Abstract
The major causes of fragile X syndrome are mutational expansion of the CGG repeat in the FMR1 gene, hypermethylation, and transcriptional silencing. Most fragile X embryos develop somatic mosaicism of disease-causing “full” expansions of different lengths. Homogeneity of the mosaic patterns among multiple tissues in the same individual indicates that these previously unstable expansions acquire mitotic stability early in fetal life. Since mitotic stability is found strictly associated with hypermethylation in adult tissues, current theory has fixed the time of instability to developmental stages when fully expanded CGG repeats exist in an unmethylated state. We used murine embryocarcinoma (EC) cells (PC13) as a model system of pluripotent embryonic cells. Hypermethylated and unmethylated full expansions on human fragile X chromosomes were transferred from murine A9 hybrids into EC cells, by means of microcell fusion. As demonstrated in the present study for the first time, even full expansion alleles that were fully methylated and stable in the donors’ fibroblasts and in A9 became demethylated, reactivated, and destabilized in undifferentiated EC hybrids. When destabilized expansions were reintroduced from EC cells into A9, instability was reversed to stability. Our results strongly support the idea that fully expanded alleles are initially unstable and unmethylated in the human embryo and gain stability upon genetic or epigenetic change of the embryonic cells.
Introduction
Fragile X syndrome [MIM 309550] is a paradigm of a human genetic disease caused by expansion of an unstable trinucleotide repeat; it is a frequent cause of inherited mental retardation and the first disorder identified to result from this previously unknown type of “dynamic” mutation (Imbert et al. 1998; Jin and Warren 2000). The unstable DNA sequence of tandemly repeated CGGs is situated in the 5′ untranslated region of the fragile X mental retardation gene (FMR1 [GenBank accession number XM_10288]; Verkerk et al. 1991; Yu et al. 1991). Normal alleles usually contain 29 or 30 trinucleotides. Repeat sizes of 60–200 trinucleotides characterize “premutation” alleles, which are carried by unaffected males and females transmitting the trait. Large disease-causing expansions occur only upon transmission by a carrier mother (Fu et al. 1991; Imbert et al. 1998). These “full-mutation” alleles include ⩾220 CGGs, but expansion size may reach 1,000 triplets. Full expansion usually coincides with excessive de novo hypermethylation spreading particularly over the large CGG repeat (Oberlé et al. 1991; Hornstra et al. 1993) and the FMR1 promoter region (Drouin et al. 1997; Schwemmle et al. 1997). Fully expanded alleles, very exceptionally, remain unmethylated, for an unknown reason. On the basis of psychometric testing, males carrying such “unmethylated full mutations” are classified as “high functioning” (Hagerman 1998); some of them may even be physically and intellectually unaffected (Wöhrle et al. 1998).
The hypermethylated state of the fragile X full mutation is found associated locally with histone deacetylation and chromatin remodeling (Coffee et al. 1999), with late DNA replication, and with transcriptional silence of the gene (Imbert et al. 1998). The latter results in loss of the FMR1 protein product, FMRP, thereby obviously causing the fragile X phenotype.
In all somatic human cells that have been studied so far, hypermethylation also coincides with mitotic stability of the repeat (Steinbach et al. 1998), despite the fact that repeat size is far beyond the threshold at which all types of triplet repeats become prone to experience large changes in their triplet numbers upon each single round of DNA replication (McMurray 1999). At least three lines of evidence substantiate an absolute association between hypermethylation and repeat stability in fragile X patients:
-
1.
Patterns of hypermethylated expansions show substantial homogeneity of repeat sizes among different tissues of fetuses with fragile X (Devys et al. 1992; Wöhrle et al. 1992) and among MZ twins with fragile X who shared circulation at embryogenesis (Devys et al. 1992). Unmethylated triplet-repeat expansions, on the other hand, were found to show marked size heterogeneity among different tissues of individuals with fragile X (Taylor et al. 1999) and of individuals with myotonic dystrophy (Wöhrle et al. 1995).
-
2.
Cloning by dilution plating of fibroblasts from patients with fragile X who present a mosaic pattern of different methylated full expansions results in the isolation of “discrete-length” alleles, thereby showing that the length of methylated expansions is stably maintained in a clonal fashion (Wöhrle et al. 1993). When similar experiments were performed with fibroblasts from a male with fragile X who had unmethylated full expansions, the clonal cell DNA samples showed multiple allele sizes with smearing of expansion patterns, a pattern that is very consistent with repeat instability during clonal cell proliferation (Gläser et al. 1999).
-
3.
When populations of cells with methylated full mutations of different allele sizes were grown to high numbers of population doublings, the pattern of expansions did not change significantly (Wöhrle et al. 1993, 1995); however, extensive changes in heterogeneous expansion patterns resulted upon continual proliferation of cells with unmethylated fragile X full-mutation alleles (Gläser et al. 1999; Salat et al. 2000). Unmethylated CTG repeat expansions in the DMPK gene of myotonic dystrophy experienced significant further expansion, which occurred with ongoing cell proliferation in vitro (Wöhrle et al. 1995).
An absolute association between mitotic stability and the hypermethylated state of the expanded allele has been found in differentiated somatic cells of fetuses and adults with fragile X, but whether this association exists in any other type of cells remains to be elucidated. Two cell types are of particular interest: somatic cell hybrids and undifferentiated embryonic cells.
Somatic cell hybrids, constructed by fusion between human fibroblasts and immortalized rodent cells, have been studied elsewhere, but the findings have been contradictory. When the human donor fibroblasts carried fully methylated full-mutation alleles, the majority of the resulting hybrid clones were reported to harbor a fully methylated and mitotically stable allele (Steinbach et al. 1998); however, examples of fully methylated but mitotically unstable fragile X expansions have also been reported (Burman et al. 1999). When unmethylated and mitotically unstable full-mutation alleles were introduced into the background of murine host cell lines, some—but not all—of the hybrid clones harbored a single unmethylated expansion that retained its unstable mitotic behavior (Gläser et al. 1999). Given these contradictory results, we decided to study the behavior of expanded fragile X repeats in murine host cells in more detail.
