Abstract
Mutations in the gene for guanylate cyclase–activating protein-1 (GCAP1) (GUCA1A) have been associated with autosomal dominant cone dystrophy (COD3). In the present study, a severe disease phenotype in a large white family was initially shown to map to chromosome 6p21.1, the location of GUCA1A. Subsequent single-stranded conformation polymorphism analysis and direct sequencing revealed an A464G transition, causing an E155G substitution within the EF4 domain of GCAP1. Modeling of the protein structure shows that the mutation eliminates a bidentate amino acid side chain essential for Ca2+ binding. This represents the first disease-associated mutation in GCAP1, or any neuron-specific calcium-binding protein within an EF-hand domain, that directly coordinates Ca2+. The functional consequences of this substitution were investigated in an in vitro assay of retinal guanylate cyclase activation. The mutant protein activates the cyclase at low Ca2+ concentrations but fails to inactivate at high Ca2+ concentrations. The overall effect of this would be the constitutive activation of guanylate cyclase in photoreceptors, even at the high Ca2+ concentrations of the dark-adapted state, which may explain the dominant disease phenotype.
Introduction
Autosomal dominant cone dystrophies are a type of hereditary macular degeneration characterized by a progressive dysfunction of the photopic system, with preservation of scotopic function. They occur in the population with a frequency of ∼1/10,000 (Small et al. 1996). The disease presents as the phenotypic triad of loss of color vision, photophobia, and reduced central visual acuity and is distinguished from the cone-rod dystrophies, in which some loss of peripheral vision also occurs. Cone dystrophy is a genetically heterogeneous disorder, with phenotype linked to loci on 17p12-13 (Balciuniene et al. 1995; Small et al. 1996), Xp21.1-p11.3 (Jacobsen et al. 1989), 6q25-q26 (retinal cone dystrophy 1 [MIM 180020]), and 6p21.1 (Payne et al. 1998). In the case of 6p21.1, mutations have been identified in GUCA1A (COD3 [MIM 602093 and 600364]) (Payne et al. 1998; Downes et al. 2001), the gene that encodes guanylate cyclase–activating protein-1 (GCAP1).
GCAP1 is a key component in the recovery of photoreceptor cells to the dark-adapted state after exposure to a light stimulus. This recovery process is mediated via a change in intracellular free calcium ions in response to a decrease in the level of cyclic guanosine monophosphate (cGMP); both calcium ions and cGMP are fundamental secondary messengers in photoreceptors. Several mechanisms, highly conserved across species, have developed to maintain a low [Ca2+]free within photoreceptors. In the dark-adapted state, the [Ca2+]free in photoreceptor outer segments rises to ∼500 nM because of the influx of Ca2+ through open cGMP-gated Na+/Ca2+ channels (Dizhoor and Hurley 1999). Exposure to light leads to the hydrolysis of cGMP by phosphodiesterase (via a rhodopsin-mediated cascade) and to a subsequent decrease in [Ca2+]free to ∼30 nM because of the continued efflux of Ca2+ through light-independent Na+/K+/Ca2+ exchangers (Baylor 1996; Koutalos and Yau 1996; Pugh et al. 1996; Sampath et al. 1998). This decrease in [Ca2+]free stimulates an increase in cGMP, synthesized by two distinct retina-specific isoforms of guanylate cyclase (RetGC1 and RetGC2), in a feedback mechanism vital to the process of light adaptation and the recovery of the dark-adapted state (Hayashi and Yamazaki 1991; Shyjan et al. 1992; Dizhoor et al. 1994; Garbers and Lowe 1994; Goraczniak et al. 1994; Lowe et al. 1995; Sitaramayya et al. 1995; Yang et al. 1995; Pugh et al. 1997).
The RetGCs are not directly sensitive to changes in [Ca2+]free; calcium exerts its regulatory effect via specialized calcium-binding proteins known as guanylate cyclase–activating proteins (GCAPs) (Dizhoor et al. 1994; Palcewski et al. 1994; Dizhoor et al. 1995; Gorczyza et al. 1995; Lowe et al. 1995; Haeseleer et al. 1999). Three mammalian isoforms have been identified, GCAP1 (Palczewski et al. 1994), GCAP2 (Dizhoor et al. 1995; Gorczyca et al. 1995) and GCAP3 (Haeseleer et al. 1999). GCAPs belong to a subgroup of the neuron-specific calcium-binding proteins (NCBPs), which includes recoverin. All NCBPs incorporate four variably functional repeats of the EF-hand domain (Polans et al. 1996), a helix-loop-helix structural domain with a selectively high affinity for Ca2+ binding (kD <10−5 M) (Nockolds et al. 1972). In the case of the GCAPs, only EF2, EF3, and EF4 are functional. Numerous studies have demonstrated that the GCAPs function to mediate the calcium-sensitive synthesis of cGMP by RetGC (Koch and Stryer 1988; Pittler and Baehr 1991; Dizhoor et al. 1994, 1995; Palczewski et al. 1994; Goryczya et al. 1995; Lowe et al. 1995; Duda et al. 1996; Frins et al. 1996; Pugh et al. 1997; Rudnicka-Nawrot et al. 1998; Haeseleer et al. 1999), activating the cyclase at [Ca2+]free <100 nM and inhibiting the cyclase at [Ca2+]free >500 nM, concentrations characteristic of light- and dark-adapted photoreceptors, respectively. This distinguishes the GCAPs from other calcium-binding proteins, which typically activate effector proteins in their calcium-loaded forms.
