To the Editor:
In this letter, we describe the Autism Genetic Resource Exchange (AGRE), a resource for the study of autism and pervasive developmental disorder (PDD). Autism presents within the first 3 years of life, is characterized by qualitative impairments in communication and social interaction—in the presence of restricted repetitive and stereotyped patterns of behavior, interests, and activities—and is part of a spectrum of disorders that includes Asperger syndrome and PDD (American Psychiatric Association 1994). Estimates of the prevalence of autism in the general population ranges from 0.04% to >0.l% (Bryson et al. 1988; Gillberg et al. 1991). Twin and family studies have demonstrated that the genetic contribution to autism and PDD is significant, with an MZ-twin concordance of 60%–90% and a 45- to 150-fold increase in risk to siblings (Ritvo et al. 1989; Jorde et al. 1990; Bailey et al. 1995). Thus, molecular genetic studies of autism-spectrum disorders are likely to contribute significantly to our understanding of this condition, as the recent results of several independent genome scans suggest (International Molecular Genetic Study of Autism Consortium 1998; Barrett et al. 1999; Philippe et al. 1999; Risch et al. 1999).
AGRE has been developed as a joint effort of the Cure Autism Now (CAN) Foundation and the Human Biological Data Interchange (HBDI), to facilitate collaborative genetic research into the etiology of autism and PDD and to make biomaterials from well-characterized families with autism widely available to the scientific community, so as to accelerate research. Since genetic studies of complex neuropsychiatric conditions are limited by the large sample sizes needed to attain adequate power (Lander and Kruglyak 1995; Risch and Merikangas 1996), the consolidation of large numbers of families into one collection that is made available to researchers at a fraction of the cost originally incurred in their ascertainment and collection is of great value to the community.
One unique feature of AGRE, which has enabled the rapid ascertainment of large numbers of families, has been the development of a protocol and the infrastructure to conduct the majority of the evaluations and blood draws in the families’ homes. This process may prove useful for more-rapid family ascertainment in studies of other neuropsychiatric conditions (AGRE Web site). To date, ∼400 multiplex families with autism and PDD are in various stages of clinical evaluation, with DNA collection completed (table 1). Both an online and a hard-copy catalogue are available, containing the pedigrees in the collection, with notations of affectation status and basic phenotypic features, such as language delay. A sample pedigree is depicted in figure 1. Biomaterials from 343 of these completed families are currently available to the scientific research community, and this resource continues to expand, with the goal of 500 families by the end of the year 2001. Family biomaterials for the AGRE program are housed at the HBDI Repository at Rutgers University, under the direction of Jay Tischfield. Quality-controlled samples, including immortalized cell lines (1 × 106 cells/ampoule), 20-μg aliquots of DNA, and 50-μl aliquots of sera, are available to the research community by simple application, which requires proof of institutional review board (IRB) approval. Samples are available to academia and industry, and significant discounts, as well as limited grants to support academic use of the resource, are available to academic researchers through the CAN Foundation. To facilitate collaboration, free samples are available to researchers who deposit their collections in AGRE through the Sharing Researcher Program.
Table 1.
Patient Recruitment and Availability
| Status | Current | Projected 2001 |
| Recruited:a | ||
| Families | 428 | 500 |
| Individuals | 1,978 | 2,250 |
| Completed:b | ||
| Families | 343 | 420 |
| Individuals | 1,595 | 1,890 |
| In process:c | ||
| Families | 85 | 80 |
| Individuals | 496 | 360 |
Consented and scheduled for ADI and blood draw.
ADI complete, biomaterials complete, quality controlled and available for distribution.
Incomplete biomaterials or ADI.
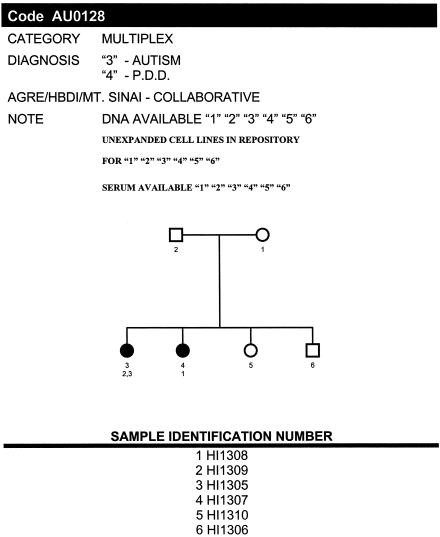
Figure 1.
Sample AGRE pedigree. A typical AGRE pedigree is depicted, with the identifying numbers given directly underneath the individuals. The second number refers to the following key, as defined by ADI scores: 1 = verbal; 2 = nonverbal; 3 = regression; and 4 = late onset. The HI numbers are actual coded database numbers that uniquely identify each individual. The availability of biomaterials is also indicated if the family was corecruited with another academic group—in this case, Mt. Sinai (AGRE Web site).
Scientific oversight for the program is provided by a Steering Committee, which includes researchers from the fields of genetics and autism. Human subjects protection oversight is provided by the IRB at the University of Pennsylvania School of Medicine. In addition to providing researchers with biomaterials, a major effort has been undertaken to develop a state-of-the-art, Internet-accessible database of detailed clinical information. Phenotypic assessment is ongoing and includes the two examinations that are completed by all of the NIH autism collaborative groups: the Autism Diagnostic Interview–Revised (ADI-R) (Lord et al. 1994) and the Autism Diagnostic Observational Schedule (ADOS) (Lord et al. 2000). All ADI and ADOS raters undergo ongoing reliability checks to prevent any drift in diagnosis (Lord et al. 1994, 2000). In addition, photographic dysmorphology, physical and neurological examination, and medical and family history are being collected by pediatric neurologists. Probands with possible secondary autism resulting from perinatal trauma, from an identified genetic syndrome, or from other medical causes are noted, although this is only a small percentage of cases. Currently, the ADI data and a subset of the ADOS data, both coded for confidentiality, are available online for researcher access through the AGRE Web site. Online phenotypic databases for the remainder of the data collected are being developed and will be available in ⩽6 mo. More information on the timeline of data and material collection is available at the Web site. We are striving to improve the utility of AGRE, and user feedback is an important element in this process.