Undifferentiated cells of embryonic origin represent a model system for investigation of the behavior of fragile X expansions during early embryonic development. Somatic mosaicism is found in the vast majority of full-mutation patients (Rousseau et al. 1994), in contrast to the mitotic stability of methylated expansions and the homogeneity in expansion size among different tissues of fetuses with fragile X (Devys et al. 1992; Wöhrle et al. 1992). Thus, there is strong, albeit indirect, evidence that expanded alleles are mitotically unstable at a very early developmental stage, during mitotic proliferation of embryonic cells that are undifferentiated, if not pluripotent. One outstanding question is whether, in these early embryonic cells, the unstable expansions are demethylated and become stabilized at embryonic cell differentiation and de novo methylation. In the present study, we investigate human fragile X–mutation alleles transferred into mouse embryocarcinoma (EC) cells PC13 (Hooper and Slack 1977), by means of microcell-mediated chromosome transfer (Fournier 1981). This enabled us to actually demonstrate, for the first time, the demethylation and reactivation of previously hypermethylated full-mutation alleles and, in particular, their destabilization in the background of embryonic cells.
Material and Methods
Cell Lines and Cell Culture
Human fibroblast cultures were established from skin biopsies from a young boy (ML) with fragile X, who was carrying a full mutation that was fully methylated, and an apparently unaffected male (GZ) with fragile X, who presented pre- and full-mutation alleles that were fully unmethylated, with the exception of a few cells with a methylated allele. Details of the latter case have been reported elsewhere (Wöhrle et al. 1998; Salat et al. 2000). The fibroblasts were maintained in Dulbecco’s modified Eagle’s medium (DMEM), following standard protocols.
The mouse EC cell line PC13TG8, a hypoxanthine phosphoribosyltransferase (HPRT)–deficient derivative of PC13, was a generous gift from M. L. Hooper of the University of Edinburgh and has been characterized in detail by Hooper and Slack (1977). In spite of their capacity for unlimited growth and their malignant properties in an abnormal environment, PC13 are embryonic cells shown to retain their developmental capacities after many generations of growth in vitro, behaving in a manner indistinguishable from that of normal embryonic cells when placed into a blastocyst (Hooper and Slack 1977). PC13 and PC13TG8 show the same pattern of predominant neural differentiation.
The EC cells were cultured on gelatinized plastic surfaces (0.2% gelatin, Sigma) without feeders and were maintained in DMEM supplemented with nonessential amino acids (GIBCO), 2 mM glutamine, 15% FCS, 0.1 mM monothioglycerol, 100 U/l penicillin, 100 μg/l streptomycin, and recombinant leukemia inhibitory factor in concentrations appropriate for the inhibition of spontaneous differentiation of embryonic stem cells. This medium is referred to as “ES-cell medium.”
Whole-Cell Fusion and Microcell-Mediated Chromosome Transfer
Human fragile X chromosomes were transferred to HPRT-deficient mouse A9 cells—which are known to make good microcell donors—by polyethylene glycol (PEG)–mediated fusion with human fibroblasts as described elsewhere (Gläser et al. 1999). Hybrid cells were grown in DMEM plus 0.1 mM hypoxanthine, 0.4 μM aminopterine, and 0.016 mM thymidine (HAT, Sigma) plus 0.05 mM Ouabain (Sigma). Clones were isolated 12–18 d after initiating selection, expanded in HAT-containing medium, harvested for DNA analysis, and cryopreserved in DMEM plus 10% dimethylsulfoxide (Sigma). Subclones were generated from parental hybrid clones by means of dilution plating, and were selected, expanded, and harvested for DNA analysis and cryopreservation.
Microcell-mediated transfer of fragile X chromosomes from whole-cell A9 hybrid clones into PC13TG8 recipient cells was performed essentially as described by Fournier (1981). In brief, donor cells were split onto bullet-shaped plastic slides, allowed to recover for 24 h in HAT-containing medium, and treated with 0.05 μg/ml colcemid (Sigma) for 48 h. Micronucleation was performed using 10 μg/ml cytochalasin B (Sigma). Microcells were attached to recipient cells by 100 μg/ml phytohemagglutinin (PHA-P, Sigma) and were fused with 44% weight/volume PEG 1500 (Boehringer). Hybrid cells were allowed to recover overnight and were then split 1:3 into HAT-containing ES-cell medium. Clones were isolated after 14–21 d and were independently analyzed. Human FMR1 alleles harbored in hybrid cells were detected by PCR amplification of exon 4, as described elsewhere (Eichler et al. 1993).
Microcell-mediated transfer of fragile X chromosomes from PC13 hybrids into A9 recipient cells was performed essentially as described above. Hybrid cells were treated with 0.05-μg/ml colcemid for 24–48 h. Cells were then transferred to Con-A–treated bullets (Islam and Islam 2000) and were allowed to attach for 90 min before micronucleation. To identify hybrids of PC13 and A9 that contained chromosomes of both murine parents, the murine markers Mt3pA+B (chromosome 8) and GA1+2 (chromosome 12) were typed using PCR primers provided by the Jackson Laboratory.
DNA Analysis
Genomic DNA was isolated from cultured cells by salt extraction (Miller et al. 1988). Aliquots (20 μg) were cleaved with restriction endonucleases EcoRI plus EagI or PstI, size-separated by electrophoresis through 0.8% agarose gels, blotted on a positively charged nylon membrane, and hybridized to α[32P]-dCTP oligolabeled probes Ox1.9 or Ox0.55, as described elsewhere (Wöhrle et al. 1993). Expansion size was measured as the CGG-repeat index, calculated by dividing the difference in size (in bp) between the normal and mutant bands by 3 and adding 30, which is the most common CGG-repeat number of normal alleles in the German population.
Sodium Bisulfite Conversion and Analysis of FMR1-Promoter Methylation
Bisulfite conversion, PCR, and DNA sequencing were performed as described elsewhere (Salat et al. 2000). Nested PCR was performed with primer pair 1F/1R, followed by M13-2F/M13-2R, designed to amplify bisulfite-converted DNA (upper strand) of the FMR1 promoter (Schwemmle et al. 1997). The primer sequences were as follows: 5′-TGA GTG TAT TTT TGT AGA AAT GGG C/T G-3′ (1F), 5′-CTC AAA AAC A/G AC CCT CCA CC A/G-3′ (1R), 5′-GGT AAC GCC AGG GGT TTC CGG TTT TC/TG C/TGA GGT AGT GTG ACT AAA ACC-3′ (M13-2F), 5′-GAA ACA GCT ATG ACC ATG A/G AA ACT AAA C A/G C CTA ACT AAA ACC-3′ (M13-2R). Products of the second PCR were purified through an agarose gel, followed by gel extraction through use of a QIAquick gel extraction kit (Qiagen) and sequencing on an ABI 377 Prism (Applied Biosystems) using fluorescent sequencing technology. For each experiment, all examined non-CpG cytosine residues were detected as T residues, indicating complete bisulfite conversion of the analyzed DNA fragments.