The genes for human GCAP1 and GCAP2 reside in a tail-to-tail array on chromosome 6p21.1 (Subbaraya et al. 1994; Surguchov et al. 1997; Howes et al. 1998) and share a common four-exon/three-intron arrangement. GCAP3 shares the same exon/intron arrangement but has been localized to chromosome 3q13.1 (Haeseleer et al. 1999). This suggests that the three GCAP genes arose as a result of the duplication and translocation of a common ancestral gene. All three proteins have been localized to the retina in various species, with GCAP1 present in higher concentrations in cone than in rod outer segments (Gorczyca et al. 1995; Cuenca et al. 1998). GCAP2 is localized to rods (Dizhoor et al. 1995), cone inner segments (Otto-Bruc et al. 1997), and layers of the inner retina (Cuenca et al. 1998; Howes et al. 1998).
To date, two mutations have been identified in GCAP1. A Y99C substitution causes a severe cone dystrophy (Payne et al. 1998), and a P50L substitution is associated with a more variable phenotype, ranging from mild cone dystrophy to pronounced cone-rod dystrophy (Downes et al. 2001). In the present article, we report the clinical and genetic characteristics of a family with a severe form of autosomal dominant cone dystrophy that segregates a novel E155G mutation in GCAP1 (GUCA1A). We also describe the functional consequences of this mutation and the ability of the mutant protein to regulate RetGC activity in response to calcium ion changes. This represents the first disease-causing mutation found in the highly conserved 12-residue Ca2+-binding motif of an EF hand.
Material and Methods
Subjects
Approval from the Institutional Review Board of the Cleveland Clinic Foundation was obtained for the present study, and informed consent was obtained from all patients. Of 67 individuals at risk for a dominantly inherited cone dystrophy in a large white kindred, 33 were found to be affected.
Clinical and Functional Investigations
A visual history was obtained from all patients, and best corrected visual acuities were assessed. Ophthalmoscopic examination was performed on all patients, and fluorescein angiography and electroretinography were performed on one patient. Blood samples were obtained by venipuncture.
After initial dark adaptation, electroretinograms (ERGs) were made, using a bipolar Burian-Allen contact lens electrode, to record response to stimuli presented within an LKC ganzfeld. The response was differentially amplified (band-pass: 0.3–500 Hz), averaged, and stored using an LKC UTAS E-2000 signal-averaging system. Responses were initially recorded from the dark-adapted eyes, using a low-intensity (−24-dB attenuation) and high-intensity (0-dB attenuation) strobe flash. A steady adapting field was then used to desensitize the rod response. After a 7-min period of light adaptation (Peachey et al. 1989), high-intensity responses were superimposed on the adapting field.
Genetic Linkage and Mutation Screening
DNA was extracted from blood samples, and genetic linkage was assessed using microsatellite marker D6S1017, which is linked to the GCAP1 locus. Linkage analysis was performed under the assumption that the disease-allele frequency is .0001 and that the mode of inheritance is autosomal dominant, with a penetrance of 90%. Each of the four exons of the GCAP1 gene was amplified by PCR and analyzed by SSCP, using the method of Orita et al. (1989). Mutational screening was then performed by direct sequencing of PCR-amplified DNA fragments corresponding to each exon of the gene. Each fragment was sequenced on both strands. Sequencing was performed with the Taq DyeDeoxy Terminator Cycle Sequencing Kit (Applied Biosystems), according to the manufacturer’s instructions.
Mutagenesis of GCAP1 and Expression in Escherichia coli
An E155G point mutation was introduced into human wild-type GCAP1, using a GeneEditor site-directed mutagenesis kit (Promega) and the following mutagenesis primer: 5′-CT TCC CTG GAA GGG TTT ATA GAG G-3′. The wild-type GCAP1 clone used for the mutagenesis carried an artificial E6S substitution to facilitate N-myristoylation of the protein, which has been found to be necessary for the activation of RetGC1 by GCAP1 (Frins et al. 1996). The mutated sequence was cloned in pET3a (Novagen) and expressed in E. coli strain BL21(DE3)pBBLysS (Newbold et al. 2001). This strain carries the pBBLysS plasmid-encoding yeast N-myristoyltransferase, which is coexpressed with the GCAP1 and effects N-terminal modification of the latter protein. Conditions for the expression and subsequent purification of the wild-type and E155G proteins were as described by Newbold et al. (2001).
Assay of RetGC1 Activation
Human RetGC1 was produced in HEK 293T cells transiently transfected with a pRC.CMV RetGC1 construct (provided by J. B. Hurley) and isolated in the form of a membrane preparation, as described by Wilkie et al. (2000). Measurement of guanylate cyclase activity was conducted in a radioassay with the use of α[32P]GTP, according to the method of Dizhoor et al. (1995), which was modified as described by Wilkie et al. (2000). Basal activity and activity stimulated by wild-type and mutant GCAP1 were measured in reaction buffers containing a range of [Ca2+]free, formulated according to the method of Tsien and Pozzan (1989).
