205/221 | A/G | 140/150 | C/T | 91/121 | 151/155 | 179/? | |
| Father of 657 | 293/297 | 205/217 | G/G | 140/144 | C/C | 91/121 | 155/155 | 179/? | |
| Patient 670 (unknown) | 301/? | 205/211 | G/G | 140/150 | C/C | 91/112 | 141/155 | 181/183 | |
| Father of 670 | 301/303 | 205/219 | A/G | 140/144 | C/C | 86/91 | 147/155 | 181/? | |
| Patient 758 (mixed northern European) | ND | 221/223 | A/G | 140/152 | C/C | 91/106 | ND | ND | |
| Patient 791 (Jewish) | ND | 217/223 | G/G | 140/144 | C/C | 91/91 | ND | ND | |
| Father of 791 (Jewish) | ND | 223/225 | G/G | 140/144 | C/C | 91/91 | ND | ND | |
Note.— The genetic markers are presented in the order in which they occur on chromosome 12. Alleles representing haplotypes segregating with either T50M or R92Q within the family are in boldface italic; alleles representing a putative haplotype cosegregating with a mutation, determined on the basis of allele similarities with other defined haplotypes, are underlined. ND = not determined; ? = results not interpretable.
However, haplotype analysis of patients with R92Q showed a high level of linkage disequilibrium within TNFRSF1A, where the 140-bp allele of the TNFR1P55 microsatellite marker cosegregated strongly with the presence of the substitution at codon 92. Using this microsatellite and the two intragenic SNPs, we have identified an intragenic haplotype, defined as G-140 bp-C, associated with the R92Q mutation on all of four independent carrier chromosomes in which phase could be assigned (table 3). In all five remaining singleton cases, in which phase could not be established, genotypes consistent with this intragenic haplotype were observed. In contrast, the frequency of this haplotype in the general northern-European population could not be >6.5%, the frequency of the 140-bp allele that we observed in 122 control chromosomes.
Genetic Heterogeneity
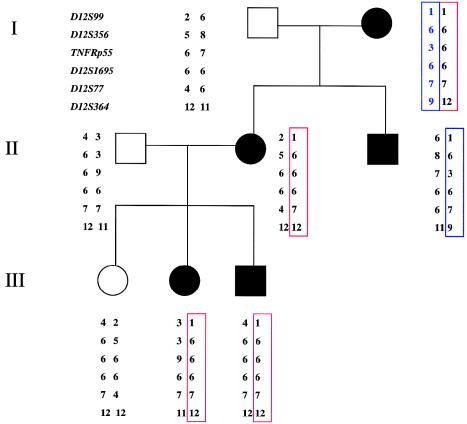
Among the families with a clinical history compatible with TRAPS who were referred for periodic fever, two multiplex families with dominant inheritance were identified. We did not find any mutation in TNFRSF1A, even after sequencing all of the exons, flanking intronic regions, and promoter regulatory sequences. One of the families was a three-generation pedigree with affected members in each generation—and, therefore, was informative for linkage analysis. Even though the first two markers were not informative in this family, from TNFRP55 to D12S364 opposite haplotypes were inherited in the two affected siblings in the second generation (fig. 5), thus ruling out TNFRSF1A mutations 3′ to this microsatellite locus in the intron following exon 1. When these results are considered together with our sequence analysis of exon 1 and the promoter region, it is highly unlikely that TNFRSF1A is the susceptibility locus. The hyperimmunoglobulinemia D with periodic fever syndrome (HIDS [MIM 260920]) and FMF were considered in the differential diagnosis, but the dominant pattern of inheritance is not consistent with either of these diagnoses. Nevertheless, the affected members were screened for a panel of 19 known FMF mutations; and they tested negative. Linkage analysis for the MWS/FCU region on distal chromosome 1q44 also identified recombinants within the family (data not shown), therefore excluding this locus as well. The second family of Irish background with parent-to-child transmission of periodic fevers was too small to be informative for linkage analysis, but no change was identified in TNFRSF1A when both the coding region and the promoter region were sequenced.
Figure 5.
Pedigree of family affected with dominantly inherited periodic-fever syndrome. One affected member of the family was screened for the TNFRSF1A mutation, throughout the coding and promoter regions; no mutation was identified. Linkage analysis was done to rule out other mutations in TNFRSF1A. Genotypes for six DNA markers from chromosome 12p are shown. Affected sibs in the second generation inherited opposite haplotypes across the interval.
