Abstract
Paget disease of bone is characterized by focal increases of the bone-remodeling process. It is the second most common metabolic bone disease after osteoporosis. Genetic factors play a major role in the etiology of Paget disease of bone, and two loci have been mapped for the disorder: PDB1 and PDB2. The gene(s) causing the typical form of the disorder remains to be characterized. To decipher the molecular basis of Paget disease of bone, we performed genetic linkage analysis in 24 large French Canadian families (479 individuals) in which the disorder was segregating as an autosomal dominant trait. After exclusion of PDB2, a genomewide scan was performed on the three most informative family nuclei. LOD scores >1.0 were observed at seven locations. The 24 families were then used to detect strong evidence for linkage to chromosome 5q35-qter. Under heterogeneity, a maximum LOD score of 8.58 was obtained at D5S2073, at θ=.1. The same characteristic haplotype was carried by all patients in eight families, suggesting a founder effect. A recombination event in a key family confined the disease region within a 6-cM interval between D5S469 and the telomere. The 16 other families, with very low conditional probability of linkage to 5q35-qter, were further used, to map a second locus at 5q31. Under heterogeneity, a maximum LOD score of 3.70 was detected at D5S500 with θ=.00. Recombination events refined the 5q31 region within 12.2 cM, between D5S642 and D5S1972. These observations demonstrate the mapping of two novel loci for Paget disease of bone and provide further evidence for genetic heterogeneity of this highly prevalent disorder. It is proposed that the 5q35-qter and 5q31 loci be named “PDB3” and “PDB4,” respectively.
Introduction
Paget disease of bone (MIM 167250) is a localized monostotic or polyostotic progressive metabolic bone disorder. The disease is characterized by an increased remodeling process in which abnormal bone resorption remains coupled to new osteoblastic bone formation. Paget disease of bone usually appears at >40 years of age (Klein and Norman 1995) and mainly affects the axial skeleton. Approximately 5% of pagetic patients present symptoms requiring treatment (Kanis 1998). The most frequent complaints are bone pain, enlargement, and deformities at the pagetic site (Kanis 1998). Other manifestations of the disease include increased susceptibility to fractures, deafness, and neurological complications, as well as an increased susceptibility to osteosarcomas (Hamdy 1995). In Western countries, Paget disease of bone is the second most common metabolic bone disorder after osteoporosis. In the United States, the disorder has an estimated frequency of 1%–3% in the population >40 years of age and of 8%–10% in those >80 years of age (Siris and Canfield 1990).
The primary defect of Paget disease seems to reside in the osteoclast, but, despite numerous studies, the pathological basis of the disorder remains unknown. Within the pagetic lesion, osteoclasts are large, multinucleated, and overactive. Pagetic cells also contain paramyxovirus-like inclusions (Mills and Singer 1976; Gherardi et al. 1980; Rebel et al. 1980b; Harvey et al. 1982; Howatson and Fornasier 1982). Detection of antigens and/or nucleic acid sequences of paramyxoviruses in symptomatic bone (Rebel et al. 1980a; Mills et al. 1984; Gordon et al. 1991; Reddy et al. 1995, 1996, 1999; Mee et al. 1998) have indicated that chronic viral infection may cause the disease (Harvey et al. 1982; Cartwright et al. 1993; Abe et al. 1995). Other studies have questioned this hypothesis, since they were unable to confirm these observations (Ralston et al. 1991; Birch et al. 1994; Helfrich et al. 2000; Ooi et al. 2000). Further experimentation is thus required in order to assess the involvement of paramyxoviral infection in the pathogenesis of Paget disease of bone.
On the other hand, there is compelling evidence that genetic factors play a major role in the etiology of Paget disease of bone. The disease is most common in western Europe (Detheridge et al. 1983), North America (Rosenbaum and Hanson 1969; Guyer and Chamberlain 1980), Australia (Barker 1984), and New Zealand (Reasbeck et al. 1983), with the highest prevalence occurring in the United Kingdom, particularly in Lancashire (prevalence >6.3%) (Barker et al. 1980). Familial risk for Paget disease of bone has been evaluated by several authors. Sofaer et al. observed a 10-fold–increased prevalence among the parents and siblings of patients, compared to spouses of patients (Sofaer et al. 1983). In the United States, Siris et al. further reported that 12% of pagetic patients had a first-degree relative affected with Paget disease of bone and calculated that first-degree relatives had a sevenfold-increased risk of developing the disease (Siris et al. 1991). In Spain, Mirales-Piga et al. observed that 40% of their index cases had at least one first-degree relative affected with Paget disease of bone (Morales-Piga et al. 1995). Familial clustering of Paget disease of bone also has been frequently documented (Sofaer et al. 1983; Siris et al. 1991; Morales-Piga et al. 1995; Haslam et al. 1998; Hocking et al. 2000). In the kindreds investigated thus far, Paget disease of bone has appeared to be transmitted with an autosomal dominant mode of inheritance with incomplete penetrance. Since the majority of patients with Paget disease of bone are asymptomatic, the incidence of a familial association is likely to be underreported. Even if multifactorial inheritance cannot be excluded, the late onset of the disease may account for the incomplete penetrance of the disorder in pedigrees with autosomal dominant inheritance.
Suggestive evidence was first reported for linkage between Paget disease of bone and the HLA locus at 6p (Fotino et al. 1977; Tilyard et al. 1982). This potential locus was named “PDB1” (MIM 167250). However, further studies did not confirm linkage to this site (Breanndan Moore and Hoffman 1988; Nance et al. 2000; Good et al. 2001), suggesting that the role of the HLA locus may be of minor importance in the etiology of Paget disease of bone.
A rare bone disorder, familial expansile osteolysis (FEO [MIM 174810]), which also displays bone lesions containing paramyxoviral-like nuclear inclusions, has been mapped to chromosome 18q21-q22 (Hughes et al. 1994). Using a candidate-locus approach and a large pagetic family, Cody et al. (1997) reported evidence for linkage between Paget disease of bone and the same 18q region, with a LOD score of 3.40 at D18S42. This locus was called “PDB2” (MIM 602080). This later study proposed that the gene(s) responsible for FEO and Paget disease of bone were either closely linked or allelic variants of the same mutant gene. Subsequently, Haslam et al. (1998) confirmed linkage to 18q in five pagetic families and observed genetic heterogeneity in three other kindreds. More recent studies confirmed genetic heterogeneity of the disorder and suggested that linkage of Paget at 18q21-q22 was relatively uncommon (Hocking et al. 2000; Nance et al. 2000; Good et al. 2001).
