Abstract
Chordoma is a rare tumor originating from notochordal remnants that is usually diagnosed during midlife. We performed a genomewide analysis for linkage in a family with 10 individuals affected by chordoma. The maximum two-point LOD score based on only the affected individuals was 2.21, at recombination fraction 0, at marker D7S2195 on chromosome 7q. Combined analysis of additional members of this family (11 affected individuals) and of two unrelated families (one with 2 affected individuals and the other with 3 affected individuals), with 20 markers on 7q, showed a maximum two-point LOD score of 4.05 at marker D7S500. Multipoint analysis based on only the affected individuals gave a maximum LOD score of 4.78, with an approximate 2-LOD support interval from marker D7S512 to marker D7S684. Haplotype analysis of the three families showed a minimal disease-gene region from D7S512 to D7S684, a distance of 11.1 cM and ∼7.1 Mb. No loss of heterozygosity was found at markers D7S1804, D7S1824, and D7S2195 in four tumor samples from affected family members. These results map a locus for familial chordoma to 7q33. Further analysis of this region, to identify this gene, is ongoing.
Chordomas (MIM 215400) are rare, slow growing, and locally invasive bone tumors thought to be derived from notochordal remnants (Malawer et al. 1997). There is a modest overall male predominance (1.7:1), which is more striking (∼3:1) among patients with sacral chordomas. The age distribution at diagnosis is unimodal, with a median of 59 years. Treatment of chordomas is predominantly surgical, followed by radiotherapy (Rich et al. 1985; Healey and Lane 1989; Tai et al. 1995). Survival rates to 5 and 10 years are 68% and 40%, respectively (McMaster et al. 2001).
Little is known about either the molecular biology of chordoma or the etiologic factors that predispose to it. No characteristic karyotypic abnormality has been described in primary chordomas (Sawyer et al. 2001). That an alteration in one or more genes may play a role is suggested by reports of five families with chordoma in two or more close relatives. The affected relatives included a middle-aged brother and sister with sacral chordoma (Foote et al. 1958), two young brothers with nasopharyngeal chordoma (Enin 1963), a man with nasopharyngeal chordoma whose mother and daughter developed the same tumor (Kerr et al. 1975), a man with sacral chordoma whose sister and niece developed clival chordomas and whose first cousin had a nasopharyngeal chordoma (Stepanek et al. 1998), and a father and daughter with clival chordoma (Dalpra et al. 1999; Miozzo et al. 2000). We have performed genomewide linkage analysis using the extended pedigree of the family reported by Stepanek et al. (1998) and have identified, on chromosome 7q33, a putative locus for familial chordoma.
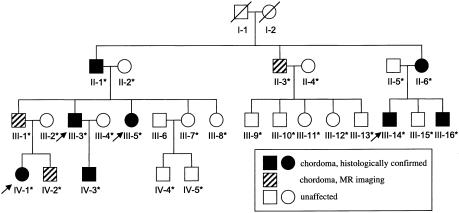
We clinically evaluated the 4 living index cases with histopathologically confirmed chordoma (individuals III-3, III-5, III-14, and IV-1 in family 1; see fig. 1) and 14 blood relatives from the family reported by Stepanek et. al. (1998), under an institutional review board–approved protocol. Magnetic resonance (MR) images of the pituitary/sella tursica, spine, sacrum, and coccyx demonstrated a clival or nasopharyngeal mass, suggestive of chordoma, in 6 of the 14 screened relatives. This diagnosis was confirmed histopathologically in three relatives who underwent tumor excision (D. M. Parry, unpublished data). Of the 10 confirmed or suspected cases of chordoma in this family, 9 involve the clivus or nasopharynx. The pattern of inheritance, including father-to-son transmission, is compatible with an autosomal dominant trait.
Figure 1.
Pedigree of family 1. The four index cases are indicated by arrows. All affected individuals had clival chordoma, except for individual III-3, who had a sacral chordoma. Deceased individuals (indicated by a diagonal line through the symbol) and individual III-6 were not genotyped. An asterisk (*) denotes that a DNA sample was obtained.