RT-PCR
Total RNA, isolated from cultured cells by means of the RNeasy mini kit (Qiagen), was treated with DNAse and reverse transcribed through use of the GeneAmp RNA-PCR Kit (Perkin-Elmer), following the manufacturers instructions. PCR primers were 5′-TTG AAT TTT CAT TTT ACA G-3′ (HuSp1) and 5′-CCT TGG TTA ATT ATC TAC A-3′ (HuSp2) for the human FMR1 allele, and 5′-AGA TGT TTT CAG CTA CTT G-3′ (MuSp1) and 5′-AAA AAA ACC CCA CAA AAA T-3′ (MuSp2) for the murine allele. These primer pairs bind to the 3′ untranslated region and were designed by R. Bauchwitz of Columbia University. Species-specific amplification was confirmed on numerous occasions, including in the present study.
Results
Mitotic Stability in A9 Host Cells
To investigate the behavior of full-mutation alleles in mouse host cells, human fragile X chromosomes bearing methylated or unmethylated full expansions were introduced into mouse A9 cells by means of whole-cell fusion with primary human fibroblasts. Results are illustrated in figure 1 and summarized in table 1.
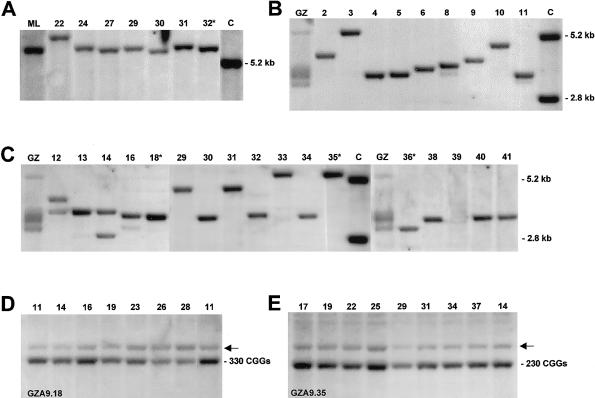
Figure 1.
Southern analysis of fragile X expansions harbored in murine A9 host cells. DNA samples isolated from fibroblasts of donors (ML and GZ) and from A9 hybrid clones were cleaved by restriction enzyme and hybridized to probe Ox1.9. A, Discrete-length alleles of methylated full expansions were isolated into different hybrid clones. DNA samples were cleaved with EcoRI plus EagI. The results for the donor’s (ML) fibroblasts and for six hybrid clones are shown with the clone numbers indicated. Because of methylation of the EagI site in the FMR1 promoter, the expanded fragments are larger than the normal 5.2-kb fragment seen in the control (lane C). Clone 32* was selected for microcell generation. B and C, Unmethylated expansions isolated into A9 hybrid clones (numbers shown above the lanes) and visualized by hybridization to EcoRI-plus-EagI–cleaved DNA. Cleavage of unmethylated EagI sites of expanded alleles results in fragments between 2.8 and 5.2 kb. Unmethylated 2.8-kb and methylated 5.2-kb-fragments are carried on the active and inactive X chromosomes of the female control (lane C). Whereas the donor’s (GZ) fibroblasts presented smears of multiple expansions, the majority of hybrids harbored a single expanded fragment with unmethylated EagI site. Clones 18*, 35*, and 36* were used in further experiments. D and E, Mitotic stability of expansions isolated from GZ into A9 clones 18 and 35 (GZA9.18, GZA9.35). Clonal DNAs were isolated at successive passages and cleaved with HindIII. The passage numbers are given above the lanes in D and E; population doublings are three to four times the passage number. The HindIII fragments of the murine gene, indicated by arrows (←), are seen in D and E above the human fragments carrying 330 and 230 CGGs, respectively.
Table 1.
Results from Southern Analysis of FMR1-Positive A9 Hybrid Clones[Note]
|
No. of Expansions |
||||
| Donor | No. of Clones | Methylation Status ofEagI Site | Single | Extra |
| ML | 34 | Methylated | 34 | 0 |
| GZ | 5 | Methylated | 4 | 1 |
| GZ | 34 | Unmethylated | 26 | 8 |
| Total | 73 | 64 | 9 | |
Note.— The majority of hybrid clones contained single mitotically stable expansions, regardless of EagI-site methylation status; however, clones with extra expansions were significantly more frequent among those harboring unmethylated FMR1 alleles (χ2=7.378, P<.0066).
The donor of methylated full expansions was an affected male with fragile X (ML). His fibroblasts showed fully methylated expansions and were completely negative for FMRP upon western analysis (Salat et al. 2000). After fusion with A9 recipient cells, 34 hybrid clones were isolated. Southern analysis of EcoRI-plus-EagI cleaved clonal DNA revealed the presence of fully expanded human FMR1 alleles in all hybrids, seen as a sharp hybridization signal of a single expanded fragment, with methylation of the EagI site maintained (fig. 1A).
The donor of unmethylated full expansions was an apparently unaffected male with fragile X (GZ). As reported elsewhere (Salat et al. 2000), these expanded alleles were mitotically unstable in proliferating fibroblasts. The primary culture used for this fusion experiment showed multiple repeat expansions. Most of them had >300 CGGs, were unmethylated, and gave a smeared pattern upon Southern analysis (fig. 1B, C). Fragments with ∼230 CGGs, with a minority of them methylated at the EagI site, were also detected. After fusion with A9 cells, discrete-length alleles were found separated into different hybrid clones. According to Southern analysis, 34 of 39 clones harbored unmethylated alleles of different lengths, and 5 clones harbored a methylated fragment with ∼230 CGGs (fig. 1B, C).