Moreover, in our mutational survey of 150 patients with periodic fever and clinical histories suggestive of TRAPS, 120 had no mutations in exons 2–5. Ninety patients were further analyzed for mutations in the remainder of TNFRSF1A (exons 1 and 6–10), by a combination of DHPLC and sequencing; no mutation was identified, except a new polymorphism (i.e., SNP) in intron 8, at position c.218 -9 T→C (fig. 6). This sequence change was evaluated in 88 control chromosomes (CEPH and Irish), and allele frequencies were .78 (T) and .22 (C). In 80 chromosomes from patients, the frequencies were the same—that is, .78 (T) and .22 (C).
Figure 6.
Genomic structure of TNFRSF1A (adapted from Fuchs et al. 1992). All 16 known TRAPS-causing mutations are clustered in exons 2–4, which encode the TNFRSF1A extracellular domain. The c.218−9 T→C polymorphism is shown in intron 8 of TNFRSF1A.
Discussion
In the present article, we have reported six novel mutations, in TNFRSF1A, as a cause of the recently described dominantly inherited periodic fever syndrome, TRAPS. These novel mutations confirm our initial report demonstrating a role for the p55 molecule in the pathogenesis of autoinflammatory disease (McDermott et al. 1999). The total number of TNFRSF1A mutations that have been described thus far is 16, including 4 mutations (C30S, R92P, C55S, and C70Y) that recently have been reported elsewhere (Dodé et al. 2000; Aganna et al. 2001; Jadoul et al. 2001; Simon et al. 2001). In contrast to our previous study, in which five of six TNFRSF1A mutations involved cysteine residues, in the present study five of six novel TRAPS mutations were identified at noncysteine residues. As a group, the noncysteine mutations are less frequently associated with amyloidosis—and perhaps are less penetrant—than the cysteine substitutions.
Our new data are consistent with our previous observation that dominantly inherited periodic fevers can be caused by structural changes in the extracellular domain of TNFRSF1A, but we now have a broader view of the mutations that can cause this phenotype. Of the 16 TRAPS mutations described thus far, 15 are missense, and all 16 are clustered within the first two cysteine–rich extracellular subdomains (i.e., CRD1 and CRD2) of the protein, spanning exons 2–4 of the genomic sequence (fig. 6). If one of the mechanisms by which TRAPS mutations cause an autoinflammatory state is interference with activation-induced receptor cleavage, then it may well be that more-drastic nonsense or frameshift mutations, resulting in a truncated or nonexpressed protein, would not have the same phenotype. Extensive analysis of coding and exon-flanking sequences of 90 patients referred for TRAPS did not identify any disease-causing mutation, either in the transmembrane or in intracellular domains of TNFRSF1A.
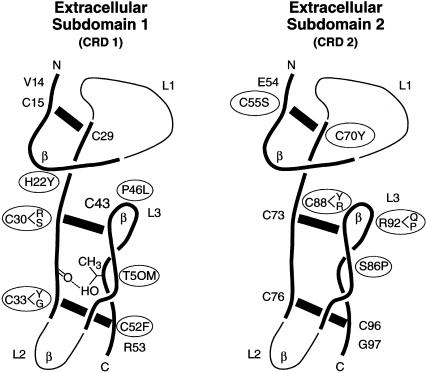
The nine cysteine substitutions thus far identified all disrupt conserved extracellular disulfide bonds (fig. 7): C30R, C30S, C33Y, C33G, and C52F would disrupt the second or third disulfide bond in the first extracellular subdomain (i.e., CRD1), whereas C55S, C70Y, C88R, and C88Y would abolish the first or second intrachain disulfide bond in the second extracellular subdomain (i.e., CRD2). The observation of multiple mutations at residues C30, C33, and C88 suggests a critical role for these sites that is not immediately apparent from the p55 crystal structure (Banner et al. 1993; Naismith et al. 1996).
Figure 7.
Crystallographically determined structure of the TNFRSF1A extracellular domains 1 and 2 (Banner et al. 1993). TRAPS mutations are shown as circled amino acids. The three disulfide bonds of CRD1 and CRD2 are depicted by thick black bars. Structurally conserved regions of the CRDs are represented by thicker lines. The β-turn positions are indicated by “β.” Loop domains are denoted as “L1”–“L3.”
