Abstract
Multiple lines of evidence have implicated the short arm of chromosome 8 as harboring genes important in prostate carcinogenesis. Although most of this evidence comes from the identification of frequent somatic alterations of 8p loci in prostate cancer cells (e.g., loss of heterozygosity), studies have also suggested a role for 8p genes in mediation of inherited susceptibility to prostate cancer. To further examine this latter possibility, we performed linkage analyses, in 159 pedigrees affected by hereditary prostate cancer (HPC), using 24 markers on the short arm of chromosome 8. In the complete set of families, evidence for prostate cancer linkage was found at 8p22-23, with a peak HLOD of 1.84 (P=.004), and an estimate of the proportion of families linked (α) of 0.14, at D8S1130. In the 79 families with average age at diagnosis >65 years, an allele-sharing LOD score of 2.64 (P=.0005) was observed, and six markers spanning a distance of 10 cM had LOD scores >2.0. Interestingly, the small number of Ashkenazi Jewish pedigrees (n=11) analyzed in this study contributed disproportionately to this linkage. Mutation screening in HPC probands and association analyses in case subjects (a group that includes HPC probands and unrelated case subjects) and unaffected control subjects were carried out for the putative prostate cancer–susceptibility gene, PG1, previously localized to the 8p22-23 region. No statistical differences in the allele, genotype, or haplotype frequencies of the SNPs or other sequence variants in the PG1 gene were observed between case and control subjects. However, case subjects demonstrated a trend toward higher homozygous rates of less-frequent alleles in all three PG1 SNPs, and overtransmission of a PG1 variant to case subjects was observed. In summary, these results provide evidence for the existence of a prostate cancer–susceptibility gene at 8p22-23. Evaluation of the PG1 gene and other candidate genes in this area appears warranted.
Introduction
The short arm of chromosome 8, specifically 8p22-23, may harbor a prostate cancer–susceptibility gene(s) for the following reasons. First, multiple loci on 8p are the sites of frequent loss of heterozygosity (LOH) in a variety of human cancers, including prostate (Macoska et al. 1995; Bova et al. 1996; MacGrogan et al. 1996; Vocke et al. 1996; Deubler et al. 1997; Prasad et al. 1998), colon (Cunningham et al. 1993), breast (Chuaqui et al. 1995), ovarian (Cliby et al. 1993), liver (Emi et al. 1992), lung (Wistuba et al. 1999), bladder (Knowles et al. 1993), and head and neck cancer (Ransom et al. 1996). In prostate cancer, LOH for markers on 8p was found to be one of the most frequent somatic alterations, occurring in >60% of prostate cancers (Cunningham et al. 1996), and multiple homozygous deletions have been mapped to this chromosomal arm (Bova et al. 1996; Prasad et al. 1998). In addition, alterations of cancer-related genes in the region, such as LZTS1, have been identified in prostate cancer specimens and cell lines (Ishii et al. 1999).
Second, genomewide scans for prostate cancer–susceptibility genes in pedigrees affected with hereditary prostate cancer (HPC) have provided some evidence for prostate cancer linkage on 8p (Smith et al. 1996; Gibbs et al. 2000). In the 66 pedigrees affected by HPC ascertained by our group (Smith et al. 1996), there were positive linkage scores at 8p, with a two-point parametric LOD of 0.7 at D8S550, a multipoint LOD assuming heterogeneity (HLOD) of 0.81 (P=.05) and a multipoint nonparametric linkage score (NPL) of 2.02 (P=.02). Similarly, Gibbs et al. (2000) reported evidence for linkage at the marker D8S1106, ∼5 cM from the marker D8S550. The maximum multipoint NPL score was 2.02 in 44 pedigrees with late age at onset (⩾66 years).
Third, a candidate prostate cancer–susceptibility gene located at 8p22-23, PG1, was cloned by a haplotype-based association study conducted by Geneset (Cohen et al. 1999). In their study, a high-density array of biallelic markers, around D8S262 and D8S277 in the 8p23 region, was used to build haplotypes in case and control samples. By comparing 281 prostate cancer case subjects with 130 unaffected control subjects ascertained in France, they found significant differences in allele, genotype, and haplotype frequencies of several SNPs in the PG1 gene between case and control subjects. The allele frequencies of G of SNP 477, T of SNP 99217, and A of SNP 467 in case subjects (control subjects) were 0.33 (0.24), 0.31 (0.23), and 0.26 (0.16), respectively, in their study. In their study, the haplotype frequencies of G-T-A for the three SNPs were 0.25 and 0.13 in case and control subjects, respectively, with an odds ratio (OR) of 2.17 (P=.0002). A single protein sequence, designated as the PG1 gene, was identified in this candidate region. The function of this gene is unknown, and no follow-up studies have been presented.
We have three major objectives in the current study: first, evaluate evidence for linkage at 8p22-23, using densely spaced markers in 159 HPC families ascertained at Johns Hopkins Hospital; second, evaluate evidence for association in the PG1 region using both the family-based approach in the 159 HPC families and the case-control approach in 249 case subjects with sporadic prostate cancer and 211 unaffected male control subjects; and third, screen the PG1 gene for segregating mutations, using the single-strand conformation polymorphism (SSCP) method.
Methods
Family Collection
All 159 families with HPC were collected and studied at the Brady Urology Institute at Johns Hopkins Hospital in Baltimore. Families were ascertained from three resources. A majority of them were ascertained through referrals generated as a response to a letter by one of us (P.C.W.) to 8,000 urologists throughout the country. The second source was identified from family history records of the patient population seen at Johns Hopkins Hospital for treatment of prostate cancer. The remaining families came from the respondents to articles published in a variety of lay publications describing our studies of families affected with prostate cancer. Prostate cancer diagnosis was verified by medical records for each affected man studied. Age at diagnosis of prostate cancer was confirmed either through medical records or from two other independent sources. The mean age at diagnosis was 64.3 years for the case subjects in these families. Of the families, 84% are non-Jewish whites, 6.9% are Ashkenazi Jews, and 8.8% are black.