We isolated genomic DNA from peripheral lymphocytes and performed genomewide genotyping in 22 individuals (II-1–II-6, III-1–III-3, III-5, III-7–III-16, IV-1, and IV-2) of family 1, using the 365 autosomal- and 2 sex-chromosome simple tandem repeat (STR) markers of The Cooperative Human Linkage Center's version 8 marker set (average spacing 10 cM; average heterozygosity .76), according to a standard PCR protocol (Research Genetics), and a Perkin-Elmer ABI 310 genetic analyzer and GENESCAN and GENOTYPER software. We performed two-point LOD-score linkage analysis, using the MLINK and ILINK programs from the LINKAGE package, version 5.10 (Lathrop et al. 1984), under the assumption of autosomal dominant inheritance of a rare allele with population frequency .0001. Because estimates of penetrance for familial chordoma are unknown, we performed the analysis under four different scenarios. In the first analysis, we used phenotype data on only the 10 affected individuals (7 with biopsy–proved chordoma and 3 with MR-imaging findings suspicious for chordoma), in an affecteds-only analysis in which we coded all clinically unaffected individuals as “unknown” with respect to disease status. The remaining three analyses included all 22 family members (affected, unaffected, and spouses), and we assumed non–age-dependent penetrances of 100%, 75%, and 50%. In these preliminary analyses, marker loci were assumed to be codominant, with n equally frequent alleles, where n = (no. of observed alleles) + 1. The additional allele at the marker locus was included to account for the fact that individuals I-1 and I-2 were not available to be genotyped. We also performed the nonparametric affected-sib-pair (ASP) test and the Haseman-Elston (HE) sib-pair test (Haseman and Elston 1972), using the S.A.G.E program SIBPAL (SAGE 1994).
Results of the genomewide screen in 22 individuals from family 1 suggested possible linkage of the disease gene to chromosomes 4, 7, 17, or 19, on the basis of either two-point LOD scores ⩾1.0 or P⩽.01, for sib-pair analyses at each of two adjacent marker loci. The greatest two-point LOD scores for each of the four chromosomes were 1.85 at marker D4S1627 (recombination fraction [θ] 0 under the assumption of 50% penetrance), 2.21 at marker D7S2195 (θ=0 under affecteds-only analysis), 1.51 at marker ATC6A06 (θ=.09 under the assumption of 100% penetrance), and 1.03 at marker D19S714 (θ=0 when all affection-status scenarios considered), respectively. Additional STR markers at ∼2-cM intervals were genotyped in each of these four regions. The greatest evidence for linkage was found on chromosome 7 in the affecteds-only analysis, where the maximum two-point LOD score was 2.26 at θ=0 at marker D7S500. The nonparametric P values were ∼.01 for the ASP test and were not significant for the HE test. Multipoint analysis using LINKMAP (Lathrop et al. 1984) gave the greatest evidence for linkage in the affecteds-only analysis, a maximum LOD score of 2.34 at marker D7S2195. All affected family members shared a common haplotype at 12 markers in a 23.6-cM region on chromosome 7q32-34. In contrast, linkage analysis of additional markers in the studied regions on chromosomes 4, 17, and 19 did not result in higher two-point LOD scores and did not reveal a common haplotype shared by all 10 affected individuals. Specifically, no additional markers on chromosomes 17 or 19 and only one new marker on chromosome 4 (marker D4S1553; LOD score 1.49 at θ=0 in affecteds-only analysis) yielded a LOD score >1 in the 2-cM screen. These three regions no longer showed consistent evidence for linkage.
