To the Editor:
Our comment concerns a recent letter to the Journal, in which de los Rios et al. (2001) reported the complete lack of BRCA2 mutations in 23 families with breast cancer and found only a single BRCA2 mutation in 66 Polish families with breast and ovarian cancer (Gorski et al. 2000). These data rule out BRCA2 from a portion of familial breast cancers, and they are reminiscent of data on the sporadic form of the disease, in which BRCA2 mutations are also absent (Miki et al. 1996; Teng et al. 1996; Brody et al. 1998).
A correlation of BRCA2 mutations and deletions with breast cancer is well established in a minority of families with breast cancer. At the biochemical level, BRCA2 is believed to function in DNA damage survey. The notion that BRCA2 is a tumor suppressor, however, is contradicted by its high expression in proliferating cells (Vaughn et al. 1996), its expression in sporadic breast cancer (Bieche et al. 1999), its positive correlation with a high mitotic index (Bieche et al. 1999), and the lack of mammary-gland cancer phenotype in BRCA2-truncated knockout mice (Connor et al. 1997). Moreover, in breast cancers with losses in the 13q12.3 region, the expression of the intact BRCA2 transcript (Bieche et al. 1999) and of the BRCA2 protein (Edwards et al. 1998) strongly suggest that the deletions targeted another gene in the area (a “cryptic” suppressor), a conclusion shared by others in the field.
Our search for genes involved in the repression of cell proliferation revealed a marker misannotation in the BRCA2 area. Our analysis shows that the correct annotation, in fact, identifies a new proliferation-arrest gene, known as “androgen-shutoff gene 3” (AS3), in this region and implicates it in cancer. Correct annotation also changes the interpretation of BRCA2 allelic imbalance data, which may ultimately explain the negative results reported by de los Rios et al. and others.
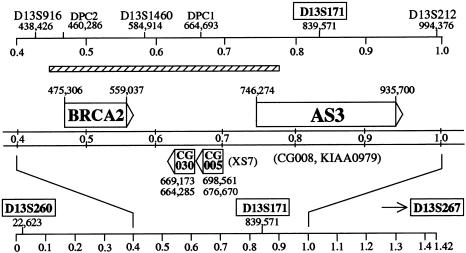
We mapped AS3 on chromosome 13 and found inconsistencies in the positions of critical markers in the databases of microsatellite markers (Cooperative Human Linkage Center, version 4), STS data (Unified Data Base, Integrated Chromosome 13 Map), radiation hybrid data (GeneMap99), and other sources. The nucleotide sequence and the individual contigs of the 13q12-13 area are now available through the Human Genome Project (Homo sapiens 13q12-q13 contig, 1,416,908 bp, accession number NT_000625). This allowed us to establish the precise nucleotide sequence–based positions, not only for AS3 (accession number NM_015928 or U95825) but for other markers and genes as well (see fig. 1)—for instance, the D13S260 (S260), D13S171 (S171), and D13S267 (S267) markers that have been the main focus of clinical studies. The S267 marker is telomeric to the sequence depicted in figure 1.
Figure 1.
Map positions of markers and genes on the nucleotide sequence of the 13q12-13 contig. Boxes indicate markers that have been the main focus of clinical studies. Top, Microsatellite markers. Markers are shown with numbers that indicate the positions of the first nucleotides of the amplicons. The scale refers to that of the 13q12-13 contig. The hatched bar represents the homozygous deletion in a pancreatic adenocarcinoma that partially removed the AS3 coding region. The left border of the deletion is centromeric to DPC2 (at ∼450,000 position), and its right border is telomeric to DPC1 (at ∼780,000–830,000 positions) (Schutte et al. 1995). Middle, the identified coding sequences in the area. Sequences are shown on the same scale as in the top panel. The arrowheads indicate the directions of transcription. The names in parentheses indicate alternative names or alternative transcripts. Bottom, the nucleotide sequence of the 13q12-13 contig in 0.1-Mb units.
The assignment of the S171 marker (accession number Z17151) is of particular importance, because it has been annotated to BRCA2 and widely used as an intragenic marker of BRCA2 (accession number XM_007138). Allelic imbalance data directly linked the marker to a suppressor gene. Increased proliferation rates in invasive ductal breast carcinoma (Beckmann et al. 1996) and lung carcinoma (Gorgoulis et al. 2000), deletions in sporadic breast cancer (Cleton-Jansen et al. 1995), unfavorable prognosis in prostate cancer (Edwards et al. 1998), and lymph node metastases in esophageal carcinoma (Harada et al. 1999) were positively correlated with losses in the S171 marker. In a study of hepatocellular carcinoma, the smallest common deleted region (SCDR1) was also the S171 marker (Lin et al. 1999). These losses point to a critical gene in the S171 position, as documented in the studies of patients with cancer mentioned above.