All 249 unrelated case subjects were recruited from among patients who underwent treatment for prostate cancer at the John Hopkins Hospital. The diagnosis of prostate cancer for all these subjects was confirmed by pathology reports. Preoperative prostate-specific antigen (PSA) levels, Gleason score, and pathological stages were available for 92, 244, and 245 of the 249 case subjects, respectively. Mean age at diagnosis for these case subjects was 58.6 years (range 37–73 years, SD 6.85). Family-history information was not obtained. Over 93% of the case subjects are white, and 3.2% are black.
From among men participating in screening programs for prostate cancer, 222 control subjects not affected with prostate cancer were selected. By applying the exclusion criteria of abnormal digital rectal examination (DRE) and abnormal PSA level (i.e., ⩾4 ng/ml), 211 were eligible for the study. The mean age at examination was 58 years (range 40–80 years, SD 8.01). Of the eligible control subjects, >86% are white, and 7.1% are black. On the basis of interviews of eligible control subjects, 5.6% have a brother or father affected with prostate cancer.
Marker Genotyping
Twenty-one microsatellite markers spanning ∼35 cM at 8p22-23 were genotyped in 159 families with HPC. These markers were selected from Marshfield comprehensive human genetic maps (Broman et al. 1998). Multiplex PCR, using fluorescently labeled primers (either fam, hex, or ned), was performed, and the resulting PCR fragments were separated using capillary electrophoresis performed with an ABI 3700 sequencer. The genotypes were scored using ABI software (GENOTYPER). A modified version of the program Linkage Designer was used to bin the alleles and check inheritance. The output from Linkage Designer was then analyzed further for any inconsistencies by running the LINKAGE software (Lathrop et al. 1984; Cottingham et al. 1993) without disease phenotype information. Marker allele frequencies were estimated from the 214 independent individuals in the data set (among them, 13 are Ashkenazi Jews and 19 are black). The marker order and distances estimated from the data using CRIMAP (Lander and Green 1987) were similar to the results in the Marshfield database. Thus, the intermarker distances of the Marshfield database were used in the analyses.
Three SNPs in the PG1 gene were genotyped in all 159 HPC pedigrees, in the 249 unrelated case subjects affected with prostate cancer, and in 211 unaffected control subjects. All information (e.g. sequence, nomenclature, and designation of SNPs) for PG1 was obtained from Cohen et al. (1999). SNP 477 (C→ G) is in intron 3, SNP 99217 (C→T) is in intron 5, and SNP 467 (G→C) is in the 3′ untranslated region. Marker D8S561 is an intragenic marker. Direct sequencing of PCR products was used to genotype the three SNPs. All the PCRs were performed in a 10-μl volume consisting of 30 ng genomic DNA, 0.2 μM each primer, 0.2 mM each dNTP, 1.5 mM MgCl2, 20 mM Tris-HCl, 50 mM KCl, and 0.5 U Taq polymerase (Life Technologies). The primers for the SNP 477 were 5′-TGTTGATTTACAGGCGGC-3′ and 5′-GGAAAGGTACTCATTCATAG-3′. The primers for the SNP 99217 were 5′-GGTGGGAATTTACTATATG-3′ and 5′-GTTTATTTTGTGTGAGCTTTG-3′. The primers for the SNP 467 were 5′-AAGTTCACCTTCTCAAGC-3′ and 5′-TGAAAGAGTTTATTCTCTGG-3′ (Cohen et al. 1999). These primers amplified 429-bp, 430-bp, and 420-bp fragments for SNP 477, SNP 99217, and SNP 467, respectively. PCR cycling conditions were as follows: 94°C for 4 min; followed by 28 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 20 s with a final extension of 72°C for 2 min, except annealing temperature for SNP 477 was 60°C. All PCR products were purified using QuickStep PCR purification kit (Edge BioSystems) to remove dNTPs and excess primers. All sequencing reactions were performed using dye-terminator chemistry (BigDye) and then were precipitated using 63%±5% ethanol. Samples were loaded onto an ABI 3700 DNA Analyzer after adding 7 μl of formamide.
Mutation Screening
Probands from 92 families affected by HPC were screened for sequence variations in the eight exons of PG1 using SSCP analysis. All eight exons were screened using 10 primer sets (see table 1) based on intronic sequence, as described by Cohen et al. (1999). Primers for this analysis were chosen with a minimum distance of 4 bp between primer 3′ base and exon boundaries. Four different electrophoresis and gel conditions were used to maximize detection of sequence variations: mutation detection enhancement (MDE) at room temperature, MDE supplemented with 5% glycerol at room temperature, MDE at 4°C, and MDE supplemented with 5% glycerol at 4°C. SSCP gels were loaded immediately after completion of the PCR reactions incorporating 33P dATP, then subjected to electrophoresis at 4 W for ⩾16 h. Fragment detection was accomplished by autoradiography. Abnormally migrating products were directly sequenced as described above. Exons containing sequence variations in HPC probands were analyzed in control individuals as well.
Table 1.
Primers Used for Mutation Screening of PG1 Exons
| Exon | Primer | AnnealingTemperature(°C) |
| 1a-F | GCCGAGCTGAGAAGATGCTG | 62 |
| 1a-R | CGGGAGCTCGGGTGGACGCC | |
| 1c-F | CGCTGCCGCCGAGCTGAG | 63 |
| 1c-R | GGCTCACCTGGACCCCGG | |
| 2-F | CAACATCATTCGTCAGTTTC | 57 |
| 2-R | ACCTAGGTTTCATGCAAATG | |
| 3-F | CTGTGAAGAGCCTCATGTAC | 62 |
| 3-R | AGAGAGAAAAGCATGGAAAC | |
| 4-F | CTGGCCAATTGTTATTTTAA | 53 |
| 4-R | AATTTAGAAACTGAGAGCTG | |
| 5-F | ACCAAATTTGCTCTATGTCC | 60 |
| 5-R | AAAGTATCTTTTCCAGGAAG | |
| 6-F | TTAATGACGGCACTGATTG | 53 |
| 6-R | AGGTGCGTGAACACACTTAC | |
| 7-F | CTTTATATGACCATGAGTTC | 46 |
| 7-R | CTGGAACTGTTGTTACTCAC | |
| 8a-F | CAGCGTGTAATAGCTACCTG | 62 |
| 8a-R | CACATACAGCTTCCTTCCAG | |
| 8c-F | CCATCAATGTTGATCTTAAGTGG | 50 |
| 8c-R | AATGTAGCACATCCCACTGTCTG |
Statistical Analyses
Tests for Hardy-Weinberg equilibrium (HWE) for all the markers and for linkage disequilibrium (LD) between all pairs of markers were performed using independent individuals (pedigree founders and spouses of family members) of families with HPC and all sporadic case subjects and control subjects not affected with prostate cancer (computer program GDA; Weir et al. 1996). The HWE tests were based on exact tests, where a large number of the possible arrays was generated by permuting the alleles among genotypes and the proportion of these permuted genotypic arrays that have a smaller conditional probability than the original data were calculated. The LD tests were based on an exact test assuming multinominal probability of the multilocus genotype, conditional on the single-locus genotype (Zaykin et al. 1995). A Monte Carlo simulation was used to assess the significance, by permuting the single-locus genotypes among individuals in the sample to simulate the null distribution. The empirical P values of both the HWE and LD tests were based on 10,000 replicate samples.