Recently, the FEO disease gene has been identified as the TNFRSF11A gene (MIM 603499) that encodes the receptor activator of nuclear factor–κB (RANK) (Hughes et al. 2000). The same heterozygotic insertion (84dup18) was detected in exon 1 of TNFRSF11A in three families with either FEO or FEO-related symptoms. One pedigree of Japanese origin that had atypical Paget disease of bone also carried a 27-bp insertion (75dup27) in the TNFRSF11A gene. Their uncommon symptoms included early onset and dental problems, suggesting that these patients may suffer from either a milder form of FEO or a particular early-onset form of Paget disease of bone (Leach et al. 2001). No RANK mutations have yet been reported for patients manifesting typical symptoms of Paget disease of bone (Hughes et al. 2000; Sparks et al. 2001).
To decipher the molecular basis of Paget disease of bone, we conducted genetic-linkage and haplotype studies in French Canadian families. Kindreds originating from this population are particularly well suited for such studies (Morissette et al. 1995, 1998). Indeed, for social and linguistic reasons, this population has maintained, for the past 3 centuries, a demographic growth without immigration and, until recently, a high birth rate with large sibships (i.e., 10–15 sibs/generation) (Bouchard and Braekeleer 1991). Pedigree reconstruction is also facilitated by this population's conservation of genealogical records, including birth, marriage, and death certificates that go back to the first immigrants. To counteract the genetic heterogeneity of Paget disease of bone, we exploited these attributes and undertook the systematic screening of large French Canadian families affected by the disorder. A total of 554 members of 24 families, each of which included at least two affected members, were recruited. We report here both the exclusion of the PDB2 locus as an important disease locus in these families and the mapping, at 5q35-qter and at 5q31, of two novel loci for Paget disease of bone. Since one very large family showed LOD scores <−2.00 at PDB2, 5q35-qter, and 5q31 and had <.01 conditional probability of being linked to these two novel loci, our data also suggest the presence of at least one additional locus for Paget disease of bone and confirm its genetic heterogeneity.
Patients, Material, and Methods
Clinical Investigation and Phenotypic Classification
This research has been approved by the Centre Hopsitalier de l'Université Laval (CHUL) Research Center Ethics Committee. All participants, affected or not, signed an informed-consent document before entering the study. Clinical assessment comprised complete bone evaluation, including measurement of total serum alkaline phosphatase, skull and pelvis X-rays (enlarged view including the last three lumbar vertebrae, the pelvis, and the femoral heads), and total-body bone scan. Up to 10% of adult and up to 20% of elderly patients with symptomatic Paget disease of bone have levels of total serum alkaline phosphatase that are within the laboratory reference range (Kanis 1998). The most common sites for involvement are the pelvis, lumbar spine, and proximal femur. Pelvis X-rays with enlarged view have thus been associated with a sensitivity of 85% (Kanis 1998) to 91% (Rénier et al. 1995) in detection of Paget disease of bone. As a general rule, scintigraphy is more sensitive than radiography, identifying 97%–98% of the pagetic lesions (Meunier et al. 1987).
Criteria for diagnosis of Paget disease of bone were abnormal-bone characteristics with monostotic or polyostotic increased bone uptake associated with typical radiographic lesions in the affected sites. Individuals >40 years of age were considered normal when they presented with normal bone scan, normal skull and pelvis X-rays, and normal levels of total serum alkaline phosphatase. Subjects <40 years of age who had normal levels of total serum alkaline phosphatase and/or normal bone scan/X-rays were considered to be of unknown status, as were all subjects for whom the data were insufficient to establish a diagnosis.
Genealogical Procedures
Pedigree genealogies were reconstituted, after permission has been obtained from the families, on the basis of registers compiled from Catholic parish records, which systematically list births, marriages, and deaths of 98% of the Québec population. Validations of the family trees were obtained through interviews with key family members. The Archives Nationales du Québec, the Québec and the Québec Civil Register, were also consulted. Most of the families came from the same region of the Province of Québec, ∼120 km from Québec City.
Venipunctures and DNA Extraction
Two blood samples (10 ml each) were obtained by venipuncture from consenting individuals. DNA was extracted from leukocytes by the guanidine hydrochloride–proteinase K method (Jeanpierre 1987). One additional 10-ml blood sample was drawn from selected subjects, to establish lymphoblastoid cell lines (Anderson and Gusella 1984).
Chemiluminescent Genotyping at the PDB2 Locus
A total of 11 microsatellite markers at the PDB2 locus, which were selected from the Généthon database (Dib et al. 1996), were analyzed. Four markers were tested within 19 families, whereas 11 microsatellites were genotyped in kindreds DD and TH. Genotyping at the PDB2 locus was done by a protocol similar to the procedure reported by Vignal et al. (Vignal et al. 1993; Morissette et al. 1995). In brief, PCR amplifications were performed in a 50-μl reaction mixture containing 100 ng of genomic DNA, 31.3 μM each dNTP, 10–50 pmol of each primer, 1 × PCR buffer (50 mM KCl, 10 mM Tris [pH 9.0], 1.5 mM MgCl2, 0.01% gelatine, and 0.1% Triton X-100), and 1 U of Taq polymerase (Perkin-Elmer Cetus). Samples were processed through 35 cycles of denaturation (95°C at 30 s), annealing (55°C at 30 s), and extension (72°C at 30 s). PCR products were diluted with 95% formamide, 0.1% xylene cyanol, and 10 mM NaOH and were resolved by electrophoresis on 6% polyacrylamide gels. Samples were then transferred onto Hybond N+ nylon membrane (Amersham), hybridized with a (CA)20 oligomer 3′-labeled with digoxigenin-11-ddUTP, and detected by chemiluminescence using the DIG system (Roche) with Kodak BioMax MR-1 film. Genotypes were scored relative to reference alleles of CEPH-family individual 134702. Inheritance of all alleles was verified by PedCheck (O'Connell and Weeks 1998). Genotyping was repeated on individuals presenting with allele-size incompatibilities.
Automated Fluorescent Genomewide Scan
After exclusion of the PDB2 locus, a genomewide scan was performed at the Centre National de Génotypage (CNG) (Evry, France). Automated multiplex genotyping was done by use of ABI PRISM Linkage Mapping Set version 2, comprising 400 microsatellite markers spaced at an average distance of 10 cM. Four primer sets were usually amplified together, and PCR reactions were performed as recommended by the manufacturer. PCR products were pooled and were separated on a 377ABI DNA sequencer. Twenty-one markers that gave inconsistent results were removed from the study, leaving 379 microsatellites for statistical analyses. Almost all markers were processed in combination panels; some of them were repeated singly. Raw genotypic data consisting of allele sizes, estimated by GENOTYPER (ABI), were transferred to the CHUL Research Center for data analysis.