Results
Clinical Findings
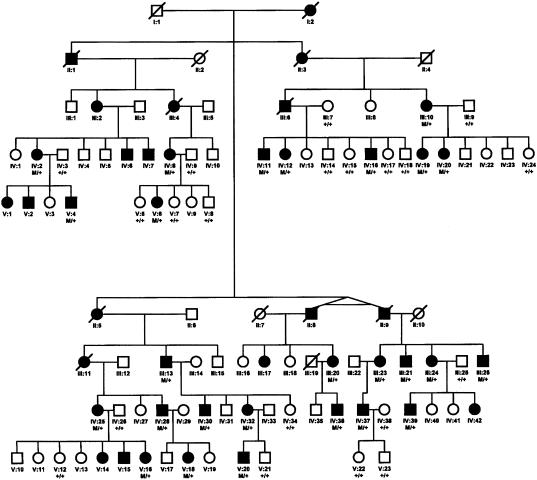
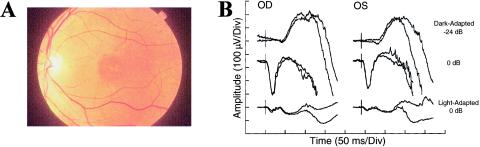
Clinical evaluations of a large kindred of five generations revealed an autosomal dominant pattern of inheritance (fig. 1). On the basis of the presence of decreased visual acuity, abnormalities of color vision, and macular lesions, 33 of 67 individuals at risk for inheriting the disease were found to be affected. The age at onset of decreased visual acuity, color vision defects, and photophobia is 8–24 years, with a mean of 16 years. Visual loss progresses, during the next two decades, until visual acuity is between 20/200 and 20/400. Fundus examination revealed a central macular lesion with pigmentary changes (fig. 2A)
Figure 1.
Autosomal dominant inheritance of cone dystrophy. Individuals are identified by generation and pedigree number; squares indicate males, and circles indicate females. Blackened symbols indicate affected individuals; open symbols, indicate unaffected individuals.
Figure 2.
Clinical evaluation. A, Fundus photograph from a patient (IV:37) with cone dystrophy, aged 26 years, with visual acuity 20/200, showing central macular pigmentary changes. B, ERGs recorded from the left (OS) and right (OD) eye of patient V:6 (aged 43 years), showing responses to ganzfeld stimuli (times of flash indicated by vertical bars) presented to the dark-adapted eye with a low-intensity strobe flash (top row) and with a high-intensity strobe flash (middle row) and to the light-adapted eye with a high-intensity strobe flash (bottom row). Two successive responses are shown for each stimulus condition. Note that the ERG of the light-adapted eye (cone) is diminished below the normal range, whereas responses of the dark-adapted eye (rod-dominated) fall within normal limits.
Figure 2B presents ERGs recorded, from the two eyes of an affected patient (V:6), under standard stimulation and recording conditions. In each stimulation condition, two successive responses are overlain to demonstrate reproducibility. The top row shows the rod-mediated ERGs obtained from a dim flash presented to the dark-adapted eye. The amplitude of the β-wave falls within the range of normal. The middle row shows the response of the dark-adapted eye to a high-intensity stimulus flash. The negative-polarity α-wave also falls within the normal range, and the positive β-wave was at or slightly below the normal range. The bottom row shows the response obtained under light-adapted conditions to isolate the cone response. For both eyes, the amplitude of the cone ERG falls well below the lower limit of the normal range.
Genetic Analysis
An initial genotype analysis with marker D6S1017 yielded a significant LOD score of 3.3 at a recombination fraction of 0.1, indicating positive linkage to the GCAP1 locus. SSCP analysis, followed by direct sequencing, revealed an A→G transition in nucleotide 464, resulting in an E155G amino acid substitution in the EF4 domain of GCAP1 (fig. 2). This change segregated with the disease phenotype in the entire family (fig. 1), with a maximum LOD score of 7.0, and it was not found in 200 normal control subjects. The primary sequences of the three functional EF hands in GCAP1 identifying the E155G mutation present in the pedigree is shown in figure 3.
Figure 3.
Alignment of the three functional EF-hand domains in GCAP1. The α-helix and loop regions of the domain are identified above the sequences. Residues in the loop that coordinate the Ca2+ ion are marked with an asterisk. The invariant Glu residues at position 12 of each Ca2+-binding loop (E75, E111, and E155) are highlighted in bold. The positions of the E155G mutation in EF4 present in our pedigree and of the previously reported Y99C mutation in EF3 are boxed.
Functional Analysis
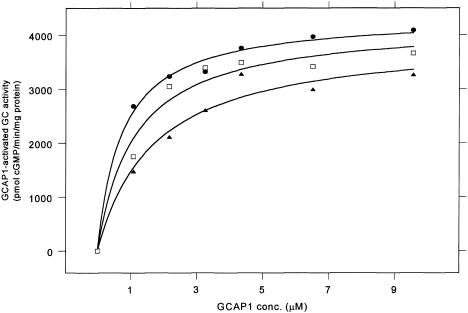
An E155G substitution was introduced into wild-type human GCAP1 by site-directed mutagenesis. This was cloned in the vector pET3a and expressed in E. coli cells. The GCAP1 protein was isolated in a cytosolic extract from the cells and purified in a single-column gel-filtration procedure, as described by Newbold et al. (2001). The purified protein was assayed for its ability to activate RetGC1, as described elsewhere (Dizhoor et al. 1995; Wilkie et al. 2000). A titration of RetGC1 activity against increasing concentrations of GCAP1 in the presence of a high substrate (GTP) concentration demonstrated that the initial rate of cGMP formation reached a maximum (saturated) value at high GCAP1 concentrations, with both wild-type and E155G GCAP1 (fig. 4). The maximum specific activation (Vmax) obtained with the mutant GCAP1 was slightly lower than that obtained with the wild type. However, the GCAP1 concentration required for half-maximal activation (Km app[GCAP1]) was approximately double that of the wild type (table 1), indicating a lower apparent affinity of RetGC1 for mutant than for wild-type GCAP1. An equimolar mixture of wild-type and mutant protein, representing the situation in an individual heterozygous for the mutation, yielded intermediate results.