Multipoint linkage analyses were performed using both parametric and nonparametric methods, implemented by the computer program GENEHUNTER-PLUS (Kruglyak et al. 1996; Kong and Cox 1997). For the parametric analysis, the same autosomal dominant model that was used by Smith et al. (1996) was assumed. Under this model, disease-gene frequency of .003, incomplete penetrance, and phenocopies were assumed. Specifically, affected men were assumed to be disease-gene carriers, with a fixed phenocopy rate of 15%, whereas all unaffected men aged <75 years and all women were assumed to be of unknown phenotype. In men aged ⩾75 years, the lifetime penetrance of gene carriers was estimated to be 63%, and the lifetime risk of prostate cancer for noncarriers was 16% in this age class. Linkage in the presence of heterogeneity was assessed by use of Smith’s admixture test for heterogeneity (Ott 1998). In this test, two types of families are assumed, one type linked to the disease locus with a proportion of α, and the other type is not linked, with the proportion 1−α. A maximum-likelihood approach was used to estimate α by maximization of the admixed LOD score (HLOD).
For the nonparametric analysis, the estimated marker identical-by-descent (IBD) sharing of alleles for the various affected relative pairs was compared with its expected values under the null hypothesis of no linkage. A statistic “Zall” in the program was used (Whittemore and Halpern 1994). Allele-sharing LOD scores were then calculated, on the basis of Zall, with equal weight assigned to all families, using the computer program ASM (Kong and Cox 1997).
Both HLOD and allele-sharing LOD can be converted to a χ2 (χ2=4.6 × HLOD). Although the true distribution of the χ2 under the null hypothesis of no linkage is unknown—especially in multipoint analysis—we assume that the distribution is a mixture of one that is degenerate at 0 and one that can be approximated by the distribution of the maximum of two independent χ2 variables, each with 1 df (Faraway 1993). P values were thus calculated by 0.5×[1-(1-P1)(1-P1)], where P1 is the P value of χ2 with 1 df.
Family-based association tests were performed for all six markers in the 159 families affected with HPC, using a software package FBAT (Laird et al. 2000). Unlike the classic transmission/disequilibrium test (TDT), which is limited to a specific pedigree structure (one genotyped proband and two genotyped parents per pedigree), the FBAT utilizes data from nuclear families, sibships, or a combination of the two to test for linkage and linkage disequilibrium between traits and genotypes. The test for linkage is valid when multiple affected members per pedigree are used, and the power to detect linkage is increased if there is an association. The test for association is valid if one affected member per pedigree is used (the genotypes of all the affected members can be included) or if the empirical variance is used to account for correlation between transmissions in families when linkage is present. In brief, the FBAT determines, from the data, an S statistic that is the linear combination of offspring genotypes and phenotypes. The distribution of the S statistic is generated by treating the offspring genotype data as random and conditioning on the phenotypes and parental genotypes. When the marker is biallelic, a Z statistic and its corresponding P value is calculated. When the marker is multiallelic, a χ2 test is performed, with number of df equal to the number of alleles.
Population-based association tests were performed for the two polymorphisms in case and control subjects. An unconditional logistic regression was used to test for association between genotypes and affection status, adjusting for potential confounders such as age. The association tests were also performed for whites only, to decrease potential population stratification. The reported P values were not adjusted for multiple testing.
Haplotype frequencies in unrelated individuals were estimated for the three SNPs by maximum-likelihood estimation, using the best state of haplotype composition (see The Haplotype Estimation Help Page). The assumption of equal prior probabilities was made as a starting point for the expectation maximization (EM) algorithm.
Results
Linkage Results at 8p22-23 in 159 Pedigrees Affected by HPC
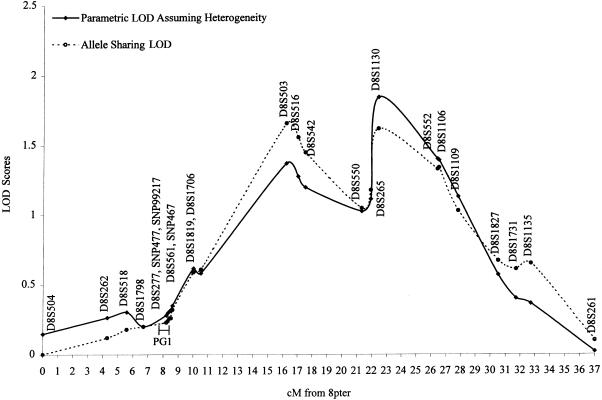
Both parametric and nonparametric multipoint linkage analyses provided evidence for linkage between a prostate cancer–susceptibility locus and markers on chromosome 8p in the complete 159 HPC pedigrees (fig. 1). The highest parametric HLOD was 1.84 (P=.004) with α=0.14, observed at D8S1130, 22 cM from 8pter at 8p22. HLOD scores ⩾0.5 extended across ∼22 cM, flanked by markers D8S1819 at 10 cM and D8S1135 at 32 cM from 8pter. The number of pedigrees that had LOD scores >0.3, >0.5, and >1 in the 22 cM region were 66, 33, and 4, respectively. In the first 66 pedigrees that were included in our previous genomewide screen (Smith et al. 1996), the highest HLOD increased from 0.7 at D8S550 (21 cM) to 1.67 (P=.005; α=0.24) at D8S1130 (22 cM), because of the inclusion of fine-mapping markers. The 93 new pedigrees also provided evidence for linkage, with the highest HLOD of 0.77 (P=.06; α=0.12) at D8S552 (26 cM). For the nonparametric analyses, the highest allele-sharing LOD was 1.66 (P=.006) observed at D8S503, ∼16 cM from 8pter in the complete family set. The highest allele-sharing LODs were 1.99 (P=.002) at D8S1130 and 0.34 (P=.21) at D8S552, respectively, in the first 66 and new 93 pedigrees.