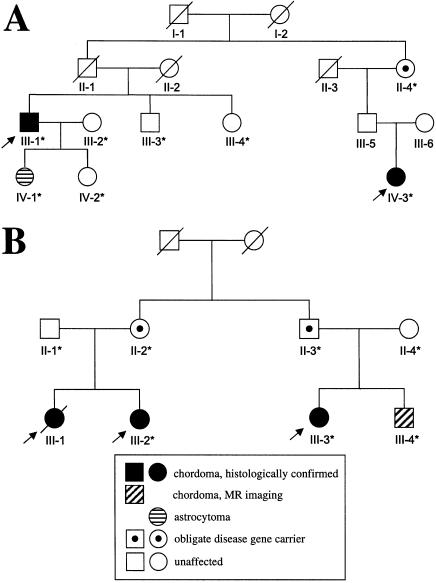
To increase the power of the linkage analysis and to narrow the suspected disease-gene region, we examined additional members of family 1 (individuals IV-3–IV-5; see fig. 1) and ascertained two smaller, independent families with chordoma, families 2 and 3 (fig. 2). Individual IV-3 in family 1 had a clival mass that was excised and confirmed as chordoma. In family 2, the two index cases (individual III-1 and his paternal first cousin once removed, individual IV-3) had histologically confirmed clival chordoma. In addition, individual IV-1, the daughter of individual III-1, had a pilocytic astrocytoma. Of the living relatives in family 3, the index cases (individual III-3 and her first cousin, individual III-2) had histopathologically confirmed clival chordoma, and, on MR imaging, individual III-4 (the brother of individual III-3) had a clival mass consistent with chordoma. Family 3 is a branch of a three-generation family with chordoma that has been reported elsewhere (Kerr et al. 1975; E.S., unpublished data). In subsequent linkage analyses, we extended the definition of “affected” to include the patient in family 2 who had astrocytoma. This decision was based on the similar occurrence of juvenile astrocytoma in a child whose sister and father had clival chordoma (Dalpra et al. 1999; Miozzo et al. 2000).
Figure 2.
Pedigrees of families 2 (A) and 3 (B). Symbols are as described in the legend to table 1.
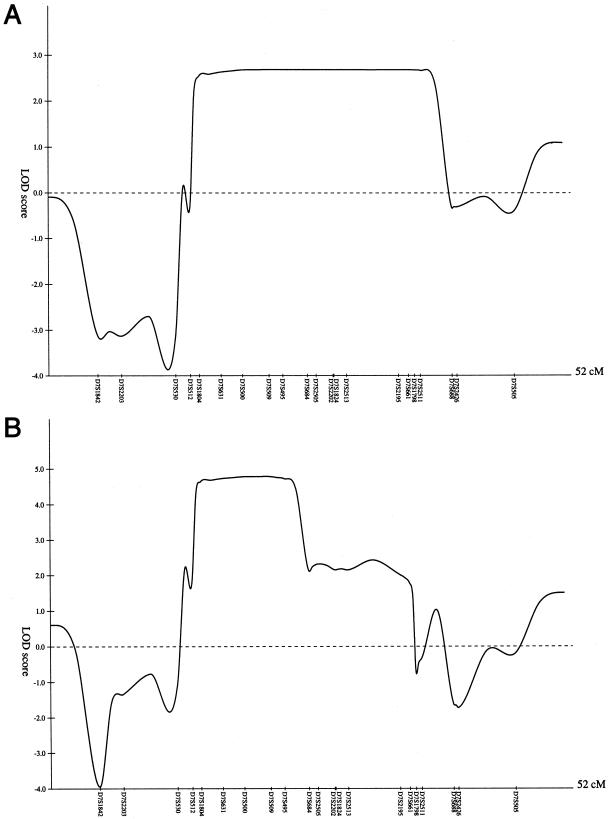
We genotyped a total of 41 individuals from the three families, for 21 chromosome 7 markers over a span of ∼47 cM; for 19 of these markers, we used available allele-frequency estimates from CEPH (Fondation Jean Dausset) and the Center for Human Genetics (Duke University Medical Center), and for the remaining 2 markers we used alleles that we assumed to be equifrequent. For family 1 alone, the highest two-point LOD score was 2.58 at θ=0 at marker D7S2195, in the affecteds-only analysis. Multipoint linkage analysis using GENEHUNTER (Kruglyak et al. 1996) and considering only the affected individuals in family 1 revealed an approximate 2-LOD support interval from D7S512 to D7S688 (fig. 3A). When the three families were analyzed together, under the assumption of locus homogeneity, two-point LOD scores >3 were found at markers D7S1804, D7S500, D7S509, D7S2505, D7S2202, and D7S2513, in the affecteds-only analysis (table 1). Multipoint linkage analyses of markers D7S1842 to D7S505 revealed a maximum LOD score of 4.78 when GENEHUNTER was used. An ∼2-LOD support interval extended from D7S512 to D7S684, with the peak being obtained when the disease gene was coincident with D7S509 (fig. 3B). Similar results were obtained when the programs LINKMAP and VITESSE (O'Connell and Weeks 1995) were used (data not shown).
