Abstract
We have shown functional complementation of a genetic deficiency in human cultured cells, using artificial chromosomes derived from cloned human genomic fragments. A 404-kb human-artificial-chromosome (HAC) vector, consisting of 220 kb of alphoid DNA from the centromere of chromosome 17, human telomeres, and the hypoxanthine guanine phosphoribosyltransferase (HPRT) genomic locus, was transferred to HPRT-deficient HT1080 fibrosarcoma cells. We generated several cell lines with low-copy-number, megabase-sized HACs containing a functional centromere and one or possibly several copies of the HPRT1 gene complementing the metabolic deficiency. The HACs consisted of alternating alphoid and nonalphoid DNA segments derived only from the input DNA (within the sensitivity limits of FISH detection), and the largest continuous alphoid segment was 158–250 kb. The study of both the structure and mitotic stability of these HACs offers insights into the mechanisms of centromere formation in synthetic chromosomes and will further the development of this human-gene-transfer technology.
Introduction
Several groups have had success in generating stable human artificial chromosomes (HACs) in mammalian cells either by telomere-directed fragmentation of endogenous human chromosomes or from the introduction of cloned human centromeric and telomeric sequences (Harrington et al. 1997; Ikeno et al. 1998; Henning et al. 1999; Mills et al. 1999; Ebersole et al. 2000; Yang et al. 2000). Both strategies have determined (1) that α-satellite, or alphoid, DNA, the major sequence component at human centromeres, can generate a functional de novo centromere in human cells and (2) that a minimal size is important for minichromosome stability (Yang et al. 2000). In addition, transfection of defined constructs into human cells has shown that in the absence of telomeric repeats a large array of alphoid DNA alone is sufficient to generate stable circular artificial minichromosomes (Ebersole et al. 2000). In human cells, characterization of neocentromeres devoid of normal centromeric sequences has led to the question of whether sequence alone is sufficient to determine a centromere; however, de novo centromere activation resulting from an epigenetic mechanism has yet to be identified (Karpen and Allshire 1997; Willard 1998; Choo 2000).
The development, via either strategy, of HACs as gene-transfer vectors is now a possibility. Owing to their potential to carry entire genomic loci, HACs should mediate long-term, tissue-specific expression of transgenes. Stable extrachromosomal maintenance of HACs would overcome the problems of gene silencing resulting from random integration into the host genome and should produce a regulated level of gene expression throughout the lifetime of the cell. As a nonviral approach based on human functional elements, the use of HACs may offer a safer alternative to viral therapeutic gene-transfer methods currently being investigated, particularly in light of a major setback in a recent clinical gene-therapy trial with adenoviral vectors (Willard 2000). Because of the size of HACs, however, methods for their efficient delivery to human cells will have to be developed.
A recently described approach to integrate one or a number of genes into HACs involves the manipulation of minichromosomes in chicken DT40 cells and the introduction of defined regions containing human genes by Cre-mediated recombination at loxP sites (Kuroiwa et al. 2000). An alternative strategy is to incorporate, into defined HAC vectors, either P1 artificial chromosome (PACs) or bacterial artificial chromosomes (BACs) with large genomic fragments spanning an entire gene locus and distant regulatory sequences and to introduce the constructs into human cells. We constructed a HAC vector for the transfer of a human marker gene, containing alphoid DNA from the centromere of chromosome 17, telomeric sequences, and, from human Xq26.2, a large genomic segment spanning the hypoxanthine guanine phosphoribosyltransferase (HPRT) locus, HPRT1 (MIM 308000; Project Ensembl) (Mejía and Larin 2000). HPRT1 encodes a purine-salvage enzyme, and mutations in this gene result in Lesch-Nyhan syndrome (MIM 300322), a neurodevelopmental disorder characterized by self-injurious and abnormal motor behavior (Caskey and Kruh 1979). In this study, we introduced the HAC vector into HPRT-deficient human fibrosarcoma cells and obtained stable artificial minichromosomes that complemented the HPRT− phenotype of the host cells. Our structural and functional characterization of the artificial minichromosomes offers insights into the requirements for centromere function and the prospects of using HACs as vectors for gene transfer.
Material and Methods
HAC Construct, Enzymes, and Media
The assembly of the HAC construct used in this study, JM2860, has been described in detail elsewhere (Mejía and Larin 2000). β-Agarase and endonucleases—except I-SceI, which was from Roche—were from New England Biolabs. Cell-culture media, selective medium supplements, and transfection reagents were purchased from Gibco BRL unless otherwise indicated.
Lipofection of Cultured Cells
JM2860 DNA was prepared in plugs of low-melting-point agarose (SeaPlaque; FMC), as described elsewhere (Larin 1995). The DNA (1–3 μg/agarose plug) was linearized by digestion by I-SceI endonuclease, washed in 10 mM Tris-HCl, 1 mM EDTA buffer at pH 8, and then equilibrated in the same buffer, containing 50 μM low-molecular-weight (∼2,000) polyethylenimine (PEI) (Aldrich) (Boussif et al. 1995; Marschall et al. 1999). The plugs were melted at 65°C, treated with β-agarase, and diluted, to a DNA concentration of 0.5 μg/ml, with OptiMEM. Lipofectin—7.5 μg in 2 ml of OptiMEM—was added to 2 ml of the DNA dilution, and 8 ml of this transfection medium was added to subconfluent cells of an HPRT− HT1080 subline (Rasheed et al. 1974; Lugo and Baker 1983), in a 10-cm petri dish. The next day, transfection medium was replaced with normal growth medium composed of Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Globepharm). Confluent cells were split into six dishes, and selection was applied, after the cells had again reached confluence, for 9 d. Selective medium supplements were used as follows: puromycin (Sigma), 0.22–0.25 μg/ml; G418, 125–160 μg/ml; and hypoxanthine, aminopterin, and thymidine (HAT) supplement composed of 0.1 mM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine. Colonies were picked into 24-well plates, expanded, and screened by PCR and FISH.