Our sequence-based mapping unambiguously demonstrated, however, that this gene is not BRCA2. Our data show that the S171 marker is not intragenic to BRCA2 (see fig. 1) and that losses in the marker cannot be directly interpreted as BRCA2 losses. The new gene, AS3, which is located in the S171 position, has recently been identified in prostate cancer cells undergoing androgen-induced proliferative arrest and shows the expression pattern of a negative regulator (Geck et al. 1997). Our sequence analysis identified the S171 microsatellite repeat as part of intron 10 of the genomic region of AS3. The 34 exons in a 200-kb area around the S171 marker code for a protein of ∼1,400 residues with various consensus domains (Geck et al. 1999). The 165-kD AS3 protein is localized in the cell nucleus. Expression of the sense and antisense AS3 sequence from a tetracycline-regulated retroviral construct showed that AS3 is a powerful negative regulator of cell proliferation (Geck et al. 2000). Recently, clinical studies have directly implicated AS3 in the development of esophageal cancer, through polymorphic variations and loss of heterozygosity that linked AS3 to lymph node metastases (Harada et al. 2001).
These data and the corrected map positions clearly indicate that (a) the exact location of the S171 marker is at the center of the 200-kb AS3 gene and is not intragenic to BRCA2, (b) the S171 microsatellite instability data link AS3 to a variety of cancers, (c) the AS3 gene product is a powerful negative regulator of cell proliferation, and (d) clinical data directly implicate AS3 in cancer. Unfortunately, on the basis of the public database annotations, the recent literature consistently assigns the S171 marker to BRCA2 (Edwards et al. 1998; Lin et al. 1999; Gorgoulis et al. 2000). In the light of our analysis, it is highly misleading to assign the S171 allelic instability data unconditionally and exclusively to BRCA2. Our results correct this misperception and reveal that AS3 is the real cognate gene of the marker, a prime candidate for a role in the negative regulation of cell proliferation.
Electronic-Database Information
URLs for data in this article are as follows:
- Cooperative Human Linkage Center, http://lpg.nci.nih.gov/CHLC/ (for databases of microsatellite markers)
- GeneMap99, http://www.ncbi.nlm.nih.gov/genemap99/ (for radiation-hybrid data)
- Human Genome Project, http://research.marshfield.org/genetics (for 13q12-q13 contig)
- Unified Data Base, http://bioinformatics.weizmann.ac.il/udb/ (for STS data)
References
- Beckmann MW, Picard F, An HX, van Roeyen CR, Dominik SI, Mosny DS, Schnurch HG , Bender HG, Niederacher D (1996) Clinical impact of detection of loss of heterozygosity of BRCA1 and BRCA2 markers in sporadic breast cancer. Br J Cancer 73:1220–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieche I, Nogues C, Lidereau R (1999) Overexpression of BRCA2 gene in sporadic breast tumours. Oncogene 18:5232–5238 [DOI] [PubMed] [Google Scholar]
- Brody LC, Biesecker BB (1998) Breast cancer susceptibility genes: BRCA1 and BRCA2. Medicine 77:208–226 [DOI] [PubMed] [Google Scholar]
- Cleton-Jansen A-M, Collins C, Lakhani SR, Weissenbach J, Devilee P, Cornelisse CJ, Stratton MR (1995) Loss of heterozygosity in sporadic breast tumours at the BRCA2 locus on chromosome 13q12-q13. Br J Cancer 72:1241–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor F, Bertwistle D, Mee PJ, Ross GM, Swift S, Grigorieva E, Tybulewicz VL, Ashworth A (1997) Tumorigenesis and a DNA repair defect in mice with a truncating BRCA2 mutation. Nat Genet 17:423–430 [DOI] [PubMed] [Google Scholar]