Figure 4.
GCAP1 titrations of RetGC1 activity with wild-type GCAP1 (blackened circles), E155G mutant (blackened triangles) and an equimolar mixture of wild-type and E155G mutant (open squares). Curves were fitted to the data points on the basis of the relation v=[GCAP1]Vmax/(Kmapp[GCAP1]+[GCAP1]), where Km app[GCAP1] is the GCAP1 concentration required for half maximal activation and where Vmax is the maximum specific activation. Values of Vmax and Km app[GCAP1] are derived from the fitted curves are given in table 1.
Table 1.
Kinetic Parameters Vmax and Km app[GCAP1] for Activation of RetGC1 by Wild-Type and E155G Mutant GCAP1
| GCAP1 | Vmax ± SEa | Km app[GCAP1] ± SEb |
| Wild type | 4350 ± 120 | 0.738 ± 0.106 |
| Wild type/E155Gc | 4219 ± 293 | 1.103 ± 0.318 |
| E155G | 3955 ± 320 | 1.688 ± 0.453 |
Vmax measured as picomoles of cGMP per minute per milligram of protein.
Km measured as micromoles.
Equimolar mixture.
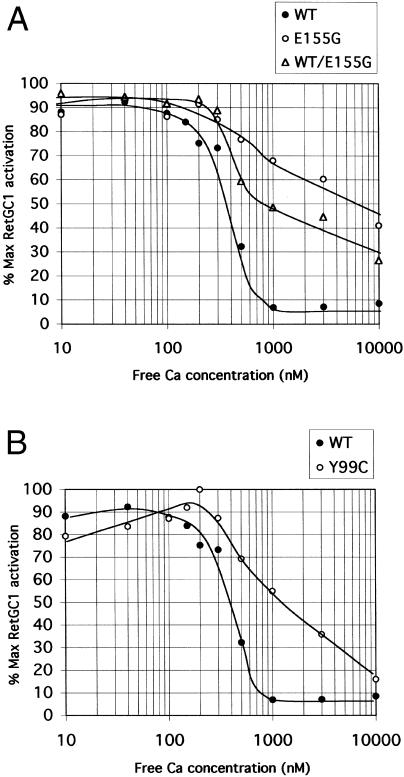
In wild-type GCAP1, binding of Ca2+ ions to the three EF hands regulates its ability to activate RetGC1. Thus, at [Ca2+]free ⩽100 nM, RetGC1 is activated by GCAP1 (Dizhoor et al. 1994; Gorczyca et al. 1994), but at micromolar concentrations it is inhibited (Rudnicka-Nawrot et al. 1998). Ca2+ titrations of RetGC1 activity in the presence of wild-type and mutant GCAP1 are shown in figure 5. With wild-type GCAP1, the [Ca2+]free for half-maximal activity was estimated to be ∼350 nM. With mutant GCAP1, however, the curve shows very little inhibition of activity at levels >350 nM. Even at 1 μM (generally regarded as above the maximum physiological [Ca2+]free in photoreceptor cells), almost 70% of maximal activity persists, compared with ∼10% with the wild type. When an equimolar mixture of wild-type and mutant GCAP1 is used, activity at [Ca2+]free levels >350 nM is again maintained; at 1 μM, almost 50% of maximal activity persists. Thus, the effect of the mutation is to maintain high levels of cyclase activity over the whole physiologically relevant range of [Ca2+]free. This effect is dominant, in that it persists even in the presence of wild-type GCAP1 protein.
Figure 5.
Ca2+ sensitivity of activation of wild-type RetGC1 by wild-type and mutant GCAP1. A, Activation by wild-type (blackened circles) and E155G mutant (open circles) GCAP1 and by an equimolar mixture of wild-type and E155G mutant (open triangles). B, Activation by wild-type (blackened circles) and Y99C mutant (open circles) GCAP1. Guanylate cyclase activity was determined in membrane preparations expressing RetGC1 activity stimulated with 8 μM GCAP1 in all cases. Results are presented as percentage of maximum RetGC1 activation (maximum RetGC1 activity minus basal activity in absence of GCAP1).
The Y99C mutation in GCAP1 has been shown to result in a similar constitutive activation of RetGC1 at high [Ca2+]free (Dizhoor et al. 1998; Sokal et al. 1998). Comparison of the magnitude of the effects of the two mutations on RetGC activity (fig. 5B) shows that the E155G mutation is more severe than Y99C, in that the failure to inhibit activity at high [Ca2+]free is more pronounced.
Discussion
A novel E155G mutation in GUCA1A, the gene encoding GCAP1, has been identified in a family affected by autosomal dominant cone dystrophy (COD3). A total of three disease-associated mutations, including this one, have been reported in GUCA1A, the other two involving Y99C (Payne et al. 1998) and P50L substitutions (Downes et al. 2001). All three disorders may present with a clinical diagnosis of cone dystrophy, but they differ in severity. E155G is similar to Y99C, in that it causes a severe phenotype. However, the mean age at onset of disease (16 years) is noticeably earlier in patients with E155G than in patients with Y99C, who do not have clinical symptoms until at least the third decade of life. In contrast, P50L is associated with a generally less-severe, but markedly variable, phenotype. This study allows us to address the question of how the identity of different mutations in the same gene results in variable expressivity of the disease.