Among the six noncysteine missense substitutions, T50M most likely prevents the formation of a highly conserved intrachain hydrogen bond (fig. 7). Three other sequence variants—P46L, S86P, and R92P—involve proline residues that would be predicted to introduce a bend into the protein secondary structure, and therefore such mutations might have an important effect on the folding of the extracellular domain. The ligand-binding domain of the p55 molecule is between amino acid 77 and amino acid 114 (Banner et al. 1993), and thus the mutations at residues 86 and 92 may affect TNF binding. H22Y, P46L, and R92Q fall within the β turns of loop 3 of CRD1 and CRD2 (fig. 7), and therefore these replacements may have a significant impact on overall loop structure.
This article is also the first report of a TNFRSF1A splice mutation associated with periodic fever. We have provided strong evidence, both from the cDNA sequence in patients and from the analysis of minigene constructs, that the c.193−14 G→A mutation results in the addition of four amino acids to CRD1. This and other mutations within CRD1 (amino acids 1–54) may affect the ligand-independent assembly of TNFRSF1A trimers on the cell membrane (Chan et al. 2000).
We have previously provided evidence that the C52F mutation is a novel type of dominant negative, in which “negative” refers not to a failure of signaling but, rather, to a failure in cytokine-receptor clearance (McDermott et al. 1999). Receptor cleavage may have a homeostatic effect both by reducing the number of available membrane sites for repeated stimulation and by producing a pool of potentially antagonistic soluble receptors, and thus a defect in cleavage could lead to a hyperinflammatory state. In the present article, we have extended the finding of impaired activation-induced shedding to other mutations. However, we were unable to demonstrate a defect in shedding in vitro for monocytes from patients with R92Q and the splicing mutation, despite the facts that (a) serum p55 levels did not increase with attacks for R92Q and (b) the splicing mutation was seen only in families with TRAPS symptoms. These data suggest that additional pathogenetic mechanisms may be operative in TRAPS.
The present study also provides important insights into genotype-phenotype correlations and related issues of penetrance. This article presents the first comprehensive analysis of TNFRSF1A genotypes seen in association with systemic amyloidosis, the most serious and life-threatening complication of TRAPS. Overall, we found that ∼14% of patients thus far reported with TRAPS have developed amyloidosis, with the risk skewed strongly toward individuals with cysteine substitutions. The prevalence of amyloidosis among patients with TRAPS is significantly higher than previously recognized. This is most likely due to the fact that, prior to the identification of TNFRSF1A as the underlying gene, it was unclear whether families with periodic fever and amyloidosis (Gertz et al. 1987; Zaks and Kastner 1997) should be grouped with the index family for FHF, in which amyloidosis was uncommon. Molecular analysis has enabled proper diagnosis and the accurate assessment of the true prevalence of this complication. We also found that, relative to individuals with cysteine substitutions, individuals with noncysteine mutations have a tendency toward lower overall penetrance, although it should be emphasized that, even for cysteine substitutions, penetrance is not 100%. At present, we have insufficient clinical data to determine whether, aside from amyloidosis, patients with cysteine substitutions have more-severe disease.
Of the TNFRSF1A substitutions reported here, P46L and R92Q are likely to have the lowest penetrance for TRAPS, with each having been found in ∼1% of control chromosomes. Despite these high frequencies in controls, we believe that both substitutions are either low-penetrance mutations or functional variants, rather than benign polymorphisms. As noted above, P46L would be predicted to have a significant effect on the loop structure of p55 molecules, and the one patient with this substitution whom we have seen has symptoms very typical of TRAPS, with a favorable response to anti-TNF therapy. Moreover, we were able to demonstrate a defect in activation-induced TNFRSF1A shedding in monocytes from a patient with P46L. In the case of R92Q, the clinical picture is somewhat more heterogeneous, but the frequency of this substitution in our cohort with periodic fever was significantly higher than that in the control population. Although we did not observe a defect in R92Q TNFRSF1A shedding in vitro, soluble p55 serum levels in vivo did not increase with attacks.