Fragile-X testing is conducted in all families (Brown et al. 1986), and cytogenetic analysis—including FISH for 15q and telomere screening—is commencing. Of 220 families tested, 3 have subjects that carry a fragile-X expansion (1.3%; W. T. Brown, unpublished data). A genome scan at an average 10-cM resolution has been completed on the first 132 families in the collection (T. C. Gilliam, personal communication), and genotype data from 188 families are available online at the AGRE Web site. As genome scans on additional families are completed, these data will be updated regularly, and investigators accessing these data will be notified of the updates automatically. More information regarding this resource, including pricing of samples and access to the resource, can be obtained from the CAN Foundation and AGRE Web sites, or by contacting the authors.
Acknowledgments
We gratefully acknowledge the families and individuals who have contributed their biomaterials, time, and financial resources to AGRE, especially Marianne Toedtman, AGRE family recruiter; Ed Berry, phlebotomist; Andrew Smith, M.D., pediatric neurologist; Paul Law, M.D., M.P.H., for database development; and Nancy Jones, AGRE Web master. We specifically thank Sallie and Tom Bernard, for their generous financial support of AGRE, and the Schering-Plough Research Institute and Pfizer Inc., for their contributions to AGRE. We also thank Maricela Alarcon, Ph.D., for her error checking and advice; Jianjun Liu, Ph.D., for updating of the genotyping data; and scientists who have started to utilize AGRE, for their helpful comments and criticism. The members of the AGRE Steering Committee are: W. Ted Brown, New York State Institute for Basic Research in Developmental Disabilities, Staten Island; Maya Bucan, University of Pennsylvania, Philadelphia; Joseph Buxbaum, Mt. Sinai School of Medicine, New York; T. Conrad Gilliam, Columbia University Genome Center, New York; David A. Greenberg, Mt. Sinai School of Medicine, New York; David H. Ledbetter, University of Chicago, Chicago; Bruce L. Miller, University of California, San Francisco; Stanley F. Nelson, UCLA School of Medicine, Los Angeles; Jonathan Pevsner, Kennedy Krieger Institute, Baltimore; Jerome I. Rotter, Cedars-Sinai Medical Center, Los Angeles; Carol Samango-Sprouse, Children’s National Medical Center, Baltimore; Gerard D. Schellenberg, University of Washington and Veterans Affairs Medical Center, Seattle; Rudolph E. Tanzi, Massachusetts General Hospital, Boston; and Kirk C. Wilhelmsen, University of California, San Francisco.
Electronic-Database Information
The URLs for data in this article are as follows:
- Autism Genetic Resource Exchange, http://www.agre.org/
- Cure Autism Now, http://www.canfoundation.org/
References
- American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders, 4th ed–revised. American Psychiatric Association, Washington, DC [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M (1995) Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25:63–77 [DOI] [PubMed] [Google Scholar]
- Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, Childress D, et al (1999) An autosomal genomic screen for autism: collaborative linkage study of autism. Am J Med Genet 88:609–615 [DOI] [PubMed] [Google Scholar]
- Brown WT, Jenkins EC, Cohen IL, Fisch GS, Wolf-Schein EG, Gross A, Waterhouse L (1986) Fragile X and autism: a multicenter survey. Am J Med Genet 23:341–352 [DOI] [PubMed] [Google Scholar]
- Bryson SE, Clark BS, Smith IM (1988) First report of a Canadian epidemiological study of autistic syndromes. J Child Psychol Psychiatry 29:433–445 [DOI] [PubMed] [Google Scholar]
- Gillberg C, Steffenburg S, Schaumann H (1991) Is autism more common now than ten years ago? Br J Psychiatry 158:403–409 [DOI] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (1998) A full genome screen for autism with evidence for linkage to a region on chromosome 7q. Hum Mol Genet 7:571–578 [DOI] [PubMed] [Google Scholar]
- Jorde LB, Mason-Brothers A, Waldmann R, Ritvo ER, Freeman BJ, Pingree C, McMahon WM, Petersen B, Jenson WR, Mo A (1990) The UCLA-University of Utah epidemiologic survey of autism: genealogical analysis of familial aggregation. Am J Med Genet 36:85–88 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, Cook EH Jr, Leventhal BL, DiLavore PC, Pickles A, Rutter M (2000) The autism diagnostic observation schedule-generic: a standardized observation of communicative and social behavior associated with the spectrum of autism. J Autism Dev Disord 30:205–223 [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A (1994) Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 24:659–685 [DOI] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, van Malldergerme L, Penet C, Feingold J, Brice A, Leboyer M (1999) Genome-wide scan for autism susceptibility genes: Paris Autism Research International Sibpair Study. Hum Mol Genet 8:805–812 [DOI] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, et al (1999) A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet 65:493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Ritvo ER, Mason-Brothers A, Freeman BJ, Pingree C, Jenson WR, McMahon WM, Petersen PB, Jorde LB, Mo A, Ritvo A (1989) The UCLA-University of Utah epidemiologic survey of autism: recurrence risk estimates and genetic counseling. Am J Psychiatry 146:1032–1036 [DOI] [PubMed] [Google Scholar]