Radioactive Genotyping for Linkage Confirmation and Disease-Locus Refinement
To confirm and to refine the regions that stood out in the genomewide scan, the three family nuclei selected for the scan were extended, and additional pedigrees were recruited, as described in the Results section (see below). These individuals were genotyped with up to 90 additional markers, by a standard radioactive protocol. In brief, markers were labelled by [35S]α-dATP incorporation in a 20-μl PCR reaction mixture containing 50 ng of genomic DNA; 1× PCR buffer; 0.2 mM each of dCTP, dTTP, and dGTP; 10 mM dATP; 4–20 pmol of each primer; and 1.5 μCi (1.5 pmol) [35S]α-dATP. PCR conditions were 35 cycles of 95°C at 30 s, 55–57°C at 30 s, and 72°C at 5–10 s. Samples were resolved by electrophoresis on 6% polyacrylamide gels. The gels were then exposed to X-ray film for 12–48 h.
Data Processing, Linkage Analysis, and Haplotype Studies
Genotypic data and family information were stored in a 4D database on a Macintosh G4 (Morissette 1992; Morissette et al. 1995). Raw genotypic data obtained from CNG were binned into odd or even values, according to reference alleles from individual 134702, by a data-import 4D script developed locally. Radioactive and fluorescent genotyping data were merged together, on the basis of reference alleles from individual 134702. Data were transferred from the 4D database to SUN computers by CAP AppleShare server software. Computations were made on either a SUN Enterprise Ultra 30 or a SUN Enterprise 450. Parametric two-point LOD scores were calculated by the MLINK program from the LINKAGE package (FASTLINK 4.1P) (Lathrop and Lalouel 1984; Schäffer et al. 1994), with a disease-allele frequency of .015 and an autosomal dominant model. On the basis of pedigree and clinical information, we defined five liability classes, with age-dependent penetrances of 0.1% (age <20 years), 10% (age 20–29 years), 10% (age 30–39 years), 50% (age 40–49 years), and 95% (age ⩾50 years) and with phenocopy probabilities of .001, .001, .005, .01, and .02, respectively. Multipoint parametric linkage analyses and haplotyping studies were done with SIMWALK 2.6 (Sobel and Lange 1996). Because allele frequencies of the ABI markers used in the genome scan had not been determined in the French Canadian population and because the number of individuals genotyped was too small to allow accurate estimation, allele frequencies were assumed to be equal for two-point linkage analyses in our genomewide scan. This procedure may lead to spurious results (Freimer et al. 1993). For locus confirmation and refinement, accurate allele frequencies were therefore estimated by ILINK (VITESSE 2.0) (O'Connell and Weeks 1995), on the basis of the genotypes obtained from all available pedigrees (Boehnke 1991). The calculation of LOD scores in the presence of heterogeneity was performed by the HOMOG program (Ott 1991).
Results
Phenotypic and Segregation Studies
We recruited 24 French Canadian families, each with at least two affected first-degree relatives, comprising a total of 554 individuals, including 56 spouses. These kindreds were of different sizes and complexities. Eight of them comprised 30–71 individuals, and 14 displayed Paget disease of bone in two generations. Twelve pedigrees came from the same geographic area.
Of these 554 individuals, 105 were diagnosed with Paget disease of bone, 303 were classified as normal, and 146 were classified as unknown, according to our classification procedure. Age at diagnosis was 29–89 years, with a mean age of 58 years. The majority (63/105 [60%]) of these patients were diagnosed at ⩾60 years of age, whereas only a small number (3/105 [2.9%]) were diagnosed at <40 years of age, confirming that the penetrance of the disease was highly age dependent. Sex distribution of the 105 patients showed an equal number of females (53) and males (52), and at least nine kindreds displayed male-to-male transmission, thus excluding X-linked dominant transmission. Among the 97 subjects >40 years of age who had one affected parent, the classifications were as follows: 35 with Paget disease of bone, 40 normal, and 22 unknown. Our observations therefore strongly suggest that, within the pedigrees that we studied, Paget disease of bone is segregating as an autosomal dominant trait with an incomplete age-dependent penetrance.
Table 1 lists the clinical characteristics of representative affected individuals selected for the genomewide scan. Four members in family LF were diagnosed with a benign autosomal dominant form of adult osteopetrosis; two of them showed concomitant Paget disease of bone.
Table 1.
Clinical Features of Representative Patients Selected for Genomewide Scan
|
Age(years) |
|||||||
| Patient | When Studied | At Diagnosis | Statusa | Total Serum Alkaline Phosphatase(×ULN)b | Radiological Features | No. of Bone Sites Affected | Total Pagetic Bonec(%) |
| DD-003 | 77 | … | N | Normal | Normal | 0 | 0 |
| DD-004 | 76 | 48 | + | 25.2 | Paget | 31 | 37 |
| DD-019 | 73 | 70 | + | .5 | Paget | 2 | 16 |
| DD-021 | 71 | 67 | + | .7 | Paget | 1 | 3 |
| DD-033 | 53 | 49 | + | .5 | Paget | 4 | 5 |
| TH-019 | 55 | 43 | + | 1.0 | Paget | 3 | 7 |
| TH-102 | 64 | 60 | + | 1.6 | Paget | 15 | 53 |
| TH-111 | 83 | 58 | + | 20.3 | Paget | 14 | 64 |
| TH-143 | 52 | 46 | + | 11.8 | Paget | 26 | 31 |
| LF-010 | 57 | 54 | + | 1.0 | Paget | 6 | 20 |
| LF-019 | 80 | … | N | Normal | Normal | 0 | 0 |
| LF-045 | 78 | 68 | + | 3.7 | Paget | 7 | 36 |
| LF-074 | 74 | 46 | + | 4.4 | Paget | 16 | 64 |
As described in the Patients, Material, and Methods section. N = normal; + = Paget disease of bone.
ULN = upper limit of normal range, as determined by each clinical laboratory.
Calculated by use of Rénier et al.'s (1995) index.