On the basis of the hypothesis that R92Q might have a broader influence on susceptibility to inflammation, we have examined the frequency of this substitution in a cohort of 135 patients with early arthritis; we found 7 patients to be positive (P=.06, Fisher exact test), and a larger study is in progress. It is possible that the phenotypic manifestations of R92Q depend on other linked or unlinked modifying genes and/or on modifying environmental factors (Dipple and McCabe 2000). These results are particularly intriguing in light of the recently reported possible susceptibility locus for rheumatoid arthritis, in the TNFRSF1A region of chromosome 12 (Jawaheer et al. 2001).
Haplotype analysis of TRAPS chromosomes bearing R92Q revealed a common intragenic haplotype (G-140 bp-C) associated with this substitution. Since this haplotype comprises only intragenic markers (and does not extend to flanking microsatellites) and is seen in a number of different populations, the data suggest a very ancient common founder. In light of the current theories of the genetics of complex diseases (Lander 1996; Risch and Merikangas 1996; Collins et al. 1997), it will be interesting to determine whether R92Q carriers with non-TRAPS early arthritis also carry this haplotype. It is also noteworthy that we did not identify a common founder haplotype associated with T50M, a mutation detected in several families of Scottish or Irish descent; our data suggest instead that T50M is a recurrent mutation arising at a CpG hotspot.
The diversity of T50M carrier chromosomes in the Scottish/Irish families is but a part of the rather unexpected genetic heterogeneity of TRAPS seen in these populations. Although TRAPS has been seen in several ethnic groups, a disproportionately large percentage of families are of Irish or Scottish descent. Altogether, we have observed 10 of 16 currently known mutations in families of Irish and/or Scottish ancestry. Although ascertainment bias remains a possibility that is difficult to exclude, it is also possible that there is a high frequency of permissive alleles at one or more interacting loci in the Irish and Scottish populations, thereby increasing the penetrance of TNFRSF1A mutations. A similar hypothesis has been proposed to explain the high frequency of limb-girdle type 2A muscular dystrophy on Reunion Island (the “Reunion Paradox”), as well as Hurler syndrome and metachromatic leukodystrophy in the lower Galilee in Israel, in the absence of high-frequency founder mutations (Beckmann 1996).
The last 5 years have witnessed striking advances in our understanding of the molecular pathogenesis of the hereditary periodic fevers (Centola et al. 1998; Drenth and van der Meer 2000). The genes underlying FMF (French FMF Consortium 1997; International FMF Consortium 1997), HIDS (Drenth et al. 1999; Houten et al. 1999), and TRAPS have all been identified, and susceptibility loci for MWS and FCU have been mapped to chromosome 1q44 (Cuisset et al. 1999; Hoffman et al. 2000; McDermott et al. 2000). Data presented in the present article suggest that yet other loci play a role in the pathogenesis of periodic fever and inflammation in man. Whether these loci encode new members of the pyrin, TNF, or mevalonate pathways or encode proteins in yet other pathways remains to be established. As is the case for the p55 TNF receptor, it is likely that these other genes will eventually be found to play an important role in a number of human inflammatory diseases.
Acknowledgments
We wish to thank Drs. Robert Baker, Stephen O’Brien, Abraham Chaiton, Harry Gewanter, David Hill, Daniel Lovel, Theresa Lu, William Mentzer, Silvia Sequiera, Seema Shah, and Stephen Straus, for referral of patients; Dr. Hani El-Gabalawy, for samples from patients with early arthritis; Dr. Kathleen A. Quane, for Irish control DNA samples; and Drs. Michael McDermott, John Compton, and Sherri Bale, for helpful discussions.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Centers for Disease Control and Prevention, http://www.cdc.gov/epiinfo/ei6.htm (for Epi Info 6 statistical software packages)