The E155G mutation occurs within the fourth EF-hand domain of GCAP1. EF-hand domains comprise a helix-loop-helix motif, the loop region of which consists of 12 residues. Several of these residues are involved in coordinating the Ca2+ ion via oxygen-containing side chains and are frequently glutamate or aspartate. Within functional EF-hand domains, an invariantglutamate residue is found at position 12, which has been shown to be essential for Ca2+ coordination (Kretsinger 1976). It is this highly conserved residue of EF4 that is mutated in the patients in this family affected with dominant cone dystrophy.
X-ray crystallographic models of EF hands in a number of calcium-binding proteins have shown that each calcium ion coordinates seven oxygen ligands (Strynadka and James 1989). One of these oxygen ligands is provided by a water molecule, and the remaining six are provided by oxygen-containing side chains of residues in the loop. The glutamate at position 12 contributes both of its side-chain oxygen atoms to the metal-ion coordination (i.e., it is bidentate), but the remaining four ligands are monodentate. Thus, substitution of Glu155 in EF4 by a glycine residue, with no oxygen-containing side chain, effectively reduces the number of coordinating oxygen ligands from seven to five. As the results presented here reveal, this has a drastic effect on calcium binding.
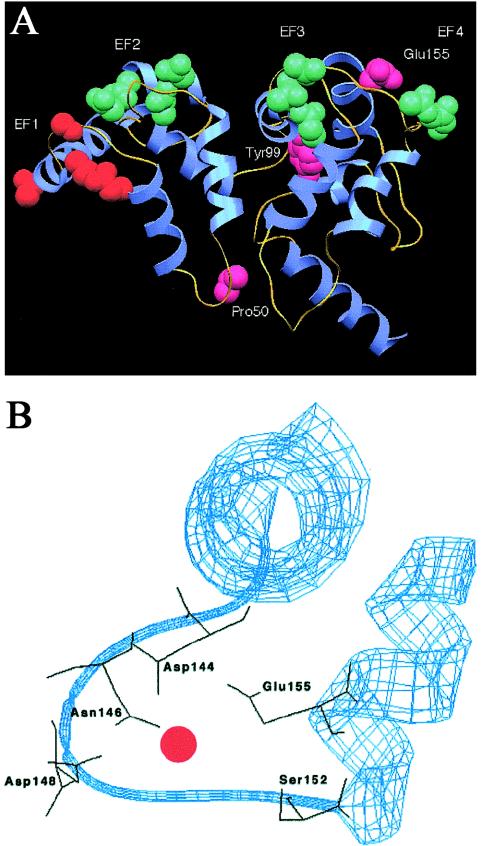
A pictorial representation of a model of GCAP1 is shown in figure 6A. This highlights the positions of the EF hands and the positions of the E155G mutation and the two previously reported mutations, Y99C and P50L. The enlargement of the EF4 domain (fig. 6B) shows the positions of the five amino acid side chains (D144, N146, D148, S152, and E155) that assist in calcium coordination. Note that there is another glutamate residue in the loop, E150, but this points away from the calcium ion and does not appear to contribute to metal binding.
Figure 6.
Model structure of GCAP1 showing the overall fold of the molecule. A, Complete structure, showing helices (blue) and loops (yellow). The key residues determining the calcium-binding ability of each EF hand are shown in space-filling mode and are labeled. The EF1 hand, which is predicted to be incapable of binding calcium, is shown in orange, to distinguish it from the calcium-binding EF hands 2–4 (green). Amino acid residues 50, 99, and 155, which correspond to the detected mutations in GCAP1, are shown in pink. The figure was produced using SETOR (Evans 1993). B, Enlargement of the EF4 domain, showing the polypeptide backbone in ribbon form (blue) and the five amino acid side chains, including E155, that assist in calcium coordination (black). The Ca2+ ion is shown as a red circle. The figure was produced using Swiss-Model (Peitsch 1996).
The E155G mutation, like the Y99C mutation, results in constitutive activation of RetGC1 at physiologically high [Ca2+]free, although the severity of the effect is somewhat greater than that produced by Y99C. Consistent with this observation, the onset of disease is earliest in patients with the E155G mutation. The E155G mutation also results in a reduced apparent affinity of the protein for RetGC1, an effect not observed with the Y99C mutant. Both these effects have been shown to persist in the presence of wild-type GCAP1 and thus act in a dominant manner. The consequence, in the cell, of the reduced apparent affinity for RetGC1 is difficult to assess, since the prevailing concentrations of both GCAP1 and RetGC1 are unknown. On the other hand, the constitutive activation of RetGC1 would be expected to result in a persistently elevated level of cGMP in the cell, above that required to keep the cGMP-gated cation channels open, resulting in maintenance of high [Ca2+]free. The mechanism of the ensuing photoreceptor degeneration is unclear, but it is known that persistently elevated calcium levels tend to disrupt the membrane potential of the mitochondrial outer membrane, leading to release of cytochrome c, with subsequent caspase activation and apoptosis (Green and Reed 1998). Previous studies have also linked elevated cGMP levels, arising from mutations in the phosphodiesterase gene, to cell death (Pittler and Baehr 1991; Farber et al. 1992).