PCR Screening of Positive Clones
Primer pair GGCCTCTTCGCTATTACGC (JNC9, vector) and CGGGAGAATCTTCACAGGAA (JNC10, insert) and primer pair CGGGCCTCTTCGCTATTAC (JNC11, vector) and ACGGGAGAATCTTCACAGGA (JNC12, insert), which generated PCR products of 293 bp and 296 bp, respectively, were used to detect the junction region between the alphoid insert and a residual fragment of the pBeloBAC11 vector (Kim et al. 1996) in JM2860 (fig. 1A).
Figure 1.
Maps of the HAC construct. A, Schematic of JM2860 HAC construct (Mejía and Larin 2000) linearized with I-SceI endonuclease. The construct contains both a 220-kb alphoid-DNA array from the human chromosome 17 centromere and a chromosome X segment with the HPRT1 gene. The ends are capped by human-telomere repeats, and the vector arms contain selectable markers for mammalian-cell transfection (PurR and neo), the ampicillin-resistance gene (AmpR) for selection in Escherichia coli, and lacZ as a reporter gene in mammalian cells. The location of the region amplified by PCR in HAC DNA derived from positive clones is indicated below the map, together with cleavage sites for the enzymes used in the structural analysis of the artificial minichromosomes (fig. 4A). B, Telomeric region of JM2860 prior to I-SceI cleavage. The I-SceI sites downstream from each 1.2-kb telomeric repeat (Tel), the position of the XhoI restriction sites, and the repE and AmpR gene probes used in the Bal31 analysis (fig. 5) are indicated below the map. The alphoid and chromosome X genomic inserts are denoted by “17 α” and “Xq26.2,” respectively.
6-Thioguanine (6-TG)–Sensitivity Assay
For each minichromosome or control cell line, 105 cells were seeded in a well of a 6-well plate and were grown in DMEM supplemented with 10% dialyzed fetal bovine serum and 5 μg 6-TG/ml (Sigma). Sensitivity to 6-TG was assessed by comparison both with identical cultures in which 6-TG had been omitted and between the minichromosome cell lines and the HPRT+ (6-TG sensitive), as well as the parental HPRT− (6-TG resistant), HT1080 cell lines.
FISH of Metaphase Chromosomes
Metaphase chromosome spreads were prepared on microscope slides and hybridized using modifications of standard FISH protocols. Artificial chromosomes in JM2860-positive clones were detected with a biotin-labeled chromosome 17 alphoid (17 α) probe (D17Z1; Oncor) and a fluorescein-streptavidin conjugate (Vector). Selected clones were analyzed by dual hybridization with the 17 α probe and the HPRT1-locus PAC from which JM2860 was derived; the latter was labeled with digoxigenin (DIG-Nick Translation Mix; Roche Diagnostics), and hybridization was performed in the presence of unlabeled human C0t1-fraction DNA. Further dual-hybridization analyses were performed with the biotin-labeled 17 α probe and a digoxigenin-labeled universal-human-telomere probe (Oncor). In dual hybridizations, the biotin-labeled probe was detected with a streptavidin-Cy3 conjugate (Sigma). In some applications, sensitivity was enhanced by the use of an additional layer (i.e., biotinylated anti-streptavidin antibody from goat [Vector]), prior to a secondary layer of fluorescent conjugate. Digoxigenin-labeled probes were detected with a mouse monoclonal anti-digoxigenin antibody followed by fluorescein isothiocyanate (FITC)–labeled sheep antibody anti–mouse immunoglobulins (Ig) (Roche). When it was necessary, a final layer of donkey anti–sheep IgG antibody and FITC conjugate (Sigma) was used for increased sensitivity. The stringency of hybridization conditions and washes was optimized for every probe or probe combination, by varying the concentration of formamide in the buffers. For microscopy, slides were mounted by use of Vectashield mounting medium (Vector) containing, as fluorescent DNA counterstains, propidium iodide or TO-PRO-3 iodide (Molecular Probes). Fluorescence microscopy was performed with an MRC-1024 laser-scanning confocal microscope and with LaserSharp software from Bio-Rad.
Chromosome Painting
A panel of 24 chromosome-specific, FITC-labeled chromosome paint probes (Chromoprobe Multiprobe Hybridisation System; Cytocell) were used in FISH, according to the supplier’s protocol, and each paint was combined with the aforementioned biotin-labeled 17 α probe. Biotin detection and microscopic analysis of slides were as described in the subsection “FISH of Metaphase Chromosomes.”
Fiber FISH
HT1080 cells were dissociated with trypsin and washed in phosphate-buffered saline (PBS). Stretched DNA fibers were prepared by first drying 10 μl of the cell suspension in PBS, at 106 cells/ml, near the edge of a microscope slide at 45–50°C. The slide was then placed vertically in a slide–cover-plate assembly, and the cells were lysed down the slide with 150 μl of 0.05 M NaOH in 28.6% ethanol. This resulted in extended chromatin fibers, which were affixed onto the slide with 200 μl of methanol, dehydrated by successive washes in increasingly concentrated ethanol, and, finally, air-dried. DNA fibers were denatured for 3 min in 70% formamide, 2× standard sodium citrate buffer, at 70°C. FISH was performed with the 17 α and HPRT1-locus probes as described in the subsection “FISH of Metaphase Chromosomes,” and slides were mounted for microscopy with 4',6-diamidino-2-phenylindole as the DNA counterstain.