- HGSBASE, http://hgbase.cgr.ki.se (for allele frequencies of SNP in exon 1 [accession number SNP000005194] and SNP in intron 5 [accession number SNP000005195])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for FCU [MIM 120100], FHF [MIM 142680], FMF [MIM 249100], HIDS [MIM 260920], and MWS [MIM 191900])
References
- Aganna E, Aksentijevich I, Hitman GA, Kastner DL, Hoepelman AIM, Posma FD, Zweers EJK, McDermott MF (2001) Tumor necrosis factor receptor-associated periodic fever syndrome (TRAPS) in a Dutch family: evidence for a TNFRSF1A mutation with reduced penetrance. Eur J Hum Genet 9:63–66 [DOI] [PubMed] [Google Scholar]
- Banner DW, D'Arcy A, Janes W, Gentz R, Schoenfeld H-J, Broger C, Loetscher H, Lesslauer W (1993) Crystal structure of the soluble human 55 kd TNF receptor-human TNFb complex: implications for TNF receptor activation. Cell 73:431–445 [DOI] [PubMed] [Google Scholar]
- Bazzoni F, Gatto L, Lenzi L, Vinante F, Pizzolo G, Zanolin E, DeGironcoli M (2000) Identification of novel polymorphisms in the human TNFR1 gene: distribution in acute leukemia patients and healthy individuals. Immunogenetics 51:159–163 [DOI] [PubMed] [Google Scholar]
- Beckmann JS (1996) The Réunion paradox and the digenic model. Am J Hum Genet 59:1400–1402 [PMC free article] [PubMed] [Google Scholar]
- Centola M, Aksentijevich I, Kastner DL (1998) The hereditary periodic fever syndromes: molecular analysis of a new family of inflammatory diseases. Hum Mol Genet 7:1581–1588 [DOI] [PubMed] [Google Scholar]
- Centola M, Wood G, Frucht D, Galon J, Aringer M, Farrell C, Kingma DW, Horwitz M, Mansfield E, Holland SM, O’Shea JJ, Rosenberg HF, Malech HL, Kastner DL (2000) The gene for familial Mediterranean fever, MEFV, is expressed in early leukocyte development and is regulated in response to inflammatory mediators. Blood 95:3223–3231 [PubMed] [Google Scholar]
- Chan FKM, Chun HJ, Zheng L, Siegal R, Bui KL, Lenardo MJ (2000) A domain in TNF receptors that mediates ligand independent receptor assembly and signaling. Science 288:2351–2354 [DOI] [PubMed] [Google Scholar]
- Collins FS, Guyer MS, Chakravarti A (1997) Variations on a theme: cataloging human DNA sequence variation. Science 278:1580–1581 [DOI] [PubMed] [Google Scholar]
- Cuisset L, Drenth JPH, Berthelot J-M, Meyrier A, Vaudour G, Watts RA, Scott DGI, Nicholls A, Pavek S, Vasseur C, Beckmann JS, Delpech M, Grateau G (1999) Genetic linkage of the Muckle-Wells syndrome to chromosome 1q44. Am J Hum Genet 65:1054–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipple KM, McCabe ERB (2000) Phenotypes of patients with “simple” Mendelian disorders are complex traits: thresholds, modifiers, and systems dynamics. Am J Hum Genet 66:1729–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodé C, Papo T, Fieschi C, Pêcheux C, Dion E, Picard F, Godeau P, Bienvenu J, Piette J-C, Delpech M, Grateau G (2000) A novel missense mutation (C30S) in the gene encoding tumor necrosis factor receptor 1 linked to autosomal-dominant recurrent fever with localized myositis in a French family. Arthritis Rheum 43:1535–1542 [DOI] [PubMed] [Google Scholar]
- Drenth JP, Cuisset L, Grateau G, Vasseur C, van de Velde-Visser SD, de Jong JG, Beckmann JS, van der Meer JW, Delpech M (1999) Mutations in the gene encoding mevalonate kinase cause hyper-IgD and periodic fever syndrome: International Hyper-IgD Study Group. Nat Genet 22:178–181 [DOI] [PubMed] [Google Scholar]
- Drenth JP, van der Meer JW (2000) Periodic fevers enter the era of molecular diagnosis. BMJ 320:1091–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskadele J, Turestkaya RL, Armstrong C, Kuprash DV, Nedospasov SA, Gallagher G (2000) A polymorphic microsatellite marker in the human p55 TNF receptor, CD 120a. Genes Immun 1:228–230 [DOI] [PubMed] [Google Scholar]