As summarized in table 1, 64 (88%) of 73 A9 hybrid clones obtained from ML and GZ donor fibroblasts gave one sharp expansion signal, indicating mitotic repeat stability independent of the EagI sites’ methylation status. Among the unmethylated full expansions from GZ fibroblasts, 26 (76%) of 34 still gave a single sharp signal in the clonal DNAs. To demonstrate mitotic stability directly, two hybrid clones derived from the cells of GZ were analyzed at successive passages. Clone GZA9.18 (fig. 1D) harbored an unmethylated expansion of ∼320 CGGs, and clone GZA9.35 (fig. 1E) contained a methylated 230-CGG fragment. Southern analysis gave no evidence of any change in repeat size through >50 further doublings of both clonal cell populations.
Demethylation and Destabilization in Undifferentiated PC13 EC Cells
To investigate the behavior of expanded fragile X repeats in undifferentiated embryonic cells, we used mouse EC cells derived from cell line PC13. Fragile X chromosomes bearing methylated and unmethylated full fragile X expansions were introduced by microcell fusion. Microcells were generated from previously established A9 hybrid clones harboring expanded FMR1 repeats that were mitotically stable. Fusion to recipient EC cells resulted in a number of hybrid clones (table 2), with a proportion of them harboring a human FMR1 allele, as revealed by PCR analysis. The EC hybrids retained their undifferentiated morphology (fig. 2A, B) that changed when differentiation was induced by retinoic acid (fig. 2C, D). The hybrid cells then aggregated to form embryoid bodies and subsequently acquired the morphology of epitheloid and neuron-like cells.
Table 2.
Hybrid Clones of A9 and PC13 Cells Harboring Human FMR1 Genes with Expanded CGG Repeats[Note]
| A9 |
EC |
|||
| Donor | Clone | Methylation Status (Harbored Alleles) | Clone | Methylation Status (Harbored Alleles) |
| GZ | GZA9.18 | Unmethylated (330 CGGs) | GZEC21.4 | Unmethylated (200, 240 CGGs, blurred) |
| GZEC21.1 | Unmethylated (ND) | |||
| GZ | GZA9.36 | Unmethylated (215 CGGs) | GZEC25.2 | Unmethylated (ND) |
| GZ | GZA9.35 | Methylated* (230 CGGs) | GZEC24.16 | Demethylated (230 CGGs) |
| ML | MLA9.32 | Methylated (530 CGGs) | MLEC30.1 | Demethylated (110–570 CGGs, smear) |
| MLEC30.2 | Demethylated (165 CGGs, smear) | |||
Note.— Methylation status established upon sequencing of bisulfite-converted genomic DNA. Methylated = fully methylated (i.e., all examined promoter CpGs methylated); Methylated* = partially methylated (i.e., all but three promoter CpGs methylated); ND = repeat size not detected, and no signal obtained on Southern analysis.
Figure 2.
Morphology and cytogenetics of undifferentiated and differentiated EC hybrids of PC13 harboring human fragile X alleles. A, Undifferentiated morphology of hybrid MLEC30.1. B, Undifferentiated morphology of a subclone isolated from MLEC30.1. C and D, Differentiated morphology of hybrid MLEC30.1. Upon treatment with 1 μM retinoic acid, the cells aggregated to form embryoid bodies (EB) and, subsequently, epitheloid (E) and neuron-like (N) cells. Bars = 100 μm. E and F, Mouse chromosomes harboring a fragment translocated from a human fragile X chromosome, indicated by arrows (→), in MLEC30.1 (E) and GZEC25.2 (F). Metaphase chromosomes were prepared from EC hybrids and analyzed by whole-chromosome painting of the human X chromosome with digoxigenin-labeled P5222-DG.5 (Oncor Appligene).
Cytogenetic analysis was performed by whole-chromosome painting of the human X chromosome on hybrid-cell metaphases. Mouse chromosomes were identified to carry a painted fragment of the human X—apparently encompassing the HPRT and FMR1 loci—translocated to the mouse chromosome during different events of interspecific chromosome rearrangement (fig. 2E, F). During proliferation in selection medium, most clones were prone to deletion of the human FMR1 gene while retaining the active human HPRT gene. The introduced human FMR1 gene was retained in only one hybrid—MLEC30.1—after continual cell proliferation.
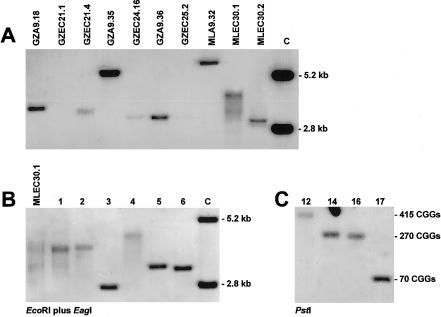
Fragile X repeat expansions in EC hybrid clones were analyzed on Southern blots of EcoRI-plus-EagI–cleaved DNA, and methylation of the promoter region was further examined by sequencing of bisulfite-converted clonal DNA. Results are illustrated in figure 3A and summarized in table 2. Fusion of microcells from GZA9.18, harboring an unmethylated FMR1 allele with 330 CGGs, resulted in two PC13 hybrid clones (GZEC21.1 and GZEC21.4), which were shown to be FMR1-positive by PCR. Although no fragile X expansions were visualized on Southern analysis of clone 21.1, careful inspection of X-rays revealed a bipartite signal of two unmethylated expansions (200 and 240 CGGs) in clone 21.4 (fig. 3A). This finding suggested mitotic instability and contraction of the transferred 330 CGG repeats to smaller alleles. Fusion of microcells from GZA9.35 harboring an expanded allele with 230 CGGs and a methylated EagI site resulted in EC clone GZEC24.16 presenting an expanded FMR1 fragment of similar size, but with an unmethylated EagI site. GZA9.36-derived microcells gave a single EC clone (GZEC25.2) with a positive PCR result but with no expansion signal detectable on Southern analysis.
Figure 3.