Figure 3.
Multipoint LOD-score plot of the region, in chromosome 7, of the gene for familial chordoma, in family 1 alone (A) and in the three families combined (B), under an affecteds-only analysis using GENEHUNTER (Kruglyak et al. 1996).
Table 1.
Chromosome 7 Linkage Analysis: Two-Point Parametric LOD Scores and Corresponding θ's for Three Families with Chordoma
|
LOD Score (θ) for |
|||||
| Penetrance |
|||||
| Marker | Genetic-MapPositiona(cM) | Affecteds Only | 100% | 75% | 50% |
| D7S1842 | 170.3 | .59 (.2) | .20 (.4) | .29 (.3) | .41 (.3) |
| D7S2203 | 172.7 | .38 (.2) | .23 (.3) | .39 (.2) | .42 (.2) |
| D7S530 | 178.2 | 1.29 (.1) | .89 (.2) | 1.03 (.2) | 1.17 (.1) |
| D7S512 | 179.7 | .92 (.1) | .29 (.2) | .68 (.1) | .83 (.1) |
| D7S1804b | 180.6 | 3.62 (0) | .98 (.2) | 2.02 (.01) | 3.08 (0) |
| D7S631 | 182 | 1.47 (0) | 1.24 (.1) | 1.13 (.05) | 1.19 (.05) |
| D7S500 | 184.2 | 4.05 (0) | 1.29 (.2) | 2.19 (.05) | 3.17 (0) |
| D7S509 | 186.9 | 4.01 (0) | 2.73 (.1) | 3.11 (.05) | 3.67 (0) |
| D7S495 | 188.3 | 1.54 (0) | .57 (.2) | .80 (.1) | 1.05 (.05) |
| D7S684 | 190.8 | 1.69 (.05) | .27 (.3) | .67 (.2) | 1.20 (.1) |
| D7S2505 | 191.7 | 3.53 (0) | 1.18 (.2) | 1.70 (.1) | 2.57 (0) |
| D7S2202 | 193.5 | 3.20 (0) | .77 (.2) | 1.48 (.1) | 2.31 (0) |
| D7S1824 | 193.5 | 2.20 (.05) | .49 (.3) | 1.16 (.1) | 1.75 (.05) |
| D7S2513 | 194.8 | 3.14 (.05) | 1.03 (.2) | 1.84 (.1) | 2.45 (.05) |
| D7S2195 | 200.1 | 2.85 (.05) | 1.39 (.2) | 2.04 (.1) | 2.46 (.05) |
| D7S661 | 202.1 | 1.62 (.05) | .15 (.3) | .54 (.2) | 1.05 (.1) |
| D7S1798 | 202.7 | .47 (.2) | .05 (.3) | .21 (.2) | .35 (.2) |
| D7S2511 | 203.3 | 2.51 (.05) | 1.59 (.2) | 1.94 (.1) | 2.16 (.1) |
| D7S688b,c | 206.5 | .54 (.2) | .35 (.2) | .43 (.2) | .49 (.2) |
| D7S2426 | 207 | 1.28 (.1) | .27 (.3) | .36 (.3) | .60 (.2) |
| D7S505 | 217.5 | .38 (.2) | .09 (.4) | .16 (.3) | .24 (.3) |
Based on the Center for Medical Genetics (Marshfield Medical Research Foundation) gender-averaged genetic map. Marker order was confirmed by reference to a physical map (Bouffard et al. 1997).