Why mutations in GCAP1 result in selective cone degeneration with preservation of rod function is unclear. Cytohistochemical studies have indicated that the concentrations of both RetGC1 and GCAP1 are higher in cone photoreceptors than in rod photoreceptors (Liu et al. 1994; Kachi et al. 1999). Thus, a mutation in GCAP1 that affects the cyclase activity might be expected to cause a more serious imbalance in cones than in rods. However, mutations in the GUCY2D gene, encoding RetGC1, which also result in constitutive RetGC1 activity, have been linked to cone-rod dystrophy (Kelsell et al. 1998; Perrault et al. 1998; Weigell-Weber et al. 2000; Wilkie et al. 2000). It is therefore difficult to explain why severe mutations in GCAP1 with similar functional consequences do not also affect the rods. One difference in the effects of the mutations is that the cone-rod mutations in RetGC1 lead to an increased apparent affinity for GCAP1, but the E155G mutation in GCAP1 leads to a decrease in this property. An increased apparent affinity of the cyclase for GCAP1 will tend to exacerbate the failure to abolish cyclase activation at high levels of [Ca2+]free, but a reduced apparent affinity should compensate for it. In this way, the overall severity of the effect of the E155G mutation may be lessened in comparison with the RetGC1 mutations, with the sparing of the rods.
Rudnicka-Nawrot et al. (1998) have examined the effects of inactivation of individual EF hands on the function of bovine GCAP1 in vitro. This was accomplished through the substitution of glutamate with aspartate (conserving the negative charge) at position 12 of the three functional EF motifs in GCAP1, thus altering the affinity of the domains for Ca2+ without disrupting their structure. Rudnicka-Nawrot et al. demonstrated that only EF3 and EF4 contribute to the Ca2+-dependent inhibition of RetGC1 by GCAP1. The mutants with a single substitution, E111D or E155D, resulted in ∼50% inhibition of cyclase activity at a [Ca2+]free of 500 mM, but double mutants (E111D and E155D) and triple mutants (E75D, E111D, and E155D) activated cGMP synthesis in a manner completely independent of [Ca2+]free. In contrast, the results presented here demonstrate that the nonconservative substitution of the position 12 glutamate by the small neutral residue glycine has a more drastic effect on calcium binding, with the consequence that a single mutation in EF4 results in <25% inhibition of the cyclase activity at a calcium concentration of 500 μM.
Finally, an understanding of the functional effects of GCAP1 mutations on signaling within the photoreceptor is important, not only for the elucidation of the pathophysiology of these cone dystrophies but also, in view of the sequence similarity and conserved structural domains of all NCBPs, for the study of the pathogenesis of retinal dystrophies arising from mutations in other intracellular Ca2+- and cGMP-mediated signaling systems.
Acknowledgments
S.E.W., E.C.D., and R.J.N. are supported by Wellcome Trust grant 003303. Y.L. is a Helen Keller Fellow. K.Z. is supported by National Institutes of Health grant EY00401, the Heed Ophthalmic Foundation, and the Grant Ritter Fund. The authors are indebted to the members of the family for their help with this study. We thank Dr. Neal Peachey, for his help with the ERG analysis, and Drs. Peachey and John Crabb, for critical review of the manuscript.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for autosomal dominant retinal cone dystrophy, COD3, and GCAP1 [OMIM 180020, 602093, and 600364])
- Swiss-Model, http://www.expasy.ch/swissmod/SWISS-MODEL.html (for protein modeling)
References
- Balciuniene J, Johanssen K, Sandgren O, Wachmeister L, Homgren G, Forsman G (1995) A gene for autosomal dominant progressive cone dystrophy (CORD5) maps to chromosome 17p12-p13. Genomics 30:281–286 [DOI] [PubMed] [Google Scholar]
- Baylor D (1996) How photons start vision. Proc Natl Acad Sci USA 93:560–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca N, Lopez S, Howes K, Kolb H (1998) The localization of guanylyl cyclase-activating proteins in the mammalian retina. Invest Ophthalmol Vis Sci 39:1243–1250 [PubMed] [Google Scholar]
- Dizhoor AM, Boikov SG, Olshevskaya EV (1998) Constitutive activation of photoreceptor guanylate cyclase by Y99C mutant of GCAP-1. J Biol Chem 273:17311–17314 [DOI] [PubMed] [Google Scholar]
- Dizhoor AM, Hurley JB (1999) Regulation of photoreceptor membrane guanylate cyclases by guanylate cyclase activator proteins. Methods 19:521–531 [DOI] [PubMed] [Google Scholar]
- Dizhoor AM, Lower DG, Olshevskaya EV, Laura RP, Hurley JB (1994) The human photoreceptor membrane guanylyl cyclase, RetGC, is present in outer segments and is regulated by calcium and a soluble activator. Neuron 12:1345–1352 [DOI] [PubMed] [Google Scholar]
- Dizhoor AM, Olshevskaya EV, Henzel WJ, Wong SC, Stults JT, Ankoudinova I, Hurley JB (1995) Cloning, sequencing, and expression of a 24-kDa Ca2+-binding protein activating photoreceptor guanylyl cyclase. J Biol Chem 270:25200–25206 [DOI] [PubMed] [Google Scholar]