Figure 1.
Results of multipoint parametric and nonparametric linkage analyses of prostate cancer–susceptibility loci, using 24 markers (21 microsatellite markers and 3 SNPs) on chromosome 8p22-23 in 159 families affected by HPC. The solid line represents parametric LOD under the assumption of heterogeneity. The dotted line represents allele-sharing LOD. Each diamond and circle represents a marker.
Linkage analyses stratified by pedigree characteristics show that the pedigrees linked to 8p tend to have late onset, larger numbers of affected family members, and male-to-male disease transmission. Since the results from parametric and nonparametric were similar, only the results from nonparametric analyses are presented (table 2). The peak allele-sharing LOD was 2.64 (P=.0005) at D8S503 in the 79 pedigrees with mean age at onset ⩾65 years, 1.41 (P=.01) at D8S503 in the 90 pedigrees with five or more affected family members, and 1.31 (P=.01) in the 99 pedigrees with male-to-male disease transmission. Evidence for linkage in this region was observed in non-Jewish white pedigrees (n=133) and in the 11 Ashkenazi Jewish pedigrees (2 from the first 66 families), but not in the 14 black pedigrees. It is worth noting that 7 of the 11 Ashkenazi Jewish pedigrees had LOD scores ⩾0.3 in the region and that, as a group, the 11 Ashkenazi families contributed disproportionately to the overall LOD score (table 2). By combining the non-Jewish white pedigrees with Ashkenazi pedigrees, we observed a LOD of 1.99 (P=.002) in the region.
Table 2.
Nonparametric Allele-Sharing LOD
|
Nonparametric Allele-Sharing LOD |
||||||||||
| Age at Onset |
No. of Affected Subjects |
Male-to-Male Transmission |
Ethnicity |
|||||||
| Markers | cMa | <65(n=79) | ⩾65(n=80) | <5(n=69) | ⩾5(n=90) | Yes(n=99) | No(n=60) | Black(n=14) | Ashkenazi(n=11) | Non-JewishWhite(n=133) |
| D8S504 | 0 | 0 | .44 | .00 | .11 | .2 | 0 | 0 | .67 | 0 |
| D8S262 | 4.3 | 0 | .67 | .00 | .36 | .11 | .02 | 0 | .78 | .13 |
| D8S518 | 5.6 | 0 | .75 | .00 | .43 | .15 | .03 | 0 | .93 | .18 |
| D8S1798 | 6.7 | 0 | .91 | .00 | .43 | .17 | .04 | 0 | 1.01 | .16 |
| D8S277 | 8.2 | 0 | 1.01 | .00 | .52 | .14 | .09 | 0 | .92 | .18 |
| SNP 477 | 8.3 | 0 | 1 | .00 | .55 | .16 | .09 | 0 | .92 | .19 |
| SNP 99217 | 8.4 | 0 | 1.02 | .00 | .59 | .18 | .09 | 0 | .92 | .2 |
| D8S561 | 8.5 | 0 | .99 | .00 | .58 | .17 | .09 | 0 | .92 | .2 |
| SNP 467 | 8.6 | 0 | 1.13 | .00 | .6 | .24 | .09 | 0 | .92 | .2 |
| D8S1819 | 10 | 0 | 1.58 | .00 | .89 | .47 | .14 | 0 | .93 | .45 |
| D8S1706 | 10.5 | 0 | 1.64 | .00 | .88 | .58 | .09 | 0 | .96 | .47 |
| D8S503 | 16.2 | 0 | 2.64 | .31 | 1.41 | 1.31 | .39 | 0 | 1.27 | 1.38 |
| D8S516 | 17 | 0 | 2.64 | .24 | 1.41 | 1.16 | .42 | 0 | 1.3 | 1.28 |
| D8S542 | 17.5 | 0 | 2.61 | .19 | 1.38 | .98 | .48 | 0 | 1.31 | 1.18 |
| D8S550 | 21.3 | 0 | 1.96 | .12 | 1.03 | .69 | .35 | 0 | 1.3 | .81 |
| D8S265 | 21.9 | 0 | 1.89 | .13 | 1.17 | .85 | .34 | 0 | 1.26 | .91 |
| D8S1130 | 22.4 | .1 | 1.97 | .15 | 1.67 | 1.07 | .56 | 0 | 1.12 | 1.39 |
| D8S552 | 26.4 | 0 | 2.32 | .28 | 1.09 | .79 | .54 | 0 | .92 | 1.11 |
| D8S1106 | 26.5 | 0 | 2.32 | .28 | 1.09 | .8 | .54 | 0 | .93 | 1.12 |
| D8S1109 | 27.8 | 0 | 1.97 | .11 | 1.03 | .71 | .33 | 0 | 1.07 | .82 |
| D8S1827 | 30.5 | 0 | 1.56 | .01 | .9 | .84 | .02 | 0 | 1.27 | .36 |
| D8S1731 | 31.7 | 0 | 1.55 | .00 | 1.09 | .9 | 0 | 0 | 1.51 | .28 |
| D8S1135 | 32.7 | 0 | 1.35 | .00 | 1.13 | .8 | .02 | 0 | 1.6 | .31 |
| D8S261 | 37 | 0 | .62 | .00 | .41 | .11 | .01 | 0 | .85 | .02 |
Based on the Marshfield map.