Exclusion of the PDB2 Locus at 18q21-q22
Linkage to the PDB2 locus was tested by genotyping six Généthon markers spanning 11.5 cM at chromosome 18q21-q22: AFM212xg5 (D18S64), AFMb330zc5 (D18S1134), AFMa052va9 (D18S1148), AFMa049ze5 (D18S1147), AFM248yb9 (D18S68), and AFM122xc1 (D18S55). At least four of these markers were assessed in each of the 19 largest pedigrees, in a total of 343 individuals. Table 2 shows that five of the microsatellites displayed LOD scores <−4.0 at recombination fraction (θ) .01. Only one of them, AFMa052va9 (D18S1148), gave a LOD score >−2.00 at all θ values tested. Two large pedigrees, DD and TH, were further genotyped with seven additional markers to cover a 50-cM interval at 18q: AFMa074yd9 (D18S1157), AFM295xh1 (D18S474), AFM057xb4 (D18S1127), AFMb317zc1 (D18S1129), AFM260yh1 (D18S465), AFM193yf8 (D18S61), and AFMa085yf1 (D18S1161). For nine markers, LOD scores <−1.00 were obtained at θ=.05, and only one marker, AFMA052va9 (D18S1148), showed an isolated positive LOD score, 0.41 at θ=.00 (data not shown). No characteristic haplotype segregating with the disease was identified in any of the 19 families. These data therefore excluded, in a major proportion of the population that we studied, the PDB2 region as a locus for Paget disease of bone.
Table 2.
Summed Two-Point LOD-Scores for Markers Spanning the PDB2 Locus at Chromosome 18q21-q22
|
LOD Score at θ = |
|||||||||
| Marker (Locus) | .0 | .01 | .05 | .1 | .2 | .3 | .4 | Distance(cM) | No. of Families |
| AFM212xg5 (D18S64) | −6.41 | −5.73 | −3.87 | −2.36 | −.73 | −.12 | .00 | 3.6 | 19 |
| AFMb330zc5 (D18S1134) | −5.27 | −4.74 | −3.28 | −2.13 | −.83 | −.24 | −.03 | 2.2 | 7 |
| AFMa052va9 (D18S1148) | −1.36 | −1.30 | −1.10 | −.82 | −.34 | −.08 | .00 | .0 | 14 |
| AFMa049ze5 (D18S1147) | −10.25 | −9.14 | −6.49 | −4.38 | −1.91 | −.64 | −.14 | 6.5 | 18 |
| AFM248yb9 (D18S68) | −7.54 | −6.41 | −3.96 | −2.34 | −.82 | −.28 | −.06 | .1 | 8 |
| AFM122xc1 (D18S55) | −4.61 | −4.22 | −3.15 | −2.20 | −.92 | −.26 | −.03 | … | 14 |
Genomewide Scan
To define the genetic basis of Paget disease of bone within the population that we studied, we performed a genomewide scan after selecting the three most informative family nuclei. As depicted in figures 1 and 2, these three family nuclei belonged to the much larger kindreds, DD, TH, and LF. They represented a total of 44 individuals, all >40 years of age. In these kindreds, 18 persons had been diagnosed with Paget disease of bone, 18 others were normal, and 5 were classified as unknown. Within the family-LF nucleus, two pagetic patients selected for our genome scan, LF-010 and LF-011, also displayed a benign adult form of osteopetrosis. These two individuals were considered to be affected. A third person, LF-070, with osteopetrosis only, was classified as unknown, since any association between Paget disease and osteopetrosis remains elusive. To estimate the expected average LOD score of our sample, we used SLINK. In case of homogeneity, if all three family nuclei were linked to the same locus, we estimated an average LOD score of 2.2 for a six-allele marker system at 5 cM of the disease gene. In case of heterogeneity, the expected average LOD score for each individual family would be 0.70 at θ=.05.
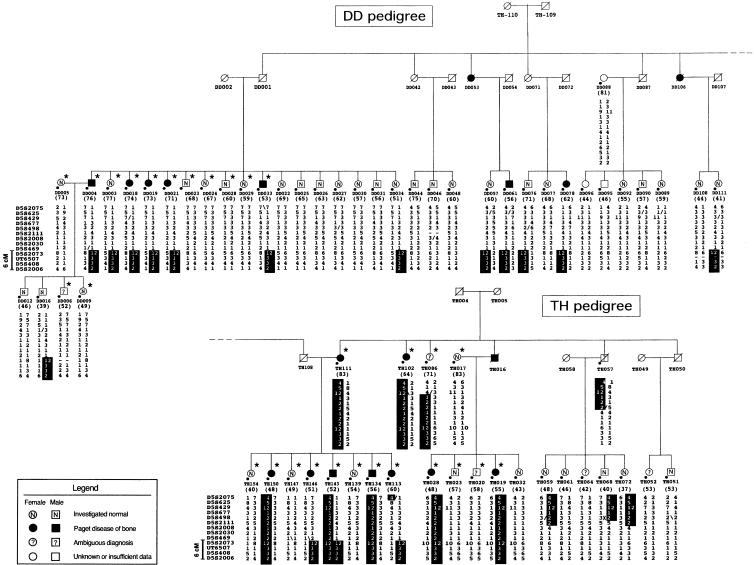
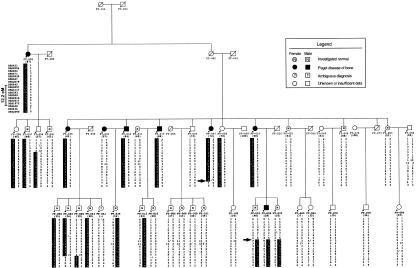
Figure 1.
Pedigree structure and haplotypes of the extended pagetic families DD and TH, showing linkage to chromosome 5q35-qter. Individuals selected for the genomewide scan are indicated by an asterisk (*). Circles and squares represent females and males, respectively; blackened symbols denote individuals affected by Paget disease of bone; empty white symbols denote individuals whose status is unknown or for whom the data were insufficient for phenotypic classification; an “N” within a white symbol denotes that the individual is normal; a question mark (?) within a white symbol denotes that the diagnosis is ambiguous; and a diagonal line through a symbol denotes that the individual is deceased. The number in parentheses below each symbol denotes the individual's age at the time of the study. “D5S” numbers for each marker genotyped are shown on the left of the third generation of each pedigree; refinement of the disease locus is shown on the left of these “D5S” numbers. The disease-associated haplotypes are denoted by the vertical black bars; within the haplotypes, maternal crossover is denoted by a slash (/), paternal crossover by a back slash (\).
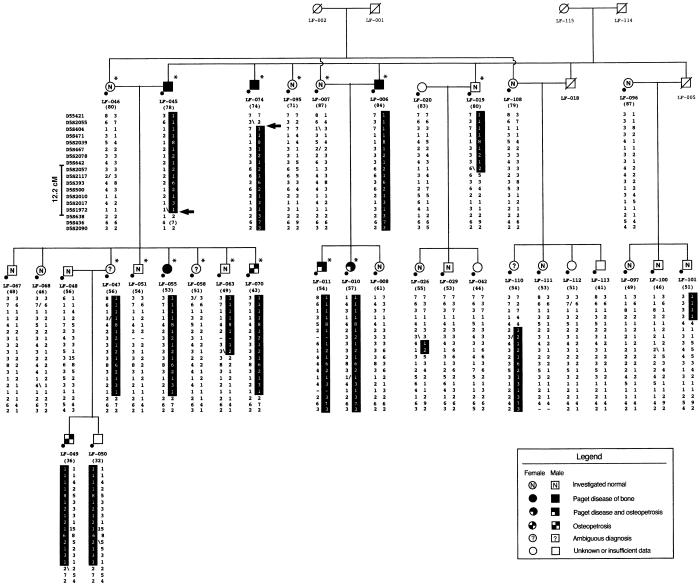
Figure 2.