Allele frequencies were not available from either CEPH (Fondation Jean Dausset) or the Center for Human Genetics (Duke University Medical Center) and, therefore, are assumed to be equal.
Family 1 only.
In the additional members of family 1, there were no recombination events in the disease-gene region. Thus, the minimal disease-gene region for this family remained the interval from D7S512 to D7S688 (table 2). Haplotype analysis of family 2 showed that the three affected individuals and the obligate carrier (individual II-4) share a common haplotype from marker D7S2203 to D7S661 (table 2), so that the minimal disease-gene region in this family extends from marker D7S1842 to marker D7S1798, a genetic distance of 32.4 cM on the gender-averaged Center for Medical Genetics (Marshfield Medical Research Foundation) genetic map. In family 3, the three affected individuals and two obligate carriers (individuals II-2 and II-3) share a common haplotype from marker D7S1842 to marker D7S495 (table 2). Thus, the corresponding minimal disease-gene region extends from centromeric of marker D7S1842 to marker D7S684. Under the assumption of locus homogeneity in the three families, haplotype analysis identified the minimal disease-gene region as extending from marker D7S512 to D7S684, a genetic distance of ∼11 cM. We confirmed the marker order by reference to a physical map of the region, consisting of YAC clones (Bouffard et al. 1997). The physical size of the minimal disease-gene region is ∼7.1 Mb. There are 14 known or predicted genes in this region (not including potential genes with a single expressed-sequence tag).
Table 2.
Disease Haplotypes of Affected Individuals and Obligate Carriers, by Nuclear Family
|
Allele inb |
||||||||||||||||||||
| Family 1 |
Family 2 |
Family 3 |
||||||||||||||||||
| Markera | II-1 | III-1 | III-3 | III-5 | IV-1 | IV-2 | IV-3 | II-3 | II-6 | III-14 | III-16 | III-1 | IV-1 | II-4 | IV-3 | II-2 | III-2 | II-3 | III-3 | III-4 |
| D7S1842 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 2 | 2 | 6 | 2 | 6 |
6 |
6 |
3 |
3 | 3 | 3 | 3 | 3 |
| D7S2203 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| D7S530 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 2 | 2 | 3 | 2 | 3 | 3 | 3 | 3 | 6 | 6 | 6 | 6 | 6 |
| D7S512 | 2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
6 |
7 |
6 |
2 | 2 | 2 | 2 | 8 | 8 | 8 | 8 | 8 |
| D7S1804 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| D7S631 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 |
| D7S500 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 1 | 1 | 1 | 1 | 9 | 9 | 9 | 9 | 9 |
| D7S509 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 5 | 5 | 5 | 5 | 4 | 4 | 4 | 4 | 4 |
| D7S495 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | 5 | 5 | 5 | 2 |
2 |
2 |
2 |
2 |
| D7S684 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 4 | 4 | 4 | 5 | 5 | 3 | 3 | 3 |
| D7S2505 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| D7S2202 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 6 | 6 | 6 | 6 | 6 | 6 | 5 | 5 | 5 |
| D7S1824 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 1 | 1 | 1 | 1 | 6 | 6 | 3 | 3 | 3 |
| D7S2513 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 9 | 9 | 9 | 9 | 1 | 1 | 2 | 2 | 2 |
| D7S2195 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 1 | 1 | 1 | 1 | 2 | 2 | 3 | 3 | 3 |
| D7S661 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
4 |
4 |
4 |
4 | 4 | 6 | 6 | 6 |
| D7S1798 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 2 | 2 |
| D7S2511 | 4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 | 4 | 6 | 6 | 3 | 3 | 3 | 3 | 3 |
| D7S688 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 4 | 4 | 4 | 4 | … | … | … | … | … | … | … | … | … |
| D7S2426 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 4 | 4 | 4 | 4 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| D7S505 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | … | 2 | 2 | 4 | 4 | 1 | 1 | 1 |
Marker order was confirmed by reference to a physical map of chromosome 7 (Bouffard et al. 1997).
The haplotype regions shared within each family are boxed. An ellipsis (…) indicates that the marker was not studied.