- de los Rios P, Jack E, Kuperstein G, Lynch H, Lubinski J, Narod SA (2001) Founder mutations of BRCA1 and BRCA2 in North American families of Polish origin that are affected with breast cancer. Am J Hum Genet 68:546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SM, Dunsmuir WD, Gillett CE, Lakhani SR, Corbishley C, Young M, Kirby RS, Dearnaley DP, Dowe A, Ardern-Jones A, Kelly J, Spurr N, Barnes DM, Eeles RA (1998) Immunohistochemical expression of BRCA2 protein and allelic loss at the BRCA2 locus in prostate cancer. CRC/BPG UK Familial Prostate Cancer Study Collaborators. Int J Cancer 78:1–7 [DOI] [PubMed] [Google Scholar]
- Geck P, Maffini MV, Szelei J, Sonnenschein C, Soto AM (2000) Androgen-induced proliferative quiescence in prostate cancer: the role of AS3 and its mediator. Proc Natl Acad Sci USA 97:10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geck P, Szelei J, Jimenez J, Lin TM, Sonnenschein C, Soto AM (1997) Expression of novel genes linked to the androgen-induced, proliferative shutoff in prostate cancer cells. J Steroid Biochem Mol Biol 63:211–218 [DOI] [PubMed] [Google Scholar]
- Geck P, Szelei J, Jimenez J, Sonnenschein C, Soto AM (1999) Early gene expression during androgen-induced inhibition of proliferation of prostate cancer cells: a new suppressor candidate on chromosome 13, in the BRCA2-Rb1 locus. J Steroid Biochem Mol Biol 68:41–50 [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Kotsinas A, Zacharatos P, Mariatos G, Liloglou T, Tsoli E, Kokotas S, Fassoulas C, Field JK, Kittas C (2000) Association of allelic imbalance at locus D13S171 (BRCA2) and p53 alterations with tumor kinetics and chromosomal instability (aneuploidy) in nonsmall cell lung carcinoma. Cancer 89:1933–1945 [DOI] [PubMed] [Google Scholar]
- Gorski B, Byrski T, Huzarski T, Jakubowska A, Menkiszak J, Gronwald J, Pluzanska A, Bebenek M, Fischer-Maliszewska L, Grzybowska E, Narod SA, Lubinski J (2000) Founder mutations in the BRCA1 gene in Polish families with breast and ovarian cancer. Am J Hum Genet 66:1963–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Tanaka H, Shimada Y, Shinoda M, Imamura M, Ishizaki K (1999) Lymph node metastasis is associated with allelic loss on chromosome 13q12-13 in esophageal squamous cell carcinoma. Cancer Res 59:3724–3729 [PubMed] [Google Scholar]
- Harada H, Uchida N, Shimada Y, Kumimoto H, Shinoda M, Imamura M, Ishizaki K (2001) Polymorphism and allelic loss at the AS3 locus on 13q12-13 in esophageal squamous cell carcinoma. Int J Oncol 18:1003–1007 [DOI] [PubMed] [Google Scholar]
- Lin YW, Sheu JC, Liu LY, Chen CH, Lee HS, Huang GT, Wang JT, Lee PH, Lu FJ (1999) Loss of heterozygosity at chromosome 13q in hepatocellular carcinoma: identification of three independent regions. Eur J Cancer 35:1730–1734 [DOI] [PubMed] [Google Scholar]
- Miki Y, Katagiri T, Kasumi F, Yoshimoto T, Nakamura Y (1996) Mutation analysis in the BRCA2 gene in primary breast cancers. Nat Genet 13:245–247 [DOI] [PubMed] [Google Scholar]
- Schutte M, da Costa LT, Hahn SA, Moskaluk C, Hoque AT, Rozenblum E, Weinstein CL, Bittner M, Meltzer PS, Trent JM, Yeo CJ, Hruban RH, Kern SE (1995) Identification by representational difference analysis of a homozygous deletion in pancreatic carcinoma that lies within the BRCA2 region. Proc Nat Acad Sci USA 92:5950–5954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng DH, Bogden R, Mitchell J, Baumgard M, Bell R, Berry S, Davis T, Ha PC, Kehrer R, Jammulapati S, Chen Q, Offit K, Skolnick MH, Tavtigian SV, Jhanwar S, Swedlund B, Wong AK, Kamb A. (1996) Low incidence of BRCA2 mutations in breast carcinoma and other cancers. Nat Genet 13:241–244 [DOI] [PubMed] [Google Scholar]
- Vaughn JP, Cirisano FD, Huper G, Berchuck A, Futreal PA, Marks JR, Iglehart JD (1996) Cell cycle control of BRCA2. Cancer Res 56:4590–4594 [PubMed] [Google Scholar]