Pedigree structure and haplotypes spanning the 5q31 region in extended family LF. Blackened symbols with a white lower-left corner denote individuals diagnosed with concomitant Paget disease of bone and benign osteopetrosis; tiled symbols denote individuals diagnosed with osteopetrosis only; arrows (→) indicate key recombinant events; all other symbols are as described in figure 1. A spontaneous mutation was detected in marker AFM263wg5 (D5S436) in individual LF-045 and is enclosed within parentheses.
Genotypes of 379 microsatellite markers covering the whole genome, at an average spacing of 11.5 cM, were analyzed in these 44 individuals. As depicted in figure 3, two-point LOD scores were plotted versus map distance, for all chromosomes. No evidence for linkage was obtained at the PDB1 locus, with LOD scores <0.26 for markers spanning the whole chromosome 6, suggesting that none of the three family nuclei had linkage to this locus.
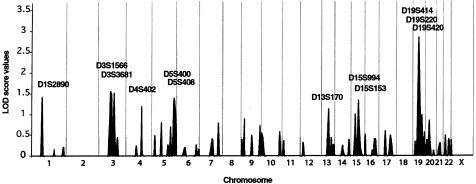
Figure 3.
Summary of genomewide scan for susceptibility loci for Paget disease of bone, in three French Canadian family nuclei. Two-point LOD scores are shown, and markers with LOD scores >1.0 are indicated. Gray vertical lines denote chromosome boundaries.
The maximum two-point LOD score detected by our genomewide scan was at chromosome 19q—a value of 2.79 for AFM295xg9 (D19S414) at θ=.00. Positive LOD scores were observed over a 54-cM interval between AFM295xg9 (D19S414) and AFM347ze1 (D19S571). To better assess these linkage results, all available members from the three extended families (139 individuals) were then genotyped with four microsatellites covering the interval of positive values; AFMa058xc5 (D19S931), AFM295xg9 (D19S414), AFM248zc1 (D19S225), and AFMa154xg1 (D19S868). After these procedures, the LOD score for AFM295xg9 (D19S414) decreased to 0.11 at θ=.00, for the three extended pedigrees (data not shown). When families were analyzed individually, only pedigree TH continued to have a positive LOD score, 0.96 at θ=.01 for AFM295xg9 (D19S414) (data not shown). However, haplotype studies within this kindred demonstrated that only seven of the nine pagetic subjects shared the same haplotype. To further test this region, we genotyped 19 additional families (343 individuals), using the same four markers. Summed LOD scores were <−2.00 at θ=.00, for all markers (data not shown). No haplotype was shared between the affected individuals within these kindreds. These investigations therefore did not support the presence, within the vicinity of AFM295xg9 (D19S414) in the French Canadian population, of a major locus for Paget disease of bone.
Mapping and Refinement of a PDB Locus at 5q35-qter
In addition to chromosome 19, LOD scores >1.0 were observed at chromosomes 1p, 3p, 4q, 5q, 13q, and 15q (fig. 3). To evaluate presence of linkage within these regions, the large extended pedigrees DD, TH, and LF were genotyped with additional markers covering these areas. Linkage and haplotype analyses led to exclusion of chromosomes 1p, 3p, and 15q (data not shown). As regards chromosome 5q, two additional markers—AFM238xe11 (D5S498) and AFM323yh1 (D5S2073)—were genotyped, in order to cover a 19-cM gap, present in the ABI version 2 panel, between AFM112yb6 (D5S400) and AFM164xb8 (D5S408). Interestingly, after examination of the distribution of the alleles, a common haplotype segregating with the disorder was detected in all affected members of families DD and TH. Genealogical studies further revealed that these two kindreds shared several founders. These observations led us to thoroughly test for linkage to chromosome 5q35.
To confirm linkage between 5q35 and Paget disease of bone, all available members of families DD and TH (i.e., 81 individuals) were genotyped for the region between AFM324yf9 (D5S2075) and AFMb005wf9 (D5S2006), with 13 markers spaced at an average distance of 1.5 cM. A maximum LOD score of 5.88 was observed at θ=.05, for AFM238xe11 (D5S498), in the extended families DD and TH (data not shown). All affected subjects shared one characteristic haplotype comprising four markers between AFM323yh1 (D5S2073) and AFMb005wf9 (D5S2006), a region that covered 6 cM (fig. 1). To fully assess this region, we then genotyped the 21 other families. Table 3 presents the combined linkage analysis of the 24 pedigrees, for five markers spanning this region. A maximum two-point LOD score of 7.98 was obtained, for AFM323yh1 (D5S2073) at θ=.16. Genotype analysis revealed that affected members of six additional families carried a haplotype identical to that identified between AFM238xe11 (D5S498) and AFMb005wf9 (D5S2006) in family TH (table 4). Therefore, of the 24 families, 8 harbored a unique disease haplotype between AFM137xf6 (D5S469) and AFMb005wf9 (D5S2006), and common ancestors were found in 5 of these 8 families. Penetrance rates of the disease were estimated in 101 carriers in these 8 kindreds. Of the 26 carriers <50 years of age, only 5 manifested Paget disease of bone, for a penetrance rate of 19%. Among the 28 carriers 50–59 years of age, 13 were diagnosed as affected, for a penetrance rate of 46%, whereas, among the 40 carriers ⩾60 years of age, 31 were affected, for a penetrance rate of 78%; in this last age group, 6 carriers >70 years of age were asymptomatic.
Table 3.
HOMOG Analysis for Markers Spanning the 5q35-qter Region in 24 French Canadian Families
|
Maximum LOD Score (θmax) |
||||
| Marker (Locus) | Homogeneity | Heterogeneity | α | χ2 |
| AFM238xe11 (D5S498) | 3.27 (.2) | 4.07 (.08) | .54 | 3.70a |
| AFMb307za9 (D5S2030) | .62 (.28) | 1.05 (.12) | .28 | 2.01 |
| AFM323yh1 (D5S2073) | 7.98 (.16) | 8.58 (.10) | .64 | 2.77 |
| AFM164xb8 (D5S408) | 3.82 (.20) | 4.66 (.00) | .35 | 3.86a |
| AFMb005wf9 (D5S2006) | 3.44 (.22) | 3.64 (.16) | .59 | .91 |
Significant evidence for heterogeneity, at P<.05.