Southern analysis of EC hybrids and their parental A9 clones. A, Lanes grouped such that each A9 clone is followed by its derivative EC clone(s). Clonal DNAs were cleaved by EcoRI plus EagI. Fragments between 2.8 and 5.2 kb (see control lane C) represent unmethylated alleles. Demethylation of previously methylated EagI sites is evident in GZEC24.16, MLEC30.1, and MLEC30.2. Repeat sizes of all clones are given in table 2. B and C, MLEC30.1, subcloned by dilution plating, and subclonal DNAs, analyzed on EcoRI-plus-EagI (B) and PstI blots (C). Results for 10 subclones are indicated by numbers above the lanes. Blurred and smeared signals consistent with mitotic instability of subcloned expansions occurred as a function of repeat size.
Transfer of the mitotically stable methylated 530-CGG full-mutation allele of MLA9.32 into PC13 cells resulted in FMR1-positive clones MLEC30.1 and MLEC30.2 (table 2). Destabilization of the expanded repeat and demethylation of the EagI site was evident from Southern analysis of both clones (fig. 3A). Clone 30.1 harbored multiple expansions, with the demethylated EagI site giving a smear of hybridization signals of 110–570 CGGs. Most expanded fragments carried repeats of 530±35 triplets. Clone 30.2 presented a smear of unmethylated expansions with a major signal at 165 CGGs.
Mitotic instability of expanded CGG repeats harbored in EC hybrid clones was confirmed by subcloning of MLEC30.1 (fig. 3B). Subclones retaining the human gene showed no methylation of the EagI site. Half of them gave hybridization signals of single expansions, with repeat sizes of 30–285. The signals of the normal and premutation-sized alleles (30–230 CGGs) were notably sharp and intense, whereas those in the lower full-mutation range (240–285 CGGs) had a diffuse appearance on a PstI blot (fig. 3C). All the other subclones demonstrated more-diffuse single-expansion signals or broader smears of multiple expansions, with repeat sizes ranging from large full mutations (430–650) down to the premutation and normal sizes. This result clearly indicates repeat instability occurring in the EC background, as a function of repeat size.
The methylation status of the FMR1-promoter region, comprising the EagI site and a total of 29 CpG dinucleotides clustered on a genomic fragment of 270 bp, was also assessed by sequencing of bisulfite-converted genomic DNA isolated from A9 and EC hybrid clones (for details, see table 2). In striking contrast to the fully methylated status of MLA9.32, the derivative EC clones MLEC30.1 and MLEC30.2 had the FMR1 promoter sequences fully demethylated.
Reactivation of Previously Inactive Full-Mutation Alleles
EC and A9 hybrids were tested, by RT-PCR, for expression of human and mouse FMR1 mRNA (fig. 4). Whereas murine FMR1 was expressed in all hybrids, expression of the human alleles was correlated with the methylation pattern of the FMR1 promoter.
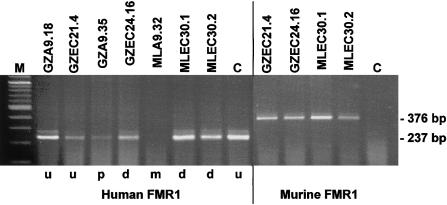
Figure 4.
Expression of human and murine FMR1 genes in EC hybrids of PC13 and their parental A9 clones. In the indicated cell lines and in human control fibroblasts (lane C), expression of the human allele was visualized by human-specific RT-PCR (237-bp products). GZA9.18, GZEC21.4, and the control cells carry unmethylated human FMR1 alleles (u). GZA9.35 and MLA9.32 harbor partially methylated (p) and fully methylated (m) alleles that are demethylated (d) in GZEC24.16, which is derived from GZA9.35, and in MLEC30.1 and MLEC30.2, which are derived from GZA9.35. Expression of the endogenous FMR1 alleles was shown by mouse-specific RT-PCR (376-bp product). The reduced intensities of the human RT-PCR signals of GZEC21.4 and GZEC24.16 correspond to the low proportions of cells that retained the human allele in these clones (see fig. 3A).
The unmethylated full-mutation allele of hybrid GZA9.18 was transcribed and was also found to be transcriptionally active in EC clone GZEC21.4. The partially methylated 230-CGG FMR1 allele in GZA9.35 was almost completely inactive. When introduced into clone GZEC24.16, where the allele was found to be demethylated, it produced a much higher level of transcripts. Hybrid MLA9.32 harbored a methylated full-mutation allele with no transcripts detected by RT-PCR analysis. When transferred to clones MLEC30.1 and MLEC30.2, these alleles were demethylated and were clearly transcribed in both EC hybrids. These findings indicate stable transcriptional reactivation of previously inactive fragile X full-mutation alleles associated with demethylation in the background of undifferentiated embryonic cells.
Regained Stabilization of Previously Destabilized Expansions
To elucidate whether the destabilization of full fragile X expansions that occurred in undifferentiated EC cells could be reversed to stability in the background of differentiated cells, expansions were retransferred from clone MLEC30.1 to HPRT-deficient A9 cells, by means of microcell fusion. When the microcells were generated, the EC hybrid clone harbored multiple human FMR1 alleles with repeat sizes extending from ∼550 CGGs continuously down into the normal size range (fig. 5).
Figure 5.
Morphology and Southern analysis of A9 hybrids harboring demethylated human fragile X expansions that were retransferred from EC cells. Microcells were generated from MLEC30.1 and fused to A9 cells. A, Morphology of a hybrid clone (16*). Bar = 100 μm. B, Isolated clones harboring chromosomes of both murine parents and a human FMR1 gene, analyzed on EcoRI-plus-EagI blots hybridized to Ox1.9. The results for nine clones, indicated by numbers, are shown. Notably sharp and intense signals of full-mutation fragments with unmethylated EagI sites were obtained in many clones; for example, 16* (315 CGGs), 22 (265 CGGs), and 24 (280 CGGs). C, One of the A9 hybrids (16*), subcloned to further prove mitotic stability of the expansion. The unmethylated full-mutation allele could be amplified by cloning (lanes 1–4) and did not experience any significant change of repeat size upon clonal expansion.