- Downes SM, Holder GE, Fitzke FW, Payne AM, Warren MJ, Bhattacharya SS, Bird AC (2001) Autosomal dominant cone and cone-rod dystrophy with mutations in the guanylate cyclase activator 1A gene–encoding guanylate cyclase activator protein-1. Arch Ophthalmol 119:96–105 [PubMed] [Google Scholar]
- Duda T, Goraczniak R, Surgucheva I, Rudnicka-Nawrot M, Gorczyca WA, Palczewski K, Sitaramayya A, Baehr W, Sharma RK (1996) Calcium modulation of bovine photoreceptors guanylate cyclase. Biochemistry 35:8478–8482 [DOI] [PubMed] [Google Scholar]
- Evans SV (1993) SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J Mol Graph 11:134–138 [DOI] [PubMed] [Google Scholar]
- Farber DB, Danciger JS, Aguirre G (1992) The beta subunit of cyclic GMP phosphodiesterase mRNA is deficient in canine rod-cone dysplasia 1. Neuron 9:349–356 [DOI] [PubMed] [Google Scholar]
- Frins S, Bonigk W, Muller F, Kellner R, Koch KW (1996) Functional characterization of a guanylyl cyclase-activating protein from vertebrate rods. J Biol Chem 271:8022–8027 [DOI] [PubMed] [Google Scholar]
- Garbers DL, Lowe DG (1994) Guanylyl cyclase receptors. J Biol Chem 269:30741–30744 [PubMed] [Google Scholar]
- Goraczniak RM, Duda T, Sitaramayya A, Sharma RK (1994) Structural and functional characterization of the rod outer segment membrane guanylate cyclase. Biochem J 302:455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorczyca WA, Gray-Keller MP, Detwiler PB, Palczewski K (1994) Purification and physiological evaluation of a guanylate cyclase activating protein from retinal rods. Proc Natl Acad Sci USA 91:4014–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorczyca WA, Polans AS, Surgucheva IG, Subbaraya I, Baehr W, Palczewski K (1995) Guanylyl cyclase activating protein: a calcium-sensitive regulator of phototransduction. J Biol Chem 270:22029–22036 [DOI] [PubMed] [Google Scholar]
- Green DR, Reed JC (1998) Mitochondria and apoptosis. Science 281:1309–1312 [DOI] [PubMed] [Google Scholar]
- Haeseleer F, Sokal I, Li N, Pettenati M, Rao N, Bronson D, Wechter R (1999) Molecular characterization of a third member of the guanylyl cyclase-activating protein subfamily. J Biol Chem 274:6526–6535 [DOI] [PubMed] [Google Scholar]
- Hayashi F, Yamazaki A (1991) Polymorphism in purified guanylate cyclase from vertebrate rod photoreceptors. Proc Natl Acad Sci USA 88:4746–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes K, Bronson JD, Dang YL, Li N, Zhang K, Ruiz C, Helekar B, Lee M, Subbaraya I, Kolb H, Chen J, Baehr W (1998) Gene array and expression of mouse retina guanylate cyclase activating proteins 1 and 2. Invest Ophthalmol Vis Sci 39:867–875 [PubMed] [Google Scholar]
- Jacobsen DM, Thompson HS, Bartley JA (1989) X-linked progressive cone dystrophy: clinical characteristics of affected males and carrier females. Ophthalmology 96:885–895 [DOI] [PubMed] [Google Scholar]
- Kachi S, Nishizawa Y, Olshevskaya E, Yamazaki A, Hiyake Y, Wakabayashi T, Dizhoor A, Usukura J (1999) Detailed localisation of photoreceptor guanylate cyclase activating protein-1 and -2 in mammalian retinas using light and electron microscopy. Exp Eye Res 68:465–473 [DOI] [PubMed] [Google Scholar]
- Kelsell RE, Gregory-Evans K, Payne AM, Perrault I, Kaplan J, Yang R-B, Garbers DL, Bird AC, Moore AT, Hunt DM (1998) Mutations in the retinal guanylate cyclase (RETGC-1) gene in dominant cone-rod dystrophy. Hum Mol Genet 7:1179–1184 [DOI] [PubMed] [Google Scholar]
- Koch KW, Stryer L (1988) Highly cooperative feedback control of retinal rod guanylate cyclase by calcium ions. Nature 334:64–66 [DOI] [PubMed] [Google Scholar]
- Koutalos Y, Yau KW (1996) Regulation of sensitivity in vertebrate rod photoreceptors by calcium. Trends Neurosci 19:73–81 [DOI] [PubMed] [Google Scholar]
- Kretsinger RH (1976) Calcium-binding proteins. Annu Rev Biochem 45:239–266 [DOI] [PubMed] [Google Scholar]
- Liu X, Seno K, Nishizawa Y, Hayashi F, Yamazaki A, Matsumoto H, Wakabayashi T, Usukura J (1994) Ultrastructural localisation of retinal guanylate cyclase in human and monkey retinas. Exp Eye Res 59:761–768 [DOI] [PubMed] [Google Scholar]
- Lowe DG, Dizhoor AM, Liu K, Gu Q, Spencer M, Laura R, Lu L, Hurley JB (1995) Cloning and expression of a second photoreceptor-specific membrane retina guanylyl cyclase (RetGC), RetGC-2. Proc Natl Acad Sci USA 92:5535–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold RJ, Deery EC, Walker CE Wilkie SE, Srinivasan N, Hunt DM, Bhattacharya SS, Warren MJ (2001) The destabilisation of human GCAP1 by a proline to leucine mutation might cause cone-rod dystrophy. Hum Mol Genet 10:47–54 [DOI] [PubMed] [Google Scholar]