Table 4.
Haplotypes of Eight Families Showing Linkage to the 5q35-qter Region
|
Haplotype of Familyb |
|||||||||
| Marker (Locus)a | Distance(cM) | TH* | NA* | MA* | GA* | BT | DA | LG | DD* |
| AFM324yf9 (D5S2075) | 2.1 | 4 | 4 | 2 | 5 | … | … | … | 1 |
| AFM210vf12 (D5S625) | 2.2 | 5 | 2 | 2 | 1 | … | … | … | 1 |
| AFM242xb10 (D5S429) | 3.3 | 12 |
2 |
1 |
4 |
… | … | … | 1 |
| AFM350xh1 (D5S677) | 2.3 | 1 | 1 | 1 | 1 | … | … | … | 3 |
| AFM238xe11 (D5S498) | 3.2 | 2 | 2 | 2 | 2 | 4 |
6 |
5 |
2 |
| AFMa083wc5 (D5S2111) | 1.9 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 4 |
| AFMb010wa5 (D5S2008) | 1.1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 |
| AFMb307za9 (D5S2030) | .7 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 |
| AFM137xf6 (D5S469) | 3.4c | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 |
| AFM323yh1 (D5S2073) | .8 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 |
| UT6507 | … | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| AFM164xb8 (D5S408) | 1.8 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| AFMb005wf9 (D5S2006) | … | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
“AFM” markers are in the order reported in the Généthon database (Dib et al. 1996); UT6507 is mapped relative to the chromosome 5 physical map of the Human Genome Project (National Center for Biotechnology Information).
Asterisks (*) indicate families that are known to share founders. The shared disease haplotype is boxed.
Estimated on the basis of all genotyped families, by ILINK (Schäffer et al. 1994).
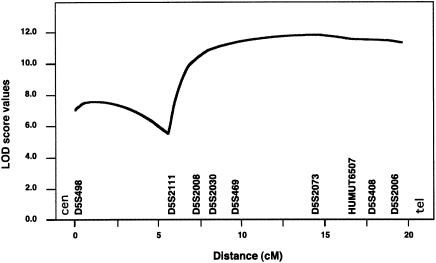
When calculated independently, LOD scores for kindred LF were between −2.54 and −4.38, for markers between AFM238xe11 (D5S498) and AFMb005wf9 (D5S2006), at 5q35. Furthermore, two other families also showed negative LOD scores, <−2.00, for these markers. These observations prompted us to test, at this locus within our set of 24 families, genetic heterogeneity of Paget disease of bone, using the HOMOG program. As depicted in table 3, significant evidence for heterogeneity (P<.05) was observed at AFM238xe11 (D5S498) (χ2=3.70) and AFM164xb8 (D5S408) (χ2=3.86). Indeed, when heterogeneity was allowed, a maximum two-point LOD score of 4.66 was obtained at θ=.00, for AFM164xb8 (D5S408), when only a fraction (α=.35) of the families were assumed to show linkage to the disease locus. Interestingly, this estimated α value of 35% was very similar to the ratio of families (8/24 [33.3%]) carrying the common disease haplotype. To better position the disease gene, multipoint parametric LOD scores were then calculated for the eight families that harbored the common haplotype, by use of the corresponding age-dependent penetrance rates, as determined above. This multipoint analysis showed a plateau with a LOD score of 11.5, between AFMb307za9 (D5S2030) and AFMb005wf9 (D5S2006), and demonstrated a maximum location score of 11.8 at AFM323yh1 (D5S2073) (fig. 4). The plateau covered a 6-cM interval that corresponded to the smallest region cosegregating with the Paget-disease-of-bone phenotype observed in family DD (fig. 1 and table 4).
Figure 4.
Multipoint parametric LOD scores at 5q35-qter in eight families showing linkage. The phenotypic model was modified according to penetrance values estimated on the basis of 101 individuals who carry the disease haplotype. Allele frequencies were estimated on the basis of all genotyped pedigrees, according to Boehnke's (1991) method. Genetic distances are from genetic maps reported elsewhere (Dib et al. 1996)—except for UT6507, which was mapped relative to other markers by use of Human Genome Project (National Center for Biotechnology Information) draft sequences.
Mapping and Refinement of a Second PDB Locus at 5q31
The disease haplotype shared at 5q35-qter by affected members of the 8 families was not found in the 16 other kindreds, including family LF. Moreover, the HOMOG test performed on marker AFM164xb8 (D5S408) estimated that the conditional probability that three of the families would show linkage to this region was <.01, suggesting the presence, within the population that we studied, of at least one additional disease-gene locus for Paget disease of bone (table 3). To identify this region, the genome-scan data were reanalyzed with use of only the results obtained with the family-LF nucleus. The highest LOD score obtained for the family-LF nucleus was 1.95 at θ=.00, for AFM263wg5 (D5S436). After additional members of the family were studied and additional markers were genotyped, maximum LOD scores of 2.98–2.94 were obtained at loci AFMa102zg1 (D5S2117), AFM042xd12 (D5S393), AFM240xg3 (D5S500), AFM203ye7 (D5S2010), and AFMb074xg1 (D5S2017), at θ=.00 (data not shown). As depicted in figure 2, a common haplotype was shared by all pagetic individuals in the pedigree. We then genotyped the 15 other families, which did not harbor the 5q35-qter disease haplotype, using five markers spanning 9 cM at 5q31: AFM042xd12 (D5S393), AFM240xg3 (D5S500), AFMb074xg1 (D5S2017), AFM263wg5 (D5S436), and AFMa052th5 (D5S2090). A LOD score of 3.70 was obtained for AFM240xg3 (D5S500), at θ=.00, with allowance for heterogeneity (table 5). Indeed, significant evidence for heterogeneity was obtained for three of the five markers, with α values of .28–.38 (table 5). These results gave evidence of a second disease locus, at 5q31. For three markers tested at 5q31 in families LF and FT, the HOMOG conditional probability for linkage was >.99 . The maximum LOD score for family FT was 2.32 at θ=.00, for both AFM203ye7 (D5S2010) and AFMa102zg1 (D5S2170) (data not shown). Genotype analysis further showed a common haplotype shared by all affected members of family FT (fig. 5). This characteristic haplotype was, however, different from the disease haplotype segregating within family LF.
Table 5.