After microcell fusion, A9 clones (n=58) were isolated, expanded, and tested, by PCR and Southern analysis. PCR tests of clonal DNAs identified 21 true hybrids containing chromosomes of both murine parents, PC13 and A9, and the human FMR1 gene. The morphology of these hybrids was typical of A9 cells and was significantly different from undifferentiated EC cells (fig. 5A)
Human FMR1 alleles were detected, by means of Southern analysis, in each clone with the EagI sites still unmethylated (fig. 5B). Discrete bands of normal and premutation alleles were found in a few clones. The majority harbored full-mutation alleles with repeat sizes of 250–415. Smears of full expansions consistent with repeat instability were detected in a few exceptional clones. Most clones presented with discrete fragments of full expansions (250–415 CGGs), giving hybridization signals strongly suggesting mitotic stability that was, notably, independent of repeat size. Subclones isolated from one of the A9 hybrids (clone 16) with a single full expansion (315 CGGs) again harbored single expansions of the same size (fig. 5C), thereby confirming that expansions previously destabilized in the EC hybrid regained stability in the A9 background.
Discussion
The majority of full-mutation fragile X patients present extensive somatic mosaicism, with homogeneity of expansion sizes among different somatic tissues (Devys et al. 1992; Wöhrle et al. 1992; Rousseau et al. 1994). Whereas the homogeneity of expansion patterns is a characteristic feature of mitotic stability of the fully methylated full-mutation alleles (Wöhrle et al. 1993, 1995), somatic mosaicism is assumed to result from unstable behavior of these expansions during a very early developmental window, before they become stabilized in their individual pattern by yet-unknown mechanisms (Steinbach et al. 1998). Fully expanded fragile X alleles have been shown to exist in oocytes of carrier females (Malter et al. 1997). Therefore, affected males with fragile X may inherit a full-mutation allele that would probably be mitotically unstable in the undifferentiated embryonic cells. To elucidate the behavior of fragile X alleles at early embryonic cell proliferation, we introduced segments of human fragile X chromosomes into the background of PC13 EC cells, by means of microcell fusion. Using PC13 hybrids as a model system of undifferentiated embryonic cells, we directly demonstrated the mitotic instability of fragile X expansions.
Exciting Properties of the PC13 Model System
Our PC13-derived EC hybrids had the typical properties of undifferentiated embryonic cells. They retained the indefinite proliferation capacity and the undifferentiated morphology of the host cell line. Upon treatment with retinoic acid, the hybrid cells aggregated to form embryoid bodies and differentiated into epitheloid and neuronlike cells. This is the typical response of PC13 cells to retinoic acid upon induction of cell differentiation (Hooper and Slack 1977). Furthermore, the EC hybrid cells had the potency to fully demethylate previously methylated full-mutation fragile X alleles. Full demethylation of fragile X alleles has not been observed to occur on any other cellular background or in any other demethylation experiment. In our hybrids, demethylation of previously methylated full-mutation alleles involved the complete FMR1 promoter region and may be associated with alterations of chromatin structure (Coffee et al. 1999).
Fragile X demethylation in EC hybrids was associated with permanent transcriptional reactivation of previously inactive FMR1 genes. Demethylation and reactivation has previously been reported to occur upon treatment of dividing cells with de-azacytidine alone or in combination with trichostatin A (Chiurazzi et al. 1999; Coffee et al. 1999); however, in striking contrast to our hybrids, these drugs induced only partial demethylation and poor reactivation that was not stably maintained upon cell proliferation.
Destabilization of Full Expansions in the PC13 Model System
Independent of their original methylation status, all expanded alleles were mitotically stable in the A9 background before they were transferred into PC13 EC cells. In the EC cells, expansions of 330 and 530 CGGs were found to have changed to different sizes upon clonal cell proliferation. Mitotic instability of sufficiently large expansions was particularly evident from the smeared pattern of expansions obtained upon Southern analysis of clone MLEC30.1; this instability also was clearly confirmed upon subcloning and was shown to occur as a function of repeat size. Instability of expanded repeats upon subclonal cell proliferation was indicated by smeared or diffuse hybridization patterns obtained from all subclones that harbored expansions of >240 CGGs. Single expansions giving a sharp and intense hybridization band, indicating increased stability, were found only in subclones harboring shorter repeats.
These findings clearly demonstrate destabilization of previously stable fragile X full-mutation alleles, but the patterns of unstable products in the undifferentiated embryonic cells differed from previous expectations. Although a few products of expansions were identified, the most prominent feature of instability was size reduction of large expansions. Since the normal endogenous murine FMR1 alleles were expressed in all EC hybrids, repeat-size reductions most likely resulted from contraction events instead of from subclonal selection (Salat et al. 2000). As demonstrated by subcloning of MLEC30.1, contraction events can create small and apparently stabilized alleles, the repeat sizes of which may correspond to a premutation or even a normal allele.
When full expansions that were destabilized and demethylated in EC hybrids were retransferred to A9, expanded alleles remained unmethylated, but most of them—independent of expansion size—regained the previous phenotype of mitotic stability. Repeat destabilization in the PC13 system of undifferentiated embryonic cells, therefore, is most likely a process that can be reversed upon genetic or epigenetic change of the cellular background. It would be worthwhile to investigate the behavior of large unstable fragile X expansions in retinoic-acid–differentiated EC hybrids, after adaptation of the culture techniques to allow for proliferation of epithelial cells and neurons.
A previous article (Burman et al. 1999) reported that, independent of methylation, instability of large expanded repeats occurred only in differentiated cells of murine origin and that a high degree of stability resulted in hybrids of “H4D2” cells that were derived from EC cell line P19. In contrast to our results for PC13, there was no demethylation of previously fully methylated expansions and, very unusually, the H4D2 hybrids retained the potency of permanent proliferation when their morphology changed upon treatment with retinoic acid. With the A9 and PC13 systems, however, we clearly demonstrated the opposite—stabilization of previously unstable expansions in the differentiated murine A9 cells and destabilization of previously stable expansions in undifferentiated EC cells. We cannot offer an explanation for the discordant findings of Burman et al. (1999), which are inconsistent with current knowledge about fragile X syndrome.