Centromeric Protein C (CENP-C) Detection
Immunochemical detection of CENP-C was performed as described elsewhere (Sullivan and Schwartz 1995), with slight modifications. In brief, colcemid-treated cells were centrifuged onto microscope slides in a Shandon Cytospin 2 apparatus and incubated with rabbit anti–CENP-C antibodies (Saitoh et al. 1992) followed by FITC-conjugated goat anti–rabbit Ig antibodies. The slides were then formalin fixed, rinsed, and, finally, treated with methanol:acetic acid (3:1) and air-dried. After a formamide denaturation step of 9 min, chromosome spreads were hybridized with the 17 α probe, as described in the subsection “FISH of Metaphase Chromosomes.” The probe was detected with streptavidin-Cy3 conjugate, with TO-PRO-3 iodide as a DNA counterstain.
Restriction-Enzyme Digestion and Hybridization Analysis
Genomic DNA from cultured cells was prepared in low-melting-point agarose plugs (Larin 1995). Restriction-enzyme digestions were performed overnight after equilibration of the plugs in the buffer recommended by the enzyme supplier, with 2 mM spermidine. The DNA was fractionated by pulsed-field gel electrophoresis (PFGE) in a CHEF Mapper apparatus (Bio-Rad). Southern blot transfer and hybridization analysis were performed with [32P]-labeled DNA probes prepared with the alphoid-DNA BAC from which the relevant construct was derived.
γ Irradiation of DNA
Genomic DNA purified in agarose plugs was exposed to a 137Cs source (Gammacell 1000 irradiator; Atomic Energy of Canada) for various times, to achieve the desired doses of γ irradiation (Taylor et al. 1996), and was fractionated by PFGE, for Southern hybridization analysis. The probes used were pCMVβ (Clontech), to detect the lacZ and ampicillin-resistance genes in the JM2860 construct, and the 17 α–DNA BAC.
Bal31 Analysis
Genomic DNA purified in agarose plugs was digested by Bal31 nuclease, as described elsewhere (Taylor et al. 1994), with BAL31 from New England Biolabs, according to the manufacturer’s instructions. The DNA was then restriction digested by XhoI and fractionated, for Southern hybridization, by agarose-gel electrophoresis. The blot was probed with the ampicillin-resistance gene (1.5 kb) and a 0.35-kb PCR fragment from the repE gene in the origin of replication of the BAC vector (Mejía and Monaco 1997).
Results
HAC Construct
The circular 404-kb HAC construct, JM2860, was generated as described in detail elsewhere (Mejía and Larin 2000). Figure 1A shows a map of JM2860 after specific linearization of the BAC with endonuclease I-SceI. The construct consists of 220 kb of human 17 α DNA, human telomeres at the end of each vector arm, and, from human chromosome Xq26.2, a large genomic fragment (162 kb) containing the entire HPRT1 genomic locus. Reporter and selectable markers (lacZ as well as neomycin- and puromycin-resistance genes) are present in each vector arm, as shown (fig. 1).
Transfection of HPRT− HT1080 Cells and Preliminary Screening of Positive Clones
DNA from JM2860 was digested by I-SceI endonuclease and then, prior to transfer into HPRT− HT1080 human fibrosarcoma cells by lipofection, condensed with 50 μM PEI (Boussif et al. 1995; Marschall et al. 1999). Positive clones were selected in the presence of either (1) G418 and puromycin, (2) HAT medium, or (3) G418, puromycin, and HAT (i.e., a triple selection). DNA from 22 clones was initially screened via PCR using primers spanning the junction region between the alphoid insert and a residual fragment of the pBeloBAC11 vector (Kim et al. 1996), 60 kb upstream from the HPRT1 gene in JM2860 (fig. 1A). Seventeen clones showed the expected PCR fragment (data not shown), suggesting that JM2860 DNA had been transferred intact into HPRT− HT1080 cells. To detect the presence of extrachromosomal elements, 14 independent clones were selected for further investigation by FISH.
Analysis of Positive Clones by FISH and by Immunofluorescence
Metaphase chromosomes prepared from 14 clones were analyzed by FISH with a 17 α–DNA probe. Eleven clones contained artificial minichromosomes present in 20%–100% of cells, with one or two copies per positive cell. This represents a de novo minichromosome-formation frequency of ∼1×10-6, relative to the number of HT1080 cells plated for transfection. Four cell lines—AG1-1, AG3-1, AG6-1, and AP1-1—contained minichromosomes only, and the remaining seven cell lines contained minichromosomes and DNA that had also either integrated into or truncated a host chromosome.
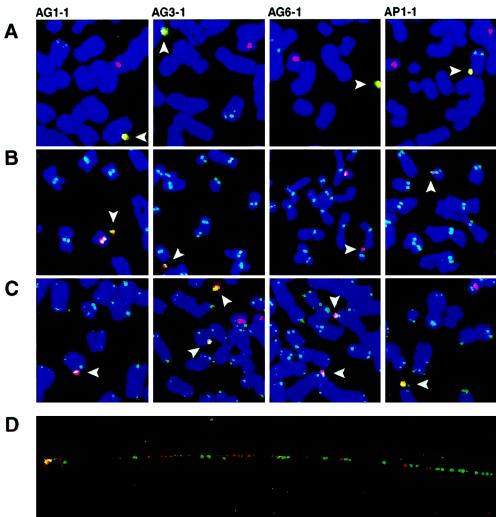
Further analysis of AG1-1, AG3-1, AG6-1, and AP1-1, with both the 17 α probe and an HPRT1 probe, showed that the two sequences colocalized on the artificial minichromosomes, and separate endogenous signals were detected, as expected, at the chromosome 17 centromeres and at the q arm of chromosome X (fig. 2A). Indirect immunofluorescence detection on metaphase chromosomes of an essential kinetochore protein, CENP-C (Saitoh et al. 1992), followed by FISH with the 17 α probe, showed colocalization of the two signals on the artificial minichromosomes and at the centromeres of the endogenous chromosomes 17 (fig. 2B). As a marker of functional centromeres, the CENP-C signal was also detected at the centromeres of all endogenous chromosomes as a doublet corresponding to the pair of sister chromatids. In a proportion of minichromosomes, CENP-C appeared as a singlet, probably because of their small size relative to the endogenous chromosomes and to the resolution of the microscopic analysis. The 17 α probe and a human-telomere probe were also shown to colocalize on the artificial minichromosomes (fig. 2C). A striking visualization of the structure of the minichromosomes in AG6-1 was provided by FISH analysis of stretched DNA fibers prepared from metaphase cells. Simultaneous hybridization with the 17 α and HPRT1 probes revealed alphoid-DNA and HPRT1 segments of varying sizes alternating over the observed length of the DNA fiber (fig. 2D).