To evaluate the impact of the marker allele frequencies on our linkage results in the black and Ashkenazi Jewish families, we repeated linkage analyses for the 14 black and 11 Ashkenazi families using marker allele frequencies estimated from 19 unrelated blacks and 13 unrelated Ashkenazi Jews, respectively. The results were similar to that using marker allele frequencies estimated from the mixed 214 unrelated subjects. In the 14 black families, the peak HLOD changed from 0.26 to 0.1 at D8S261. In the 11 Ashkenazi families, the peak HLOD changed from 1.25 to 1.24 at D8S1135. The robustness of our linkage results to the estimates of marker allele frequencies is probably due to the use of dense markers in multipoint analyses.
The evidence for linkage in and around the PG1 gene (8 cM from pter) was weak. The highest HLOD and allele sharing LOD was 0.35 (P=.18) and 0.32 (P=.20), respectively, in the five markers within and surrounding the gene (from D8S277 to SNP 467).
Analysis of PG1: Family-Based Linkage and Association Tests in 159 HPC Pedigrees
Tests for HWE were performed for all microsatellite markers and SNPs analyzed, using 214 unrelated individuals from the 159 HPC pedigrees for which genotype information was available. All the markers tested were in HWE (P>.05). Marker-marker LD was tested for the five closely spaced markers (SNPs) in the PG1 region. Markers SNP 477, SNP 99217, D8S561, and SNP 467 were in strong LD, with P<.0001 for all pairwise tests. Marker D8S277 was not in LD with these four markers (SNPs).
Family-based linkage and association tests were performed for the three SNPs. There was overtransmission of allele T of SNP 99217 from parents to affected sons, with Z=2.19 (P=.03). The observed score S was 151 for allele T, compared with the expected 139. Similar tests for SNP 477 and SNP 467 were not significant, with Z=0.85 (P=.40) and Z=0.31 (P=.76), respectively. To decrease the impact of heterogeneity among races, the family-based linkage and association tests were performed again in the 133 non-Jewish white pedigrees. The test for SNP 99217 was significant, with Z=2.70 (P=.007). The tests for the other two SNPs were not significant.
As either linkage or association in the data may lead to the significant test statistics, we performed two additional analyses to further explore the finding. The first analysis was a family-based association test using the empirical variance to account for correlation between transmissions in families when linkage is present. In this analysis, the evidence for association decreased, with Z=1.66 (P=.10) and Z=2.07 (P=.04), respectively in the complete 159 HPC pedigrees and in 133 non-Jewish white pedigrees. The second analysis is the stratified linkage analyses based on the probands’ genotype at SNP 99217. The pedigrees whose probands are T carriers contributed disproportionally to the evidence for linkage at 5 markers in the region. The 77 pedigrees whose probands are heterozygous ‘T’ and the 15 pedigrees whose probands are homozygous ‘T’ carriers had allele-sharing LODs of 0.5 (P=.12) and 1.44 (P=.01) at SNP 99217, respectively. In contrast, the 78 pedigrees whose probands are not T carriers had HLOD of 0. These data suggest that both linkage and association contribute to the significance of the family- based test.
Analysis of PG1: Population-Based Association Tests in HPC Probands, Unrelated Case Subjects, and Unaffected Control Subjects
The three PG1 SNPs were genotyped in all 159 HPC pedigrees and in 249 unrelated prostate cancer case subjects and 211 unaffected control subjects. All SNPs were in HWE in each subset. Allele frequencies of the three SNPs were compared between case and control subjects. To decrease the confounding factor of racial differences, the comparison was limited to whites only. For SNP 477, the allele frequencies of G were 0.33, 0.33, and 0.31, in the 123 HPC probands, 216 unrelated case subjects, and 178 unaffected control subjects, respectively. For SNP 99217, the allele frequencies of T were 0.32, 0.31, and 0.30, in the 131 HPC probands, 222 unrelated case subjects, and 177 unaffected control subjects, respectively. For SNP 467, the allele frequencies of A were 0.24, 0.25, and 0.24, in the 120 HPC probands, 210 unrelated case subjects, and 177 unaffected control subjects, respectively. No significant difference was observed in the allele frequencies between the probands and control subjects, between the unrelated case subjects and control subjects, or between all case subjects and control subjects in any of the three SNPs.
Genotype frequencies of the three SNPs were also compared in the white subjects only (table 3). No statistical differences in genotype frequencies were observed between case and control subjects for any of the three SNPs. There was a trend toward higher homozygous rates of the less-frequent alleles of each SNP in the case subjects with HPC and in the unrelated case subjects, compared with those in the control subjects; however, the differences were not statistically significant. For example, the odds ratio (OR) was 1.39 (95% confidence interval [CI] 0.73–2.63) when the homozygous frequencies for T/T of SNP 477 in all case and control subjects were compared.
Table 3.
Genotypes of Three SNPs in PG1 in Probands, Unrelated Case Subjects, and Unaffected Control Subjects (White Subjects Only)
|
Case Subjects (%) |
Odds Ratioa (95% CI) |
|||||
| SNP andGenotype | Control Subjects (%) | Sporadic | HPC | Sporadic Case Subjects vs. Control Subjects | HPC Case Subjects vs. Control Subjects | All Case Subjects vs. Control Subjects |
| SNP 477: | n=178 | n=222 | n=123 | |||
| C/C | .47 | .46 | .46 | 1 | 1 | 1 |
| C/G | .44 | .41 | .42 | 1.06 (.70–1.60) | .96 (.59–1.58) | 1.03 (.70–1.51) |
| G/G | .09 | .13 | .12 | 1.38 (.69–2.74) | 1.36 (.61–3.04) | 1.39 (.73–2.63) |
| Any G | 1.11 (.75–1.65) | 1.03 (.64–1.64) | 1.09 (.76–1.57) | |||
| SNP 99217: | n=177 | n=217 | n=131 | |||
| C/C | .49 | .51 | .47 | 1 | 1 | 1 |
| C/T | .42 | .36 | .41 | .97 (.64–1.48) | .99 (.61–1.61) | .99 (.67–1.44) |
| T/T | .08 | .13 | .11 | 1.20 (.59–2.45) | 1.33 (.60–2.97) | 1.25 (.65–2.41) |
| Any T | 1.01 (.68–1.51) | 1.05 (.66–1.67) | 1.03 (.72–1.48) | |||
| SNP 467: | n=177 | n=212 | n=120 | |||
| G/G | .59 | .59 | .60 | 1 | 1 | 1 |
| G/A | .34 | .32 | .32 | 1.10 (.72–1.69) | .92 (.55–1.54) | 1.03 (.69–1.53) |
| A/A | .07 | .10 | .08 | 1.30 (.60–2.84) | 1.20 (.49–2.93) | 1.28 (.63–2.63) |
| Any A | 1.14 (.76–1.71) | .97 (.60–1.57) | 1.08 (.74–1.56) | |||
All odds ratios were adjusted for age.