HOMOG Analysis for Markers Spanning the 5q31 Region after Removal of Eight Families Showing Linkage to 5q35-qter
|
Maximum LOD Score (θmax) |
|||||
| Marker (Locus) | Homogeneity | Heterogeneity | α | χ2 | P |
| AFM042xd12 (D5S393) | 1.73 (.22) | 3.64 (.00) | .34 | 8.76a | .002 |
| AFM240xg3 (D5S500) | 1.50 (.24) | 3.70 (.00) | .38 | 10.13a | .0001 |
| AFMb074xg1 (D5S2017) | 1.75 (.22) | 3.43 (.00) | .28 | 7.73a | .003 |
| AFM263wg5 (D5S436) | 1.02 (.26) | 2.09 (.04) | .35 | 4.92 | .015 |
| AFMa052th5 (D5S2090) | .52 (.26) | .55 (.16) | .44 | .14 | … |
Significant evidence for heterogeneity.
Figure 5.
Pedigree structure and haplotypes spanning the 5q31 region in extended family FT. Symbols are as described in figures 1 and 2.
To refine the 5q31 disease interval, families LF and FT were genotyped with a total of 18 markers (figs. 2 and 5). In family LF, a centromeric recombination event was observed in affected individual LF-074, between AFMb355wf5 (D5S2055) and AFM144yf4 (D5S404). In this kindred, the most telomeric recombination event occurred in affected individual LF-045, between AFMa190xb1 (D5S1972) and AFM282wd5 (D5S638). This interval corresponded to a 21-cM region at chromosome 5q31. In family FT, a recombination event in affected subject FT-028 delimited the centromeric boundary of the disease locus, at AFM286xg9 (D5S642). In recombinant FT-007, the telomeric boundary was at AFM263wg5 (D5S436). These four recombination events confined the 5q31 disease locus within a 12.2-cM interval delineated by markers AFM286xg9 (D5S642) and AFMb074xg1 (D5S2017).
Because the 5q31 disease region was 30 cM distant from our first localization at 5q35-qter, and because these two intervals were delimited by specific recombination events, we concluded that these two loci harbored two distinct disease genes. We suggest the acronyms “PDB3” and “PDB4” as names for the 5q35-qter and 5q31 loci, respectively.
Discussion
This study led to several important findings. We first reported a collection of large families in which Paget disease of bone was segregating as an autosomal dominant trait. We next excluded PDB2, at 18q21-q22, as a major locus for the disorder in these families. Completion of a genomewide scan, study of additional family members, and higher-density genotyping within regions of interest led to the mapping of two novel loci for Paget disease of bone, at 5q35-qter and 5q31. Key recombinants were then exploited to confine the 5q35-qter locus within a 6-cM interval, whereas the 5q31 locus was refined within 12.2 cM. Observation of a large family with LOD scores <−2.00 at PDB2, 5q35-qter, and 5q31 suggested the existence of at least one additional locus for Paget disease of bone, confirming genetic heterogeneity of the disorder.
PDB1 and PDB2 were excluded in three family nuclei and in 19 of the kindreds, respectively. We thus inferred that these two loci may play only a minor role, if any, in Paget disease of the bone in the French Canadian population. Since only one previously published report has shown suggestive evidence for linkage to PDB1 (Fotino et al. 1977), more investigations are required to confirm presence of a disease gene at 6p. On the other hand, six families have been reported to show linkage to PDB2 (Cody et al. 1997; Haslam et al. 1998). It is not yet known, however, whether any of these kindreds harbor mutations either in the FEO disease gene, TNFRS11A, or in a second gene within its vicinity.
Our preliminary genome-scan analysis revealed seven regions with LOD scores >1.0 (fig. 3). The highest value (2.79) was reached with AFM295xg9 (D19S414), at chromosome 19q. Further analysis of this region, with additional markers and individuals, revealed that this peak represented, in fact, a false-positive linkage result. Similar procedures also led to marked decreases of LOD scores at chromosomes 1p, 3p, and 15q. These false-positive results were caused mainly by genetic heterogeneity of the disorder within the three family nuclei selected for the genomewide scan, in which the initial, relatively small per-locus number of individuals showing linkage reduced the specificity of the linkage test. If all three family nuclei showed linkage to the same locus, then linkage to 5q35-qter would have been easier to detect; on the other hand, we would have missed the 5q31 locus. Two other factors also markedly influenced LOD scores obtained during our genomewide scan. These factors were marker informativity and genetic distances between markers composing the ABI panel. As mentioned above (see the “Mapping and Refinement of a PDB locus at 5q35-qter” subsection), a 19-cM gap located between AFM112yb6 (D5S400) and AFM164xb8 (D5S408) made it more difficult to detect linkage to 5q35. Phenotypic characterization also played a major role in the linkage analysis. A complete reevaluation of all diagnosis was therefore performed in a blind fashion before final analysis of our data. These four factors (small number of individuals, marker informativity, marker distance, and phenotypic characteristics) hindered identification of the disease loci during the first pass of our genomewide scan, and therefore only suggestive evidence for linkage was observed. Nevertheless, the large size of several of the pedigrees collected, often comprising more than 30 individuals, and the presence of founder effects in many of the families that we studied counteracted these pitfalls. In the end, these two major features allowed us to obtain convincing evidence for linkage to the disease loci, even if Paget disease of bone is a genetically heterogeneous disorder within the population that we studied.
Genetic heterogeneity of Paget disease of bone within our set of families was first suggested by exclusion, at 5q35-qter, of kindred LF. This hypothesis was subsequently confirmed by our HOMOG analysis of the 24 families. When this test was first applied to the five microsatellites genotyped at 5q35 in all families, it was significant for only two markers. This lack of significance for the three other markers may be explained by the low power of HOMOG to detect heterogeneity when θ>.1. Using 20 families, for which α, the proportion of families showing linkage to a simulated locus, was 30% (i.e., α=.30), Ott (1986) demonstrated that the probability to detect genetic heterogeneity with HOMOG decreased from .898 at θ=.01 to .54 at θ=.10. In agreement with this study, only the two 5q35 markers—AFM164xb8 (D5S408) and AFM238xe11 (D5S498)—showing a maximum θ (θmax) <.1 gave significant χ2 values. All 5q31 microsatellite markers that showed θmax=0 also gave significant χ2 values.