Figure 2.
FISH analysis of minichromosome cell lines. A–C, Details of metaphase chromosome spreads from four minichromosome cell lines—AG1-1, AG3-1, AG6-1, and AP1-1—showing 17 α and DNA probe for the HPRT1 locus (A); 17 α and antibody to CENP-C, a marker of functional centromeres (B); and 17 α– and human-telomeric–DNA probe. Note the colocalization, on artificial minichromosomes (arrowheads), of the 17 α–DNA probe (red signal) and a second DNA or antibody probe (green signal). Overlap between the red and the green signals is shown as white or yellow pseudocolor. Total DNA stained with TO-PRO-3 is shown as blue pseudocolor. In addition to doing so on minichromosomes, the probes gave expected signals on the endogenous chromosome 17 centromere (A–C), the HPRT1 locus on Xq26.2 (A), the kinetochores of sister chromatids in each chromosome (B), and the telomeric termini of chromosomes (C). D, Dual hybridization of 17 α (red signal) and HPRT1-locus (green signal) probes to stretched DNA fibers from the AG6-1 cell line. The signals show that both sequences alternate on the minichromosome, as consecutive segments of irregular size. Gaps along the minichromosome fiber are expected where sequences are not represented in the probes used—that is, in the 22-kb region of JM2860 represented in figure 1B and in repetitive sequences masked by human C0t1 DNA used in the hybridization medium.
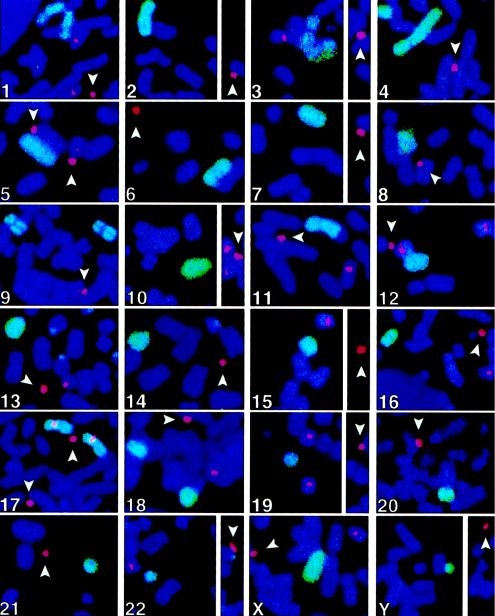
Metaphase chromosomes from each cell line were also hybridized, with a panel of 24 human-chromosome–specific paint probes (chromosomes 1–22, X, and Y) in conjunction with the 17 α probe, to determine whether the HACs contained any endogenous chromosomal DNA (fig. 3). Each paint probe detected the corresponding endogenous chromosome, but only the 17 α probe was observed to hybridize to the HACs and the endogenous chromosomes 17. We were unable to detect on the HACs the presence of a signal from the X-chromosome paint to the HPRT1 locus, presumably because the signal is not within the detection limits of the paint probe. Overall, on the basis of both the paint-probe results and the alternate structure revealed by fiber FISH, we assume that the HACs did not contain any long stretches of endogenous sequences within the sensitivity level of each probe. In the course of the FISH analyses, a truncation of the long arm of the Y chromosome was observed in a portion of the cells. Rarely, other rearrangements were also observed, such as the translocation of the q arm of chromosome 11, which is visible in figure 3; these rearrangements were not accompanied by any detectable modifications of minichromosomes.
Figure 3.
Analysis of minichromosomes by chromosome paints. Artificial minichromosomes (arrowheads), detected by the 17 α FISH probe (red signal), are negative for a set of human-chromosome–specific (chromosomes 1–22, X, and Y) paint probes (green signal). In these experiments, adequate detection of the corresponding natural chromosome is shown for each individual paint probe. The image shows details of metaphase chromosome spreads from the AG6-1 cell line, representative of the results from three other minichromosome cell lines—AG1-1, AG3-1, and AP1-1—analyzed in parallel.
Structural Characterization of Artificial Minichromosomes
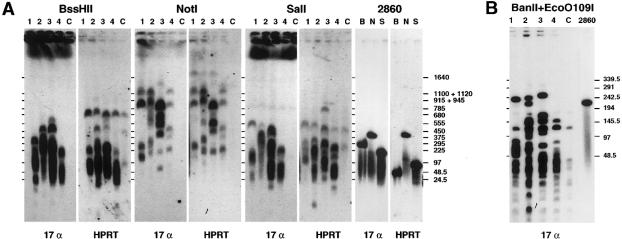
The structure of the HACs was investigated by restriction digestion of genomic DNA from AG1-1, AG3-1, AG6-1, and AP1-1, followed by PFGE and hybridization with the 17 α–cDNA and HPRT1-cDNA probes (fig. 4). Three enzymes with a known restriction pattern on JM2860—NotI, SalI, and BssHII—as well as a combination of BanII and EcoO109I were used.
Figure 4.