Haplotype frequencies of the three SNPs were also compared between case and control subjects. The estimated haplotype frequencies of G-T-A for the three SNPs (SNP 477, SNP 99217, and SNP 467) were 0.21, 0.25, and 0.22, in HPC probands, unrelated case subjects, and unaffected control subjects, respectively. No significant statistical differences in the haplotype frequencies were found between all possible pair comparisons.
Mutation Screening of PG1
SSCP mutation-screening analysis of probands from 92 families with HPC produced band patterns indicative of two different sequence variants in exon 1 and three different variants in exon 4. For exon 1, sequence analysis identified one variant as a silent polymorphism (C→G at codon 43, position 2159 in the genomic sequence reported by Cohen et al. [1999]), which was present in 14.1% of probands and in 7.9% of unaffected control subjects. The other variant was a nonsynonymous change at codon 22 (G→C at position 2095, resulting in substituting Ala for Gly), present in 4.2% of probands and 2.2% of control subjects.
Sequence analysis of the variants in exon 4 demonstrated two silent polymorphisms (T→C in codon 145 at position 25631 in the genomic sequence reported by Cohen et al. [1999], and A→G in codon 139 at position 25615) and a nonsynonymous change at position 25649 (G→A resulting in a substitution of Thr for Ala at codon 151). These variants were present at low frequencies (0.5%–3%) with no differences between case and control subjects (e.g., the Ala→Thr change was observed in one proband, one sporadic case subject, and one control individual).
Discussion
By testing for linkage and association between prostate cancer susceptibility and markers on 8p22-23 in 159 HPC pedigrees, 249 unrelated case subjects, and 211 unaffected control subjects, we obtained the following three findings. (1) There was evidence for linkage between a prostate cancer–susceptibility locus and markers on 8p22-23, with a highest HLOD of 1.84 (P=.004) at D8S1130. The region providing evidence for linkage spanned ∼22 cM at 8p22-23. The evidence for linkage was observed in the first 66 HPC pedigrees and in the 93 new HPC pedigrees. The pedigrees with late age at onset, a large number of affected family members, and male-to-male disease transmission provided stronger evidence for linkage at the region. (2) One intronic sequence variant (allele T of SNP 99217) in the putative prostate cancer–susceptibility gene (PG1) was overtransmitted from parents to affected offspring, with Z=2.19 (P=.03) and Z=2.70 (P=.007) in all 159 HPC pedigrees and in 133 non-Jewish white pedigrees, respectively. The overtransmission of allele T likely reflected evidence for both linkage and association in the data, since (a) a family-based association test that accounted for the presence of linkage provided weaker but still marginally significant test statistics, with Z=1.66 (P=.10) and 2.07 (P=.04) in all HPC pedigrees and in non-Jewish white pedigrees, and (b) families whose probands carry T are more likely to be linked to the PG1 gene region. (3) No statistical differences were found in the allele, genotype, and haplotype frequencies for the three SNPs or other sequence variants in the PG1 gene between HPC probands, unrelated prostate cancer case subjects, and unaffected control subjects. However, a trend (but not a statistically significant one) was observed toward higher homozygous rates of the less-frequent allele of each SNP in the HPC case subjects and in the unrelated case subjects, compared with those among the control subjects.
Evidence for linkage at 8p22-23 in our study did not reach the genomewide screen criteria for significant or suggestive linkage as proposed by Lander and Kruglyak (1995). However, we think our results provide a basis for further study in this region for a number of reasons. First, the prior probability that a prostate cancer–susceptibility gene lies near 8p22-23 is high as extensive evidence from LOH studies in prostate and other cancers indicates the existence of tumor-suppressor genes in the region (for review, see work by Bookstein [2001]). Therefore, the stringent criterion for significant linkage, which is used to account for the low prior probability of any pair of genes being located within a recombination fraction of <.5 in the human genome, is not appropriate in this situation (Ott 1998). Secondly, although the HLOD of 1.84 (P=.004) could represent false-positive evidence for linkage, our simulation results suggested that it is unlikely. On the basis of the same structure of 159 pedigrees with HPC (affection status and availability of genotyping) and the genetic model used in the analyses, we simulated 10,000 replicates with a six-allele marker (equally frequent) not linked to the disease gene using FASTSLINK (see D. Weeks's FTP page). We then analyzed each replicate and only observed 10 of the 10,000 replicates with a HLOD >1.84, yielding an empirical P value of .001. Thirdly, and perhaps most importantly, the same region was reported to be linked to a prostate cancer–susceptibility gene in an independent genomewide-screen linkage study. Gibbs et al. (2000) reported a maximum multipoint nonparametric linkage score of 2.02 at D8S1106 in 44 pedigrees with late age at onset (⩾66 years), using genomewide screen markers. This marker was in our linkage region, ∼5 cM from the peak marker, D8S1130. Interestingly, we observed the same trend that pedigrees with late age at onset tend to be linked to this region, with a peak allele sharing LOD of 2.64 (P=.0005) in our 80 pedigrees with age at onset ⩾65 years. Lastly, both series of our HPC pedigrees (the first 66 HPC pedigrees included in the initial genomewide screen and the 93 pedigrees ascertained later) provided evidence for linkage. The trend for this linkage to be more prominent in families with older age at diagnosis was observed in both the first and the second groups of families (allele-sharing LOD scores of 1.46, P=.009 and 1.32, P=.01 respectively).