- Nockolds CE, Kretsinger RH, Coffee CJ, Bradshaw RA (1972) Structure of a calcium-binding carp myogen. Proc Natl Acad Sci USA 69:581–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T (1989) Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA 86:2766–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto-Bruc A, Fariss RN, Haeseleer F, Hunag J, Buczylko J, Surgucheva I, Baehr W, Milam AH, Palczewski K (1997) Localization of guanylate cyclase-activating protein 2 in mammalian retinas. Proc Natl Acad Sci USA 94:4727–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Subbaraya I, Gorczyca WA, Helekar BS, Ruiz CC, Ohguro H, Huang J, Zhao X, Crabb JW, Johnson RS (1994) Molecular cloning and characterization of retinal photoreceptor guanylyl cyclase-activating protein. Neuron 13:395–404 [DOI] [PubMed] [Google Scholar]
- Payne AM, Downes SM, Bessant DAR, Taylor R, Holder GE, Warren MJ, Bird A, Bhattacharya SS (1998) A mutation in guanylate cyclase activator 1A (GUCA1A) in an autosomal dominant cone dystrophy pedigree mapping to a new locus on chromosome 6p21.1. Hum Mol Genet 7:273–277 [DOI] [PubMed] [Google Scholar]
- Peachey NS, Alexander KR, Fishman GA, Derlacki DJ (1989) Properties of the human cone system electroretinogram during light adaptation. Appl Optics 28:1145–1150 [DOI] [PubMed] [Google Scholar]
- Peitsch MC (1996) ProMod and Swiss-Model: internet-based tools for automated comparative protein modelling. Biochem Soc Trans 24:274–279 [DOI] [PubMed] [Google Scholar]
- Perrault I, Rozet J-M, Gerber S, Kelsell RE, Souied E, Cabot A, Hunt DM, Munnich A, Kaplan J (1998) A retGC-1 mutation in autosomal dominant cone-rod dystrophy. Am J Hum Genet 63:651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittler SJ, Baehr W (1991) Identification of a nonsense mutation in the rod photoreceptor cGMP phosphodiesterase beta-subunit gene of the rd mouse. Proc Natl Acad Sci USA 88:8322–8326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polans A, Baehr W, Palczewski K (1996) Turned on by Ca2+! The physiology and pathology of Ca2+-binding proteins in the retina. Trends Neurosci 19:547–554 [DOI] [PubMed] [Google Scholar]
- Pugh EN, Duda T, Sitaramayya A, Sharma RK (1997) Photoreceptor guanylate cyclases: a review. Biosci Rep 17:429–473 [DOI] [PubMed] [Google Scholar]
- Rudnicka-Nawrot M, Surgucheva I, Hulmes JD, Haeseleer F, Sokal I, Crabb JW, Baehr W, Palczewski K. (1998) Changes in biological activity and folding of guanylate cyclase-activating protein 1 as a function of calcium. Biochemistry 37:248–257 [DOI] [PubMed] [Google Scholar]
- Sampath AP, Matthew HR, Cornwall MC, Fain GL (1998) Bleached pigment produces a maintained decrease in outer segment Ca2+ in salamander rods. J Gen Physiol 111:53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyjan AW, de Sauvage FJ, Gillett NA, Goeddel DV, Lowe DG (1992) Molecular cloning of a retina-specific membrane guanylyl cyclase. Neuron 9:727–737 [DOI] [PubMed] [Google Scholar]
- Sittaramayya A, Duda T, Sharma RK (1995) Regulation of bovine rod outer segment membrane guanylate cyclase by ATP, phosphodiesterase and metal ions. Mol Cell Biochem 148:139–175 [DOI] [PubMed] [Google Scholar]
- Small KW, Syrquin M, Mullen L, Gehrs K (1996) Mapping of autosomal dominant cone degeneration to chromosome 17p. Am J Ophthalmol 121:13–18 [DOI] [PubMed] [Google Scholar]
- Sokal I, Li N, Surgucheva I, Warren MJ, Payne AM, Bhattacharya SS, Baehr W, Palczewski (1998) GCAP1(Y99C) mutant is constitutively active in autosomal dominant cone dystrophy. Mol Cell 2:1–20 [DOI] [PubMed] [Google Scholar]
- Strynadka NCJ, James MNG (1989) Crystal structures of the helix-loop-helix calcium-binding proteins. Ann Rev Biochem 58:951–998 [DOI] [PubMed] [Google Scholar]
- Subbaraya I, Ruiz CC, Helekar BS, Zhao X, Gorczyca WA, Pettenati MJ, Rao PN, Palczewski K, Baehr W (1994) Molecular characterization of human and mouse photoreceptor guanylate cyclase-activating protein (GCAP) and chromosomal localization of the human gene. J Biol Chem 269:31080–31089 [PubMed] [Google Scholar]
- Surguchov A, Bronson JD, Banerjee P, Knowles JA, Ruiz C, Subbaraya I, Palczewski K, Baehr W (1997) The human GCAP1 and GCAP2 genes are arranged in a tail-to-tail array on the short arm of chromosome 6 (p21.1). Genomics 39:312–322 [DOI] [PubMed] [Google Scholar]
- Tsien R, Pozzan T (1989) Measurement of cytosolic free Ca2+ with quin2. Methods Enzymol 172:230–262 [DOI] [PubMed] [Google Scholar]
- Weigell-Weber M, Fokstuen S, Torok B, Niemeyer G, Schinzel A, Hergersberg M (2000) Codons 837 and 838 in the retinal guanylate cyclase gene on chromosome 17p: hot spots for mutations in autosomal dominant cone-rod dystrophy. Arch Ophthalmol 118:300 [DOI] [PubMed] [Google Scholar]
- Wilkie SE, Newbold RJ, Deery E, Walker CE, Stinton I, Ramamurthy V, Hurley JB, Bhattacharya SS, Warren MJ, Hunt DM (2000) Functional characterisation of missense mutations at codon 838 in retinal guanylate cyclase correlates with disease severity in patients with autosomal dominant cone-rod dystrophy. Hum Mol Genet 9:3065–3073 [DOI] [PubMed] [Google Scholar]
- Yang RB, Foster DC, Garbers DL, Fulle HJ (1995) Two membrane forms of guanylyl cyclase found in the eye. Proc Natl Acad Sci USA 92:602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]