Structural analysis of minichromosome DNA. Genomic DNA from minichromosome clones AG1-1 (lanes 1), AG3-1 (lanes 2), AG6-1 (lanes 3), and AP1-1 (lanes 4) and from HT1080 HPRT− nontransfected cells (lanes C) was fractionated, after restriction-enzyme digestion, by PFGE, as indicated. A, Cleavage by BssHII (lanes B), NotI (lanes N), and SalI (lanes S). A filter transfer was successively hybridized with the 17 α BAC used in JM2860 and with an HPRT1-cDNA probe; the results are shown side by side. Similarities between the NotI patterns suggest detection of the same fragments by either probe. The fourth column shows fragments cleaved by BssHII, NotI, and SalI of the parental HAC construct JM2860 (fig. 1A). (The HPRT1-locus BssHII and SalI fragments of 64 and 114 kb, respectively, here display a lower apparent size.) The positions of DNA size standards (λ-DNA fragments and concatemers as well as Saccharomyces cerevisiae chromosomes) are indicated. B, BanII+EcoO109I digestion. A filter transfer was hybridized with the 17 α probe, which detected a number of fragments of different sizes in each cell line, indicating the length of the alphoid segments in the HACs. Several fragments of similar size were also shared by the four cell lines and the parental HT1080 HPRT− cells. The sizes (in kb) of the λ-DNA concatemer standards are indicated.
JM2860 is cleaved, by NotI, at two closely spaced sites flanking the boundary between the 17 α and HPRT1 genomic inserts (fig. 1A), generating two fragments—one of 404 kb (detected by both probes; fig. 4A) and one of <100 bp, which was not seen on the PFGE. Compared to the parental HT1080 control cells, the 17 α and the HPRT1 probes detected, in the four minichromosome cell lines, additional DNA fragments from 50 kb to >1 Mb in size, indicating rearrangement of the input DNA. Variation in the intensity of the bands within a cell line probably reflects differences either in copy number or in the relative contents of the target sequences detected by either probe. Each cell line exhibited a different NotI pattern.
Irregular rearrangement of input DNA from JM2860 in the minichromosome cell lines was also indicated by analysis with BssHII, SalI (fig. 1A), and BanII+EcoO109I, which do not cleave the 17 α DNA in JM2860. BssHII and SalI generate 286-kb and 222-kb fragments, respectively, in JM2860, as detected by the 17 α probe (fig. 4A), whereas the HPRT1 gene lies within a 64-kb BssHII fragment and a 114-kb SalI fragment, as detected by the HPRT1 probe (fig. 4A). In most of the minichromosome cell lines, BssHII and SalI fragments similar in size to those from JM2860 are observed, but these are part of a complex pattern including smaller and larger fragments detected with either probe and absent from the parental HT1080 cell line (fig. 4A). Although larger fragments may result from inactivation of relevant restriction sites by overlapping CpG methylation of genomic DNA, these results are compatible with the irregular, alternating pattern of alphoid and HPRT1 segments observed by fiber FISH.
Used in combination, BanII and EcoO109I generate a 220-kb JM2860 fragment detected by the 17 α probe (fig. 4B), whereas the nonalphoid moiety of the construct is digested to fragments <1 kb on average. Compared to the parental HT1080 control, the size of BanII+EcoO109I fragments from the minichromosome cell lines detected by the 17 α probe was ∼50–250 kb (fig. 4B). Both endonucleases are insensitive to CpG methylation. Under the assumptions that each of the fragments detected occurs once on the minichromosome and that together they represent an alphoid content similar to that in the parental JM2860 (55%), the order of magnitude for minichromosome size is estimated at 1–5 Mb.
To determine whether the HACs were linear or circular structures, DNA from AG1-1, AG3-1, AG6-1, and AP1-1 was digested by the exonuclease Bal31, followed by XhoI restriction cleavage. Filter transfers were hybridized with probes for the BAC vector and the ampicillin-resistance genes from either arm of JM2860 (fig. 1B), to detect truncated fragments resulting from the removal, by the exonuclease, of sequences from each telomeric end. Figure 5 shows that in each cell line fragments were generated that—compared to the I-SceI–linearized DNA from JM2860, which was entirely degraded—were resistant to Bal31 activity. When both the sizes of the fragments detected and the restriction map of JM2860 are taken into account, these results show the presence of an internal JM2860 segment stretching from the BAC vector to the ampicillin-resistance gene. This may be due to a JM2860 molecule having escaped I-SceI cleavage. Although linearization of the vector was confirmed by PFGE, residual circular molecules were not specifically detected, because these fail to fractionate into the gel. An additional, rearranged internal copy of the same segment was detected in AG6-1.
Figure 5.
Bal31 exonuclease analysis. Southern blot analysis of Bal31-digested genomic DNA shows that the telomeric ends of JM2860 (fig. 1A) are internal in the HACs. Zero (lanes 0), two (lanes 2), or four (lanes 4) units of Bal31 exonuclease were used, followed by digestion by XhoI. The 4.8-kb and 6.3-kb terminal XhoI fragments of I-SceI–cleaved JM2860 are detected, respectively, by AmpR and repE probes (fig. 1B) in the control, in which the exonuclease is omitted (lanes 0). These fragments are degraded by Bal31, whereas the fragments detected by the probes in minichromosome cell lines are resistant (notwithstanding a band-intensity decrease expected in light of the extension of Bal31 degradation to internal regions). The 6.8-kb JM2860 fragment detected by the AmpR probe probably results from incomplete digestion by I-SceI. Genomic DNA from the parental HT1080 cells was included as a negative control for the minichromosome probes used.
Further studies using controlled γ irradiation and fractionation, on a pulsed-field gel (Taylor et al. 1996), of of DNA molecules ⩽3 Mb in size suggested that the artificial minichromosomes were circular molecules (data not shown). Both the 17 α probe and a specific probe containing the lacZ and ampicillin-resistance genes detected a band within the limiting mobility of the pulsed-field gel—which was present only after γ irradiation and increased in intensity with higher doses, indicating that the circular molecules had generated linear forms after breakage.