Even though some evidence for linkage at the PG1 gene was observed in parametric and nonparametric linkage analyses and family-based linkage and association test, the rather weak linkage at PG1 gene and the distance (10–15 cM) between the PG1 gene and the highest linkage region indicated that the PG1 plays a minor role, if any, in accounting for the linkage signal at 8p22-23. One or several other genes in the region may contribute to the observed linkage. Several important candidate tumor-suppressor genes reside in the 8p22-23 region, including the N33 (Bova et al. 1996), macrophage-scavenger-receptor (MSR) (Kagan et al. 1995; Bova et al. 1996), the N-acetyltransferase genes NAT1 and NAT2 (Wang et al. 1999), LZTS1 (Ishii et al. 1999), and DLC1 (deleted in liver cancer; see Yuan et al. 1998; Wilson et al. 2000). Several mutations in LZTS1 were found in prostate cancer cell lines. Transcript analysis from several LZTS1-expressing tumors revealed truncated mRNAs, including a frameshift (Ishii et al. 1999). Mutations in DLC1 were found in colorectal and ovarian tumors (Wilson et al. 2000). Unfortunately, studies investigating possible associations between the genomic sequence variants and prostate cancer have not been published.
The interpretation of the results from our PG1 gene–association study is difficult. Although overtransmission of allele T of SNP 99217 from parents to affected offspring provides evidence that PG1 might influence prostate cancer susceptibility, the lack of statistically significant differences in the allele, genotype, and haplotype frequencies between case and control subjects is not consistent with this notion. Our results contrast with the results from the case-control study reported by Cohen et al. (1999). Although the exact reason for the difference is unknown, several of the following factors may contribute to the difference. First, there may be allele-frequency differences between the French and U.S. populations, and the former may be a more homogeneous population. This is, however, unlikely to be the major reason in this case, because the allele frequencies in the case subjects are similar in the two populations. Second, the power to detect the association in our study sample is limited. Using the point estimates of ORs and frequencies from Cohen et al. (1999), the power to detect an OR of 2.2, at the significance level of .05, with a genotype frequency of 14% in control subjects, is 72% in our combined 345 case and 177 control samples (white subjects only). Third, potential misclassification may be present in our control group. Although the unaffected control subjects in our study had normal results on digital rectal examination and normal PSA levels (i.e., <4 ng/ml), some of our control subjects are young, and they could be disease-gene carriers who will develop prostate cancer later. The ORs adjusted for age in our study may alleviate the problem but cannot remove the confounder. Last, random sampling error in control subjects in both studies could lead to the difference. In consideration of the limited power to detect a weak association and potential bias in the study, further studies utilizing larger number of control subjects may help to answer the question.
In summary, our study provides evidence for prostate cancer linkage at 8p22-23. The linkage results, along with the consistent evidence that 8p22 is the most commonly deleted region in prostate cancer cells and the discovery of mutations in some tumor-suppressor genes in the region warrant further studies. The results of the evaluation of the PG1 gene are inconclusive but interesting enough to suggest further studies of this gene as well. With the availability of more-complete sequence data for the human genome, studies to systematically evaluate all the genes in the region using an association study design (either case-control or family-based) are justified and likely to succeed.
Acknowledgments
The authors thank all the study subjects who participated in this study. This work was partially supported by National Cancer Institute SPORE grant CA58236 and by two grants from the Department of Defense (to W.B.I. and J.X.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GDA: Software for the Analysis of Discrete Genetic Data, http://lewis.eeb.uconn.edu/lewishome/gda.html
- Haplotype Information Help Page, http://www.bioinf.mdc-berlin.de/hap/ithap-help.html
- Linkage Designer, http://dnalab-www.uia.ac.be/dnalab/ld.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for prostate cancer [MIM 176807], HPC2 [MIM 605367], HPC1 [MIM 601518], PCaP [MIM 602759], PCBP/CAPB [MIM 603688], and HPCX [MIM 300147])
- Weeks FTP page, ftp://watson.hgen.pitt.edu (for FASTSLINK)
References
- Bova GS, MacGrogan D, Levy A, Pin SS, Bookstein R, Isaacs WB (1996) Physical mapping of chromosome 8p22 markers and their homozygous deletion in a metastatic prostate cancer. Genomics 35:46–54 [DOI] [PubMed] [Google Scholar]
- Bookstein R (2001) Tumor suppressor genes in prostate cancer. In: Chung LWK, Isaacs WB, Simons JW (eds) Prostate cancer, biology, genetics, and the new therapeutics. Humana Press, Totowa, NJ [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuaqui RF, Sanz-Ortega J, Vocke C, Linehan WM, Sanz-Esponera J, Zhuang Z, Emmert-Buck MR, Merino MJ (1995) Loss of heterozygosity on the short arm of chromosome 8 in male breast carcinomas. Cancer Res 55:4995–4998 [PubMed] [Google Scholar]
- Cliby W, Ritland S, Hartmann L, Dodson M, Halling KC, Keeney G, Podratz KC, Jenkins RB (1993) Human epithelial ovarian cancer allelotype. Cancer Res 53:2393–2398 [PubMed] [Google Scholar]
- Cohen D, Chumakov I, Blumenfeld M, Bougueleret L (1999) GENSET, assignee. Prostate cancer gene. US patent US005945522A. August 31, 1999 [Google Scholar]
- Cottingham RW Jr, Idury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Cunningham C, Dunlop MG, Wyllie AH, Bird CC (1993) Deletion mapping in colorectal cancer of a putative tumour suppressor gene in 8p22-p21.3. Oncogene 8:1391–1396 [PubMed] [Google Scholar]