As an alternative way to confirm and potentially narrow the region containing the putative chordoma gene, we looked for allelic loss of the non–disease-associated haplotype within microdissected tumor cells from flash-frozen chordoma tissue from individuals II-1, II-6, III-3, and III-16 in family 1. Between three and five informative microsatellite markers on chromosomes 4, 7, 17, and 19, from each of the regions initially suggested to be linked to the disease gene were analyzed for loss of heterozygosity (LOH); under the conditions utilized, we could detect LOH if (a) blood and paired-tumor-cell DNA amplification was successful and produced consistent results in at least two attempts and (b) there was >40% loss of one allele versus the other allele. No LOH was found at any of three markers on chromosomes 7, 17, and 19; however, tumor cells from a skin metastasis in individual III-3 had LOH at markers on chromosome 4 (table 3). Without LOH for the markers on chromosome 7, we could not narrow the region suspected to carry the chordoma gene. However, this absence of LOH may indicate that the disease gene exerts its oncogenic effect in a dominant fashion. Alternatively, the markers may be too far from the disease gene, or LOH may occur over a very small region.
Table 3.
LOH Analysis in Four Samples of Chordoma Tumor from Family 1
|
LOH Status ina |
||||
| Marker | II-1b | II-6b | III-3c | III-16b |
| Chromosome 4: | ||||
| D4S2639 | + | |||
| GATA10G07 | − | NI | + | NI |
| D4S2361 | − | − | + | − |
| D4S1647 | NI | NI | + | − |
| D4S1625 | + | |||
| Chromosome 7: | ||||
| D7S1804 | − | − | − | − |
| D7S1824 | − | − | − | − |
| D7S2195 | − | − | − | − |
| Chromosome 17: | ||||
| D17S1851 | − | − | − | − |
| ATC6A06 | − | − | − | − |
| D17S1820 | − | − | − | − |
| Chromosome 19: | ||||
| D19S558 | − | − | − | − |
| D19S1165 | − | − | − | − |
| D19S930 | − | − | − | − |
+ = Presence of LOH; − = absence of LOH; NI = not informative.
Sample was from the primary tumor.
Sample was from a skin metastasis of the sacral primary tumor.
Because of its role as a secreted growth-and-differentiation factor, we selected the pleiotrophin gene, PTN, as the first gene to sequence. When we began, only partial genomic-structure information was available for this gene (GenBank accession numbers S50394, S50404, S50405, S50408, and S50409). We determined additional intronic sequence by amplifying across the introns, using oligonucleotides in the coding regions and performing direct sequencing on the PCR products. Oligonucleotide primers amplifying the four coding exons of the human pleiotrophin gene were designed by use of these intronic sequences. We used DNA-sequence analysis to examine the coding region (exons 2–5) and the adjacent splice sites of PTN, for sequence variants in genomic DNA from two affected members of family 1 and from one unrelated individual. No variants were found, suggesting that PTN is not a gene for familial chordoma.
We are continuing to sequence other candidate genes in the region, in DNA from affected members of all three families. In addition, we are seeking additional families with chordoma, for linkage studies to further fine-map this chordoma locus.