Mitotic Stability of HACs
To investigate HAC stability in AG1-1, AG3-1, AG6-1, and AP1-1, these cell lines were analyzed for the persistence of artificial minichromosomes during growth under nonselective conditions. FISH was performed with the 17 α probe after 26–40 d and again, after 54–62 d of culture in the absence of G418, puromycin, and HAT. The results of this analysis are summarized in table 1. Daily rates of minichromosome loss were calculated from the number of minichromosome-positive metaphase spreads; the values were .0049–.017 (0.49%–1.7%/d), depending on the cell line, without large differences between the first and the second FISH analyses. In two cell lines, AG1-1 and AG3-1, minichromosome loss was accompanied by integration events in the natural chromosomes, affecting 8%–16% of nuclei; the proportion of these events, however, did not increase during the second off-selection month. With a generation time of ∼1 d for HT1080 cells, these rates of minichromosome loss suggest efficient segregation in 98%–99% of mitoses and, therefore, the presence of a functional centromere in all but a small minority of the minichromosomes. The number of artificial minichromosomes per nucleus was unchanged relative to the cells grown in selective medium.
Table 1.
Minichromosome Stability in the Absence of Selective Pressure
|
FISH Analysis of Cells off Selectionb(% of spreads) |
||||||||
| Cell Line | Initial Selection | Minichromosomes Observed under Selectiona (% of spreads) | Days Off Selection | Minichromosome | TerminalIntegrationd | OtherChromosomalIntegrationEvents | NoSignale | Daily Rate of Minichromosome Lossc |
| AG1-1 | HAT | 74 | 32 | 50 | 0 | 10 | 40 | .012 |
| 62 | 32 | 0 | 8 | 60 | .013 | |||
| AG3-1 | G418 + puromycin | 88 | 40 | 44 | 0 | 16 | 40 | .017 |
| 54 | 36 | 0 | 12 | 52 | .016 | |||
| AG6-1 | Triple | 96 | 32 | 82 | 0 | 0 | 18 | .0049 |
| 58 | 68 | 0 | 0 | 32 | .0059 | |||
| AP1-1 | Triple | 92 | 32 | 60 | 0 | 0 | 40 | .013 |
| 60 | 32 | 0 | 0 | 68 | .017 | |||
Control cells had been passaged under selection for 7–11 wk since transfection.
FISH analysis of metaphase chromosome spreads was performed with a probe for 17 α DNA; 50 spreads from each cell line were analyzed at the two time points indicated, after selective pressure was lifted.
Calculated by the formula Nn=N0×(1-R)n where N0 is the number of metaphase chromosome spreads showing minichromosomes in the cells cultured under selection, Nn is the number of minichromosome-containing metaphase chromosome spreads after n days of culture in the absence of selection, and R is the daily rate of loss.
Hybridization signal at the tip of a natural chromosome, suggesting a telomere-directed truncation event.
Spreads with a hybridization signal on the natural chromosomes 17 only.
Complementation of the HPRT− Phenotype
The ability of HACs to serve as vectors for gene transfer was investigated by including the HPRT1 gene (40 kb) in JM2860 and by using host cells with an HPRT− genetic background (Lugo and Baker 1983). Cell lines containing minichromosomes were successfully isolated in the presence of HAT supplement, to select for the HPRT1 gene. The presence of HPRT1 FISH signals only on the artificial minichromosomes and at the nonfunctional locus in Xq26.2 demonstrated that one or more copies of the HPRT1 gene linked to the minichromosomes in AG1-1, AG3-1, AG6-1, and AP1-1 must be responsible for the complementation of the HPRT− phenotype of the host cells. One of the cell lines, AG3-1, was isolated and expanded in the absence of selective pressure for the HPRT1 gene. Subsequent transfer to growth medium containing HAT resulted in no adverse effects for the culture, suggesting continued expression of the gene.
Further evidence of the transformation to an HPRT+ phenotype mediated by the minichromosomes was obtained by use of 6-TG, which is converted to cytotoxic thioguanine deoxynucleotide triphosphate in an HPRT-dependent pathway. Whereas the parental HT1080 cells exhibited normal growth in the presence of 5 μg 6-TG/ml, the four minichromosome lines showed complete cell death in ⩽5 d. By contrast, cells of the four lines previously grown off selection for 57 d maintained high densities and, over the same 5-d period, reached confluence in the presence of 6-TG. This is consistent with the growth of a subpopulation of minichromosome-negative cells during extended culture off selection (table 1). It also indicates that minichromosome formation was not accompanied by integrations of the HPRT1 gene into the natural chromosomes, in accordance with the data obtained by FISH analysis of the four cell lines.
Discussion
In this study, we intended to determine whether the integration of a full-length human genomic locus into a HAC construct would allow the formation of artificial chromosomes complementing the corresponding metabolic deficiency in human cultured cells. A large HAC construct containing human telomeres, 17 α DNA, and the HPRT1 gene was transferred into HPRT− HT1080 cells, and several aminopterin-resistant, positive clones were isolated. These were shown to contain HACs with a functional centromere at a consistently low copy number of 1 or 2/cell. The HACs were derived only from the input DNA and contained one or possibly several copies of the HPRT1 gene. Analysis of HAC stability showed that accurate segregation occurred in >98% of mitoses and that, in the absence of selective pressure, 35%–70% of the cells retained HACs at the end of a 2-mo period. These frequencies were comparable to rates of minichromosome loss in HT1080 cells reported by Ikeno et al. (1998). In two other studies (Harrington et al. 1997; Ebersole et al. 2000), HAC transmission was observed in 100% of mitoses of some clones, although variation in mitotic stability was noted between different cell lines.