- Cunningham JM, Shan A, Wick MJ, McDonnell SK, Schaid DJ, Tester DJ, Qian J, Takahashi S, Jenkins RB, Bostwick DG, Thibodeau SN (1996) Allelic imbalance and microsatellite instability in prostatic adenocarcinoma. Cancer Res 1996 56:4475–4482 [PubMed] [Google Scholar]
- Deubler DA, Williams BJ, Zhu XL, Steele MR, Rohr LR, Jensen JC, Stephenson RA, Changus JE, Miller GJ, Becich MJ, Brothman AR (1997) Allelic loss detected on chromosomes 8, 10, and 17 by fluorescence in situ hybridization using single-copy P1 probes on isolated nuclei from paraffin-embedded prostate tumors. Am J Pathol 150:841–850 [PMC free article] [PubMed] [Google Scholar]
- Emi M, Fujiwara Y, Nakajima T, Tsuchiya E, Tsuda H, Hirohashi S, Maeda Y, Tsuruta K, Miyaki M, Nakamura Y (1992) Frequent loss of heterozygosity for loci on chromosome 8p in hepatocellular carcinoma, colorectal cancer, and lung cancer. Cancer Res 52:5368–5372 [PubMed] [Google Scholar]
- Faraway JJ (1993) Distribution of the admixture test for the detection of linkage under heterogeneity. Genet Epidemiol 10:75–83 [DOI] [PubMed] [Google Scholar]
- Gibbs M, Stanford JL, Jarvik GP, Janer M, Badzioch M, Peters MA, Goode EL, Kolb S, Chakrabarti L, Shook M, Basom R, Ostrander EA, Hood L (2000) A genomic scan of families with prostate cancer identifies multiple regions of interest. Am J Hum Genet 67:100–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Baffa R, Numata SI, Murakumo Y, Rattan S, Inoue H, Mori M, Fidanza V, Alder H, Croce CM (1999) The FEZ1 gene at chromosome 8p22 encodes a leucine-zipper protein, and its expression is altered in multiple human tumors. Proc Natl Acad Sci USA 96:3928–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan J, Stein J, Babaian RJ, Joe YS, Pisters LL, Glassman AB, von Eschenbach AC, Troncoso P (1995) Homozygous deletions at 8p22 and 8p21 in prostate cancer implicate these regions as the sites for candidate tumor suppressor genes. Oncogene 11:2121–2126 [PubMed] [Google Scholar]
- Knowles MA, Shaw ME, Proctor AJ (1993) Deletion mapping of chromosome 8 in cancers of the urinary bladder using restriction fragment length polymorphisms and microsatellite polymorphisms. Oncogene 8:1357–1364 [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X (2000) Implementing a unified approach to family-based test of association. Genet Epidemiol Suppl 19:36–42 [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA 84:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Kruglyak L (1995) Genetic dissection of complex traits: guideline for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGrogan D, Levy A, Bova GS, Isaacs WB, Bookstein R (1996) Structure and methylation-associated silencing of a gene within a homozygously deleted region of human chromosome band 8p22. Genomics 35:55–65 [DOI] [PubMed] [Google Scholar]
- Macoska JA, Trybus TM, Benson PD, Sakr WA, Grignon DJ, Wojno KD, Pietruk T, Powell IJ (1995) Evidence for three tumor suppressor gene loci on chromosome 8p in human prostate cancer. Cancer Res 15:5390–5395 [PubMed] [Google Scholar]
- Ott J (1998) Analysis of human genetic linkage. 3d ed. Johns Hopkins Press, Baltimore [Google Scholar]
- Prasad MA, Trybus TM, Wojno KJ, Macoska JA (1998) Homozygous and frequent deletion of proximal 8p sequences in human prostate cancers: identification of a potential tumor suppressor gene site. Genes Chromosomes Cancer 23:255–262 [PubMed] [Google Scholar]
- Ransom DT, Leonard JH, Kearsley JH, Turbett GR, Heel K, Sosars V, Hayward NK, Bishop JF (1996) Loss of heterozygosity studies in squamous cell carcinomas of the head and neck. Head Neck 18:248–253 [DOI] [PubMed] [Google Scholar]
- Smith JR, Freiji D, Carpten JD, Gronberg H, Xu J, Isaacs SD, Brownstei MJ, Bova GS, Guo H, Bujnovsky P, Nusskern DR, Damber JE, Bergth A, Emanuelsson M, Kallioniemi OP, Walker-Daniels J, Baily-Wilson JE, Beaty TH, Meyers DA, Walsh PC, Collins FS, Trent JM, Isaacs WB (1996) Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science 274:1371–1374 [DOI] [PubMed] [Google Scholar]
- Vocke CD, Pozzatti RO, Bostwick DG, Florence CD, Jennings SB, Strup SE, Duray PH, Liotta LA, Emmert-Buck MR, Linehan WM (1996) Analysis of 99 microdissected prostate carcinomas reveals a high frequency of allelic loss on chromosome 8p12-21. Cancer Res 56:2411–2416 [PubMed] [Google Scholar]
- Wang CY, Debiec-Rychter M, Schut HA, Morse P, Jones RF, Archer C, King CM, Haas GP (1999) N-Acetyltransferase expression and DNA binding of N-hydroxyheterocyclic amines in human prostate epithelium. Carcinogenesis 20:1591–1595 [DOI] [PubMed] [Google Scholar]
- Weir BS (1996) Genetic data analysis II: methods for discrete population genetic data. Sinauer, Sunderland, MA [DOI] [PubMed] [Google Scholar]
- Whittemore A, Halpern J (1994) A class of tests for linkage using affected pedigree members. Biometrics 50:118–127 [PubMed] [Google Scholar]
- Wilson PJ, McGlinn E, Marsh A, Evans T, Arnold J, Wright K, Biden K, Young J, Wainwright B, Wicking C, Chenevix-Trench G (2000) Sequence variants of DLC1 in colorectal and ovarian tumours. Hum Mutat 15:156–165 [DOI] [PubMed] [Google Scholar]
- Wistuba II, Behrens C, Virmani AK, Milchgrub S, Syed S, Lam S, Mackay B, Minna JD, Gazdar AF (1999) Allelic losses at chromosome 8p21-23 are early and frequent events in the pathogenesis of lung cancer. Cancer Res 59:1973–1979 [PubMed] [Google Scholar]
- Yuan BZ, Yang Y, Keck-Waggoner CL, Zimonjic DB, Thorgeirsson SS, Popescu NC (1999) Assignment and cloning of mouse Arhgap7 to chromosome 8A4-B2, a conserved syntenic region of human chromosome 8p22→p21. Cytogenet Cell Genet 87:189–190 [DOI] [PubMed] [Google Scholar]
- Zaykin D, Zhivotovsky L, Weir BS (1995) Exact tests for association between alleles at arbitrary numbers of loci. Genetica 96:169–178 [DOI] [PubMed] [Google Scholar]