Results Applied to the Human Fragile X Embryo
Our results are in accordance with the well-known situation in patients with full-mutation fragile X, in whom large expanded repeats are stably maintained in somatic tissues, if hypermethylated, and are supposed to be mitotically unstable in an early postzygotic—and, most probably, undifferentiated—stage (Imbert et al. 1998; Steinbach et al. 1998). This justifies application of the results obtained from our PC13 model system of undifferentiated cells of embryonic origin to a human embryo who started as a pre- or a full-mutation zygote. On the basis of our results, we can make some interesting predictions:
Expanded FMR1 alleles received from a carrier mother either will be unmethylated or will become demethylated in the undifferentiated cells of the early embryo. When these cells proliferate, the expanded alleles will be transcriptionally active and mitotically unstable, as has already been deduced from the high frequency of mosaic full-mutation patterns established in early fetal life (Taylor et al. 1999). The findings for PC13 EC cells also predict contraction—rather than expansion—to occur in the early developmental window of repeat instability. Therefore, postzygotic expansion of an inherited premutation to a disease-causing full mutation would be less likely to occur than contraction of fully expanded alleles that were generated prezygotically and inherited by the embryo through an oocyte.
The concept of prezygotic transition of pre- to full-mutation alleles, followed by frequent postzygotic contraction of the inherited, mitotically unstable repeats, is in accord with the high frequency of somatic mosaicism in fragile X patients (Rousseau et al. 1994) and with patterns of somatic mosaicism consisting of a major fragment accompanied by some larger and many shorter fragments (e.g., see the report by Wöhrle et al. [1995]). In particular, this concept is not at variance with the finding that the occurrence of mosaicism with pre- and full-mutation alleles is independent of the maternal premutation size (Moutou et al. 1997), whereas transition to the full mutation is strongly dependent on the size of the maternal premutation (Heitz et al. 1992; Imbert et al. 1998).
Role of Methylation in Repeat Stabilization
A role of DNA methylation in the stabilization of expanded CGG repeats has been proposed previously, in view of an absolute association of hypermethylation with stability of fully expanded repeats in human somatic cells (Steinbach et al. 1998; Burman et al. 1999). This association apparently also exists in undifferentiated embryonic cells of murine origin, since sufficiently large unmethylated full expansions were found to be mitotically unstable in our PC13 hybrids; however, most fully expanded fragile X repeats, when introduced from human fibroblasts or from a PC13 hybrid, gained mitotic stability in the background of differentiated murine A9 cells regardless of their methylation status. These findings indicate that methylation of large expanded CGG repeats—though contributing significantly to repeat stabilization—would not be an essential factor and that the primary determinants of stabilization remain to be identified.
Fragile X Mice
So far, all attempts to generate transgenic mice with an expanded FMR1 CGG repeat have failed (Bontekoe et al. 1997; Lavedan et al. 1998). Premutation-sized repeats that were introduced into the endogenous FMR1 gene of mouse embryonic stem cells by homologous recombination did not expand to a full mutation either by mitotic instability at stem cell proliferation or upon transmission from female carrier mice to offspring. As indicated by our results for EC cells, large expanded human CGG repeats are, indeed, mitotically unstable in the background of undifferentiated stem cells of murine origin; however, mitotic instability may not lead to full expansion of a premutation allele and may create only contraction products. The generation of full-mutation fragile X mice may require the introduction of substantially large human full-mutation alleles into embryonic stem cells. This will most probably not be achieved by a conventional replacement strategy.
Acknowledgments
We are grateful to Heidrun Lindenthal, Renate Weber, and Antje Kollak for excellent technical assistance. This project is supported by the Deutsche Forschungsgemeinschaft (STE330/7). We are also grateful to Martin L. Hooper, University of Edinburgh, Western General Hospital, Edinburgh, for giving us clone PC13TG8, which was developed and characterized in his lab, and to Robert Bauchwitz, Columbia University, Department of Genetics and Development, New York, for primer sequences.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human FMR1 mRNA sequence [accession number XM_010288])
- Jackson Laboratory, The, http://www.jax.org/ (for strain-specific marker alleles and PCR primers)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for fragile X syndrome [MIM 309550])
References
- Bontekoe CJ, de Graaff E, Nieuwenhuizen IM, Willemsen R, Oostra BA (1997) FMR1 premutation allele (CGG)81 is stable in mice. Eur J Hum Genet 5:293–298 [PubMed] [Google Scholar]
- Burman RW, Popovich BW, Jacky PB, Turker MS (1999) Fully expanded FMR1 CGG repeats exhibit a length- and differentiation-dependent instability in cell hybrids that is independent of DNA methylation. Hum Mol Genet 8:2293–2302 [DOI] [PubMed] [Google Scholar]
- Chiurazzi P, Pomponi MG, Pietrobono R, Bakker CE, Neri G, Oostra BA (1999) Synergistic effect of histone hyperacetylation and DNA demethylation in the reactivation of the FMR1 gene. Hum Mol Genet 8:2317–2323 [DOI] [PubMed] [Google Scholar]
- Coffee B, Zhang F, Warren ST, Reines D (1999) Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet 22:98–101 [DOI] [PubMed] [Google Scholar]
- Devys D, Biancalana V, Rousseau F, Boué J, Mandel JL, Oberlé I (1992) Analysis of full fragile X mutations in fetal tissues and monozygotic twins indicate that abnormal methylation and somatic heterogeneity are established early in development. Am J Med Genet 43:208–216 [DOI] [PubMed] [Google Scholar]
- Drouin R, Angers M, Dallaire N, Rose TM, Khandjian W, Rousseau F (1997) Structural and functional characterization of the human FMR1 promoter reveals similarities with the hnRNP-A2 promoter region. Hum Mol Genet 6:2051–2060 [DOI] [PubMed] [Google Scholar]
- Eichler EE, Richards S, Gibbs RA, Nelson DL (1993) Fine structure of the human FMR1 gene. Hum Mol Genet 2:1147–1153 [DOI] [PubMed] [Google Scholar]