This study has also shown that the HACs obtained were several megabases in size, which is significantly larger than the input DNA derived from the HAC construct, JM2860. The studies on DNA structure and fiber-FISH analysis indicated that the HACs consist of irregular alternating stretches of alphoid and HPRT1-locus DNA. This may have resulted from recombination of the input DNA, ligation of random truncated fragments, or a combination of these mechanisms. Southern hybridization data from our Bal31 analysis indicate that certain minichromosome segments probably derive from JM2860 molecules that have escaped digestion by I-SceI endonuclease prior to transfection. Except for the relatively small restriction fragments detected in that experiment, the linear or circular origin of minichromosome DNA cannot be confirmed. Chen et al. (2001) found that linear but not circular plasmid molecules formed large concatemers, possibly by nonhomologous end-joining (Critchlow and Jackson 1998), when transferred to mouse liver cells in vivo. Head-to-tail concatemers integrated into the host-cell chromosomes, however, have been observed with both linear and circular DNA (Folger et al. 1982). The irregular structure of the minichromosomes in the present study may be related to other structural aspects of input DNA, such as the inclusion of a large noncentromeric insert in JM2860.
Under the assumption of an unbiased representation of the alphoid and nonalphoid components of the HAC vector in the minichromosomes, these may be expected to contain several copies of the HPRT1 gene, although the number of intact, potentially functional copies is difficult to determine. Given the nature of the final HAC structure, our results indicate that, in a context in which both transgene copy number and expression levels must be tightly controlled, the insertion, by recombinational methods, of the transgene into an established HAC (Kuroiwa et al. 2000) should be considered.
HACs have a distinct advantage over those gene-transfer vectors that either may be silenced following integration into the host-cell chromosomes or may incorporate cDNA inserts that may lack important sequences for long-term regulated expression of the transgene. By relying entirely on the endogenous mechanisms for chromosome maintenance, HACs also offer a valid alternative approach to viral vectors such as adenovirus. Recently, Wade-Martins et al. (2000), using an episomal vector derived from Epstein-Barr virus (EBV), reported the transfer of the HPRT1 locus. Although this is a useful strategy for both the expression and the analysis of large genes in some mammalian cell lines, the system may not be generally applicable, because EBV-based vectors rely on the presence of a viral transactivator, EBNA1, and because there are safety concerns over their use for gene therapy (Van Craenenbroeck et al. 2000). Moreover, in the aforementioned report, the rates of episome loss in the absence of selection were fourfold greater than those for HACs in the present study.
Previous studies in this field have aimed to maximize the size of the alphoid-DNA fragments used to generate artificial chromosomes, either by preparing concatemers of an alphoid insert (Harrington et al. 1997) or by working with a large alphoid yeast-artificial-chromosome clone (Henning et al. 1999). It is unclear, however, whether a large continuous array of alphoid DNA (a) is necessary to form a functional minichromosome centromere or (b) increases the probability that the centromere will initially be established. In the present study, the irregular arrangement of alternating alphoid and nonalphoid sequences observed in the HACs conflicts with the structure of natural centromeres, in which continuous alphoid-DNA arrays may be up to several megabases in size. The largest 17 α–DNA fragments in our HACs were 158–250 kb in size, providing an estimate of the minimal length that may be required to form a de novo centromere (however, two closely spaced alphoid segments could possibly act as a single functional unit [Choo 2000]).
Recent preliminary data from our laboratory, however, indicate both that mitotic stability is not strictly correlated with the size of continuous alphoid segments and that the exclusion of the HPRT1 insert from the parental HAC construct results in lower minichromosome loss (J. E. Mejía, A. Alazami, A. Willmott, and Z. Larin, unpublished data). It is possible that, in the JM2860 HACs, the boundaries between alphoid and nonalphoid DNA fail to determine the boundaries between heterochromatic and euchromatic DNA. Early S-phase origins of replication in transcriptionally active segments derived from the HPRT1 locus may initiate premature replication of neighboring alphoid DNA. Temporally decoupling the synthesis of alphoid DNA from that of centromeric proteins such as centromeric protein A (Karpen and Allshire 1997; Choo 2000) could impair the maintenance of functional centromeres in HACs. Conversely, longer stretches of alphoid DNA may preserve internal segments of late replication, by virtue of a greater distance between these and early S-phase origins of replication. Replication interference may inhibit kinetochore formation in a portion of artificial minichromosomes, leading to nonattachment to the mitotic spindle and, at anaphase, the failure to segregate with the other chromosomes. This mechanism of minichromosome loss is consistent with the conservation of minichromosome-copy number seen in the remaining positive cells. It may not be a coincidence that the lowest rate of minichromosome loss observed in the present study occurs in the cell line (AG6-1) in which the maximal-alphoid-segment size is the longest.
Future studies could address the timing of replication of both alphoid and nonalphoid DNA in artificial chromosomes—and its relation to the maintenance and stability of artificial chromosomes in human cells. Should the detrimental effect that replication-timing interference may have on centromere function be demonstrated, it might be possible to isolate the alphoid-DNA segment in the HAC construct by flanking it with replication-timing–boundary elements (Watanabe et al. 2000). The use of a shorter alphoid segment in an optimal sequence context would allow a reduction in the total size of HAC constructs, to make them compatible with more-efficient delivery vehicles, such as the HSV-1 amplicon packaging system, with a theoretical transgene capacity of ⩽150 kb (Sena-Esteves et al. 2000).
We have shown that artificial chromosomes based on the major component of natural human centromeres, alphoid DNA, can be used as vectors for the complementation of a genetic deficiency in human cells. The HACs generated in this study constitute a pertinent model to address the formation and maintenance of de novo centromeres and indicate directions for further research and improvements in HAC-vector technology for gene transfer.
Acknowledgments
This work was supported by a fellowship from the Wellcome Trust (to Z.L.), by SmithKline Beecham, by the University of Oxford Medical Research Fund, and by the Dystrophic Epidermolysis Bullosa Research Association of the United Kingdom.