Acknowledgments
We thank Deborah Zametkin, for her outstanding skills as a research nurse; Kathrine A. Allikian and Sufeng Li, for excellent technical assistance; Marguerite Adkins, for secretarial assistance; John W. Gillespie, for assistance with microdissection; and, especially, the patients and their families, for their participation. Portions of this work were supported by National Cancer Institute contract 263-MQ-013900 (to M.J.K.) and by the Cancer Epidemiology and Biostatistics Fellowship Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute (support to J.F.K.). Some of the results reported were obtained by use of the program package S.A.G.E., which is supported by U.S. Public Health Service Resource grant RR03655 from the National Center for Research Resources.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- Center for Human Genetics, Duke University Medical Center, http://wwwchg.mc.duke.edu/index.html (for marker-allele frequencies)
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- CEPH–Fondation Jean Dausset, http://www.cephb.fr/ (for marker-allele frequencies)
- Cooperative Human Linkage Center, The, http://lpg.nci.nih.gov/CHLC (for autosomal- and sex-chromosome STR markers)
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for PTN [accession numbers S50394, S50404, S50405, S50408, and S50409])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for chordoma [MIM 215400])
References
- Bouffard GG, Idol JR, Braden VV, Iyer LM, Cunningham AF, Weintraub LA, Touchman JW, Mohr-Tidwell RM, Peluso DC, Fulton RS, Ueltzen MS, Weissenbach J, Magness CL, Green ED (1997) A physical map of human chromosome 7: an integrated YAC contig map with average STS spacing of 79 kb. Genome Res 7:673–692 [DOI] [PubMed] [Google Scholar]
- Dalpra L, Malgara R, Miozzo M, Riva P, Volonte M, Larizza L, Fuhrman Conti AM (1999) First cytogenetic study of a recurrent familial chordoma of the clivus. Int J Cancer 81:24–30 [DOI] [PubMed] [Google Scholar]
- Enin IP (1963) Khordoma nosoglotki u dvukh chlenov odnoi sem'i [Chordoma of the nasopharynx in two members of the same family]. Vestn Otorinolaringol 26:88–90 [PubMed] [Google Scholar]
- Foote RF, Ablin G, Hall WMW (1958) Chordoma in siblings. California Med 88:383–386 [PMC free article] [PubMed] [Google Scholar]
- Haseman JK, Elston RC (1972) The investigation of linkage between a quantitative trait and a marker locus. Behav Genet 2:3–19 [DOI] [PubMed] [Google Scholar]
- Healey JH, Lane JM (1989) Chordoma: a critical review of diagnosis and treatment. Orthop Clin North Am 20:417–426 [PubMed] [Google Scholar]
- Kerr WA, Allen KL, Haynes DR, Sellars SL (1975) Familial nasopharyngeal chordoma. S Afr Med J 49:1584 [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malawer MM, Link MP, Donaldson SS (1997) Sarcomas of bone. In: DeVita VT, Hellman S, Rosenberg SA (eds) Cancer: principles and practice of oncology. Lippincott-Raven, Philadelphia, pp 1844–1846 [Google Scholar]
- McMaster ML, Goldstein AM, Bromley CM, Ishibe N, Parry DM (2001) Chordoma: incidence and survival patterns in the United States, 1973–1995. Cancer Causes Control 12:1–11 [DOI] [PubMed] [Google Scholar]
- Miozzo M, Dalpra L, Riva P, Volonta M, Macciardi F, Pericotti S, Tibiletti MG, Cerati M, Rohde K, Larizza L, Fuhrman Conti AM (2000) A tumor suppressor locus in familial and sporadic chordoma maps to 1p36. Int J Cancer 87:68–72 [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1995) The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet 11:402–408 [DOI] [PubMed] [Google Scholar]
- Rich TA, Schiller A, Suit HD, Mankin HJ (1985) Clinical and pathologic review of 48 cases of chordoma. Cancer 56:182–187 [DOI] [PubMed] [Google Scholar]
- SAGE (1994) Statistical analysis for genetic epidemiology, release 2.2. Department of Epidemiology and Biostatistics, Case Western Reserve University, Cleveland [Google Scholar]
- Sawyer JR, Husain M, Al-Mefty O (2001) Identification of isochromosome 1q as a recurring chromosome aberration in skull base chordomas: a new marker for aggressive tumors? Neurosurg Focus 10:Article 6 [DOI] [PubMed] [Google Scholar]
- Stepanek J, Cataldo SA, Ebersold MJ, Lindor NM, Jenkins RB, Unni K, Weinshenker BG, Rubenstein RL (1998) Familial chordoma with probable autosomal dominant inheritance. Am J Med Genet 75:335–336 [DOI] [PubMed] [Google Scholar]
- Tai PT, Craighead P, Bagdon F (1995) Optimization of radiotherapy for patients with cranial chordoma: a review of dose-response ratios for photon techniques. Cancer 75:749–756 [DOI] [PubMed] [Google Scholar]