- Fournier REK (1981) A general high-efficiency procedure for production of microcell hybrids. Proc Natl Acad Sci USA 78:6349–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YH, Kuhl DP, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJ, Holden JJ, Fenwick RG Jr, Warren ST, Oostra BA, Nelson DL Caskey CT (1991) Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 67:1047–1058 [DOI] [PubMed] [Google Scholar]
- Gläser D, Wöhrle D, Salat U, Vogel W, Steinbach P (1999) Mitotic behavior of expanded CGG repeats studied on cultured cells: further evidence for methylation-mediated triplet repeat stability in fragile X syndrome. Am J Med Genet 84:226–228 [PubMed] [Google Scholar]
- Hagerman R (1998) Clinical and diagnostic aspects of fragile X syndrome. In: Wells RD, Warren ST (eds) Genetic instabilities and hereditary neurological disorders. Academic Press, San Diego, pp 15–25 [Google Scholar]
- Heitz D, Devys D, Imbert G, Kretz C, Mandel JL (1992) Inheritance of the fragile X syndrome: size of the fragile X premutation is a major determinant of the transition to full mutation. J Med Genet 29:794–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper ML, Slack C (1977) Metabolic co-operation in HGPRT+ and HGPRT- embryonal carcinoma cells. Dev Biol 55:271–284 [DOI] [PubMed] [Google Scholar]
- Hornstra IK, Nelson DL, Warren ST, Yang TP (1993) High resolution methylation analysis of the FMR1 gene trinucleotide repeat region in fragile X syndrome. Hum Mol Genet 2:1659–1665 [DOI] [PubMed] [Google Scholar]
- Imbert G, Feng Y, Nelson D, Warren ST, Mandel JL (1998) FMR1 and mutations in fragile X syndrome: molecular biology, biochemistry, and genetics. In: Wells RD, Warren ST (eds) Genetic instabilities and hereditary neurological disorders. Academic Press, San Diego, pp 509–528 [Google Scholar]
- Islam MQ, Islam K (2000) Evidence for suppression of cellular growth in vitro and selection against the indigenous mouse X chromosome in A9 cell hybrids after microcell-mediated transfer of an X from other mammalian species. Cytogenet Cell Genet 88:110–113 [DOI] [PubMed] [Google Scholar]
- Jin P, Warren ST (2000) Understanding the molecular basis of fragile X syndrome. Hum Mol Genet 9: 901–908 [DOI] [PubMed] [Google Scholar]
- Lavedan C, Grabczyk E, Usdin K, Nussbaum RL (1998) Long uninterrupted CGG repeats within the first exon of the human FMR1 gene are not intrinsically unstable in transgenic mice. Genomics 50:229–240 [DOI] [PubMed] [Google Scholar]
- Malter HE, Iber JC, Willemsen R, de Graaff E, Tarleton JC, Leisti J, Warren ST, Oostra BA (1997) Characterization of the full fragile X syndrome mutation in fetal gametes. Nat Genet 15:165–169 [DOI] [PubMed] [Google Scholar]
- McMurray CT (1999) DNA secondary structure: a common and causative factor for expansion in human disease. Proc Natl Acad Sci USA 96:1823–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutou C, Vincent MC, Bincalana V, Mandel JL (1997) Transition from premutation to full mutation in fragile X syndrome is likely to be prezygotic. Hum Mol Genet 6:971–979 [DOI] [PubMed] [Google Scholar]
- Oberlé I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boué J, Bertheas MF, Mandel JL (1991) Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science 252:1097–1102 [DOI] [PubMed] [Google Scholar]
- Rousseau F, Heitz D, Tarleton J, MacPherson J, Malmgren H, Dahl N, Barnicoat A, Mathew C, Mornet E, Tejada I, Maddalena A, Spiegel R, Scinzel A, Marcos JAG, Schwartz C, Mandel JL (1994) A multicenter study on genotype-phenotype correlations in the fragile X syndrome, using direct diagnosis with probe StB12.3: the first 2,253 cases. Am J Hum Genet 55:225–237 [PMC free article] [PubMed] [Google Scholar]
- Salat U, Bardoni B, Wöhrle D, Steinbach, P (2000) Increase of FMRP expression, elevated levels of FMR1 mRNA, and clonal selection in proliferating cells with unmethylated fragile X repeat expansions: a clue to the sex bias in the transmission of full mutations? J Med Genet 37:842–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwemmle S, de Graaff E, Deissler H, Gläser D, Wöhrle D, Kennerknecht I, Just W, Oostra BA, Dörfler W, Vogel W, Steinbach P (1997) Characterization of FMR1 promoter elements by in vivo footprinting analysis. Am J Hum Genet 60:1354–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach P, Wöhrle D, Gläser D, Vogel W (1998) Systems for the study of triplet repeat instability: cultured mammalian cells. In: Wells RD, Warren ST (eds) Genetic instabilities and hereditary neurogical disorders. Academic Press, San Diego, pp 509–528 [Google Scholar]
- Taylor AK, Tassone F, Dyer PN, Hersch SM, Harris JB, Greenough WT, Hagerman RJ (1999) Tissue heterogeneity of the FMR1 mutation in a high-functioning male with fragile X syndrome. Am J Med Genet 84:233–239 [PubMed] [Google Scholar]
- Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DPA, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang F, Eussen BW, van Ommen GJB, Blonden H, Caskey CT, Nelson DL, Oostra BA, Warren ST (1991) Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65:905–914 [DOI] [PubMed] [Google Scholar]
- Wöhrle D, Hennig I, Vogel W, Steinbach P (1993) Mitotic stability of fragile X mutations in differentiated cells indicates early post-conceptional trinucleotide repeat expansion. Nat Genet 4:140–142 [DOI] [PubMed] [Google Scholar]
- Wöhrle D, Hirst MC, Kennerknecht I, Davies KE, Steinbach P (1992) Genotype mosaicism in fragile X fetal tissues. Hum Genet 89:114–116 [DOI] [PubMed] [Google Scholar]
- Wöhrle D, Kennerknecht I, Wolf M, Enders H, Schwemmle S, Steinbach P (1995) Heterogeneity of DM kinase repeat expansion in different fetal tissues and further expansion during cell proliferation in vitro: evidence for a casual involvement of methyl-directed DNA mismatch repair in triplet repeat stability. Hum Mol Genet 4:1147–1153 [DOI] [PubMed] [Google Scholar]
- Wöhrle D, Salat U, Gläser D, Mücke J, Meisel-Stosiek M, Schindler D, Vogel W, Steinbach P (1998) Unusual mutations in high functioning fragile X males: apparent instability of expanded unmethylated CGG repeats. J Med Genet 35:103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Pritchard M, Kremer E, Lynsch M, Nancarrow J, Baker E, Holman K, Mulley JC, Warren ST, Schlessinger D, Sutherland GR, Richards RI (1991) Fragile X genotype characterized by an unstable repeat region of DNA. Science 252:1179–1181 [DOI] [PubMed] [Google Scholar]