Electronic-Database Information
Accession numbers and the URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HPRT1 [MIM 308000] and Lesch-Nyhan syndrome [MIM 300322])
- Project Ensembl, http://www.ensembl.org/ (for HPRT1 [Ensembl gene ENSG00000101965])
References
- Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr J-P (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA 92:7297–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey CT, Kruh GD (1979) The HPRT locus. Cell 16:1–9 [DOI] [PubMed] [Google Scholar]
- Chen Z-Y, Yant SR, He C-Y, Meuse L, Shen S, Kay MA (2001) Linear DNAs concatemerize in vivo and result in sustained transgene expression in mouse liver. Mol Ther 3:403–410 [DOI] [PubMed] [Google Scholar]
- Choo KHA (2000) Centromerization. Trends Cell Biol 10:182–188 [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP (1998) DNA end-joining: from yeast to man. Trends Biochem Sci 23:394–398 [DOI] [PubMed] [Google Scholar]
- Ebersole TA, Ross A, Clark E, McGill N, Schindelhauer D, Cooke H, Grimes B (2000) Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum Mol Genet 9:1623–1631 [DOI] [PubMed] [Google Scholar]
- Folger KR, Wong EA, Wahl G, Capecchi MR (1982) Patterns of integration of DNA microinjected into cultured mammalian cells: evidence for homologous recombination between injected plasmid DNA molecules. Mol Cell Biol 2:1372–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF (1997) Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15:345–355 [DOI] [PubMed] [Google Scholar]
- Henning KA, Novotny EA, Compton ST, Guan X-Y, Liu PP, Ashlock MA (1999) Human artificial chromosomes generated by modification of a yeast artificial chromosome containing both human alpha satellite and single-copy DNA sequences. Proc Natl Acad Sci USA 96:592–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H (1998) Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol 16:431–439 [DOI] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC (1997) The case for epigenetic effects on centromere identity and function. Trends Genet 13:489–496 [DOI] [PubMed] [Google Scholar]
- Kim U-J, Birren BW, Slepak T, Mancino V, Boysen C, Kang H-L, Simon MI, Shizuya H (1996) Construction and characterization of a human bacterial artificial chromosome library. Genomics 34:213–218 [DOI] [PubMed] [Google Scholar]
- Kuroiwa Y, Tomizuka K, Shinohara T, Kazuki Y, Yoshida H, Ohguma A, Yamamoto T, Tanaka S, Oshimura M, Ishida I (2000) Manipulation of human minichromosomes to carry greater than megabase-sized chromosome inserts. Nat Biotechnol 18:1086–1090 [DOI] [PubMed] [Google Scholar]
- Larin Z (1995) Functional analysis of mammalian genomes using yeast artificial chromosomes. In: Monaco AP (ed) Pulsed field gel electrophoresis—a practical approach. IRL Press, Oxford, pp 139–157 [Google Scholar]
- Lugo TG, Baker RM (1983) Chromosome-mediated gene transfer of HPRT and APRT in an intraspecific human cell system. Somat Cell Genet 9:175–188 [DOI] [PubMed] [Google Scholar]
- Marschall P, Malik N, Larin Z (1999) Transfer of YACs up to 2.3 Mb intact into human cells with polyethylenimine. Gene Ther 6:1634–1637 [DOI] [PubMed] [Google Scholar]
- Mejía JE, Larin Z (2000) The assembly of large BACs by in vivo recombination. Genomics 70:165–170 [DOI] [PubMed] [Google Scholar]
- Mejía JE, Monaco AP (1997) Retrofitting vectors for Escherichia coli-based artificial chromosomes (PACs and BACs) with markers for transfection studies. Genome Res 7:179–186 [DOI] [PubMed] [Google Scholar]
- Mills W, Critcher R, Lee C, Farr CJ (1999) Generation of an approximately 2.4 Mb human X centromere-based minichromosome by targeted telomere-associated chromosome fragmentation in DT 40. Hum Mol Genet 8:751–761 [DOI] [PubMed] [Google Scholar]
- Rasheed S, Nelson-Rees WA, Toth EM, Arnstein P, Gardner MB (1974) Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer 33:1027–1033 [DOI] [PubMed] [Google Scholar]
- Saitoh H, Tomkiel J, Cooke CA, Ratrie H, Maurer M, Rothfield NF, Earnshaw WC (1992) CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell 70:115–125 [DOI] [PubMed] [Google Scholar]
- Sena-Esteves M, Saeki Y, Fraefel C, Breakefield XO (2000) HSV-1 amplicon vectors—simplicity and versatility. Mol Ther 2:9–15 [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S (1995) Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet 4:2189–2197 [DOI] [PubMed] [Google Scholar]
- Taylor SS, Larin Z, Tyler-Smith C (1994) Addition of functional human telomeres to YACs. Hum Mol Genet 3:1383–1386 [DOI] [PubMed] [Google Scholar]
- ——— (1996) Analysis of extrachromosomal structures containing human centromeric alphoid satellite DNA sequences in mouse cells. Chromosoma 105:70–81 [DOI] [PubMed] [Google Scholar]
- Van Craenenbroeck K, Vanhoenacker P, Haegeman G (2000) Episomal vectors for gene expression in mammalian cells. Eur J Biochem 267:5665–5678 [DOI] [PubMed] [Google Scholar]
- Wade-Martins R, White RE, Kimura H, Cook PR, James MR (2000) Stable correction of a genetic deficiency in human cells by an episome carrying a 115 kb genomic transgene. Nat Biotechnol 18:1311–1314 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Tenzen T, Nagasaka Y, Inoko H, Ikemura T (2000) Replication timing of the human X-inactivation center (XIC) region: correlation with chromosome bands. Gene 252:163–172 [DOI] [PubMed] [Google Scholar]
- Willard HF (1998) Centromeres: the missing link in the development of human artificial chromosomes. Curr Opin Genet Dev 8:219–225 [DOI] [PubMed] [Google Scholar]
- ——— (2000) Artificial chromosomes coming to life. Science 290:1308–1309 [DOI] [PubMed] [Google Scholar]
- Yang JW, Pendon C, Yang J, Haywood N, Chand A, Brown WRA (2000) Human mini-chromosomes with minimal centromeres. Hum Mol Genet 9:1891–1902 [DOI] [PubMed] [Google Scholar]