- French FMF Consortium (1997) A candidate gene for familial Mediterranean fever. Nat Genet 17:25–31 [DOI] [PubMed] [Google Scholar]
- Fuchs P, Strehl S, Dworzak M, Himmler A, Ambros PF (1992) Structure of the human TNF receptor 1 (p60) gene (TNFR1) and localization to chromosome 12p13. Genomics 13:219–224 [DOI] [PubMed] [Google Scholar]
- Gertz MA, Pettit RM, Perrault J, Kyle RA (1987) Autosomal dominant familial Mediterranean fever-like syndrome with amyloidosis. Mayo Clin Proc 62:1095–1100 [DOI] [PubMed] [Google Scholar]
- Hoffman HM, Wright FA, Broide DH, Wanderer AA, Kolodner RD (2000) Identification of a locus on chromosome 1q44 for familial cold urticaria. Am J Hum Genet 66:1693–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten SM, Kuis W, Duran M, de Koning TJ, van Royen-Kerkhof A, Romejin GJ, Frankel J, Dorland L, deBarse MMJ, Huijbers WAR, Rijkers GT, Waterham HR, Wanders RJA, Poll-The BW (1999) Mutations in MVK, encoding mevalonate kinase, cause hyperimmunoglobulinemia and periodic fever syndrome. Nat Genet 22:175–177 [DOI] [PubMed] [Google Scholar]
- International FMF Consortium (1997) Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell 90:797–807 [DOI] [PubMed] [Google Scholar]
- Jadoul M, Dodé C, Cosyns JP, Abramowicz D, Georges B, Delpech M, Pirson Y (2001) Autosomal-dominant periodic fever with amyloidosis: novel mutation in tumor necrosis factor receptor 1 gene. Kidney Int 59:1677–1682 [DOI] [PubMed] [Google Scholar]
- Jawaheer D, Seldin MF, Amos CI, Chen WV, Shigeta R, Monteiro J, Kern M, Criswell LA, Albani S, Nelson JL, Clegg DO, Pope R, Schroeder HW Jr, Bridges SL Jr, Pisetsky DS, Ward R, Kastner DL, Wilder RL, Pincus T, Callahan LF, Flemming D, Wener MH, Gregersen PK (2001) A genomewide screen in multiplex rheumatoid arthritis families suggests genetic overlap with other autoimmune diseases. Am J Hum Genet 68:927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES (1996) The new genomics: global views of biology. Science 274:536–539 [DOI] [PubMed] [Google Scholar]
- McDermott MF, Aganna E, Hitman GA, Ogunkolade BW, Booth DR, Hawkins P (2000) An autosomal dominant periodic fever associated with AA amyloidosis in a North Indian family maps to distal chromosome 1q. Arthritis Rheum 43:2034–2040 [DOI] [PubMed] [Google Scholar]
- McDermott MF, Aksentijevich I, Galon J, McDermott EM, Ogunkolade, BW, Centola M, Mansfield E, et al (1999) Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell 97:133–144 [DOI] [PubMed] [Google Scholar]
- McDermott MF, Ogunkolade BW, McDermott EM, Jones LC, Wan Y, Quane KA, McCarthy J, Phelan M, Molloy MG, Powell RJ, Amos CI, Hitman GA (1998) Linkage of familial Hibernian fever to chromosome 12p13. Am J Hum Genet 62:1446–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott EM, Smillie DM, Powell RJ (1997) The clinical spectrum of familial Hibernian fever: a 14-year follow-up study of the index and extended family. Mayo Clin Proc 72:806–817 [DOI] [PubMed] [Google Scholar]
- Mulley J, Saar K, Hewitt G, Rüschendorf F, Phillips H, Colley A, Sillence D, Reis A, Wilson M (1998) Gene localization for an autosomal dominant familial periodic fever to 12p13. Am J Hum Genet 62:884–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith JH, Devine TQ, Kohno T, Sprang SR (1996) Structures of the extracellular domain of the type I tumor necrosis factor receptor. Structure 4:1251–1262 [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Simon A, Dodé C, van der Meer JWM, Drenth JPH (2001) Familial periodic fever and amyloidosis due to a new mutation in the TNFRSF1A gene. Am J Med 110:313–315 [DOI] [PubMed] [Google Scholar]
- Williamson LM, Hull D, Mehta R, Reeves WG, Robinson BH, Toghill PJ (1982) Familial Hibernian fever. Q J Med 51:469–480 [PubMed] [Google Scholar]
- Zaks N, Kastner DL (1997) Clinical syndromes resembling familial Mediterranean fever. In: Sohar E, Gafni J, Pras M (eds) Familial Mediterranean fever. Freund, London and Tel Aviv, pp 211–215 [Google Scholar]