The estimation of genetic heterogeneity is important in the attempt to determine whether α corresponds exactly to the ratio really observed. However, estimation of θ and α by the HOMOG test is subject to several biases (Janssen et al. 1997). These biases include preferential extension of families already showing linkage to one locus, as well as misclassification and/or diagnostic errors. In our study, ascertainment biases were minimized because family recruitment was completed before the genotyping process was begun. Moreover, diagnoses of all members of the three family nuclei used in our genome scan were carefully revalidated. Disease-gene expressivity and its related penetrance may also introduce biases, if these two parameters vary between the different disease loci. In this regard, we have no evidence that age-dependent penetrance ratios of the disorder differed between the families showing linkage to the two loci. Finally, precise estimates of θ and α are obtained only when the sizes of families showing linkage are similar to those of families not showing linkage. Since several families in our study were small, linkage cannot be conclusively established for each one of them. Nevertheless, the ratio of pedigrees showing linkage at AFM164xb8 that were estimated by HOMOG was 35%, a percentage very similar to the proportion of families harboring the common disease haplotype at 5q35-qter (8/24 [33.3%]). When HOMOG was applied at 5q31 in the 16 other families, the test showed evidence for linkage, under heterogeneity, for three markers spanning this interval, and the estimated proportion of families showing linkage was 28%–38%. Two large families, LF and FT, showed linkage, at a high level of confidence, to the 5q31 locus, whereas linkage to this region was excluded in one other large pedigree. On the other hand, of the 13 remaining families, several were small and could not be demonstrated to show linkage individually at either locus; none of them harbored one of the three identified disease haplotypes. Finally, since, in the very large family, linkage to 5q35-qter and PDB2, as well as to 5q31, was excluded, at least one additional locus for Paget disease of bone remains to be mapped. Such high genetic heterogeneity, with, potentially, five disease-gene loci, may explain the large prevalence of Paget disease of bone in the general population.
Using a relatively homogeneous population, we expected to observe at least one founder effect, particularly if, in the population that we studied, the trait locus was controlled by a few major genes. In agreement with this assumption, we detected a common, 6-cM disease haplotype at chromosome 5q35-qter, in all affected individuals in eight of the pedigrees that we studied. In contrast to this observation, the two pedigrees showing linkage to 5q31 did not harbor the same disease haplotype, and, as far as we know, they do not share common ancestors. Thus, Paget disease of bone in these two families showing linkage to 5q31 may be caused by two different mutations either in the same gene or in two closely linked genes. Alternatively, these two families may share a common haplotype shorter than the distance between two of the markers genotyped in this study. Genotyping more markers within the linkage interval will help to prove this hypothesis.
One important phenotypic observation in this study was the concomitant diagnosis of both Paget disease of bone and benign osteopetrosis in two individuals in family LF (fig. 2). To our knowledge, this is the first time that these two diseases have been diagnosed within the same individual. Osteopetrosis mutations generally involve an absence or a decrease of osteoclastic functions, whereas osteoclasts in Paget disease of bone are overactive and each phase of the bone-remodeling process is accelerated. The presence of both disorders in the same person may thus appear to be contradictory, since their concomitant expression may involve potentially antagonistic mechanisms. All four osteopetrotic patients in family LF harbored the Paget-disease-of-bone haplotype, suggesting that both of these diseases may be linked to 5q31, either involving closely linked genes or being caused by the same mutation but leading to different phenotypes. Association of benign osteopetrosis with Paget disease of bone may also have occurred by chance, since this association was observed in only two patients. At present, it is difficult to assess whether both disorders are associated, with benign osteopetrosis preceding the development of Paget disease of bone, since phenotypic manifestations of osteopetrosis are usually detected in younger individuals, whereas Paget disease of bone is rarely diagnosed at <40 years of age.
This study has emphasized the importance of heredity in the pathogenesis of Paget disease of bone and has confirmed the dominant-segregation model originally proposed by McKusick (1972). However, penetrance of the disease within the families that showed linkage was lower than that expected at the beginning of our investigations and was highly age dependent. Penetrance frequencies observed here may not be applicable to other studies of linkage of Paget disease of bone, since the genetic defects involved in the French Canadian families that we studied may alter disease expressivity.
Although the etiology of Paget disease of bone remains unknown, it is likely that cytokine dysregulation may be involved in its pathogenesis. Linkage of Paget disease of bone to 5q31 is thus of particular interest, in light of the fact that many of genes located within this chromosomal segment are members of the cytokine family, including those encoding interleukin-3, -4, -5, -9, and -13; colony-stimulating factor–2; fibroblast growth factor–1; and CD14; on the other hand, no gene seems to encode osteoclast- or bone-specific protein in the 5q35-qter region.
In conclusion, we have reported the mapping and refinement of two novel loci for Paget disease of bone; the first locus was localized at 5q35-qter, the second at 5q31. We also have confirmed the genetic heterogeneity of the disorder. Further work will be necessary to identify the causative genes and to elucidate the mechanisms by which they contribute to the susceptibility of this highly prevalent disorder.
Acknowledgments
The authors would like to thank all the families who participated in this study. Special thanks go to Alan Tenenhouse and John Caminis, for their thoughtful advice at the initial steps of this research. The authors also thank Claire Brousseau, for her tremendous work in contacting the families, scheduling the confirmatory exams, and collecting all medical records; Marc Gendreau, for his outstanding support in clinical data collection and analysis; and Evelyne Lejeune and her team of research nurses, for their excellent logistical support. The authors are also grateful to the members of the Centre National de Genotypage. N.L. is a recipient of a studentship from the Fonds pour la Formation des Chercheurs et l’Aide à la Recherche and the Fonds de la Recherche en Santé du Québec (FCAR-FRSQ). V.R. is an FRSQ National Investigator. Funding to initiate this work was provided by a John G. Haddad Jr. Award granted by the Paget Foundation for Paget’s Disease of Bone and Related Disorders. Subsequent funding was provided by Canadian Institutes for Health Research grant MOP-3804.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Généthon, http://www.genethon.fr (for reference genetic map and markers used to fine-map critical regions)
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/ (for Human Genome Project draft)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PDB/PDB1 [MIM 167250], PDB2 [MIM 602080], FEO [MIM 174810], and TNFRSF11A [MIM 603499])
References
- Abe S, Ohno T, Park P, Higaki S, Unno K, Tateishi A (1995) Viral behavior of paracrystalline inclusions in osteoclasts of Paget's disease of bone. Ultrastruct Pathol 19:455–461 [DOI] [PubMed] [Google Scholar]
- Anderson MA, Gusella JF (1984) Use of cyclosporin A in establishing Epstein-Barr virus–transformed human lymphoblastoid cell lines. In Vitro 20:856–858 [DOI] [PubMed] [Google Scholar]
- Barker DJ (1984) The epidemiology of Paget's disease of bone. Br Med Bull 40:396–400 [DOI] [PubMed] [Google Scholar]
- Barker DJ, Chamberlain AT, Guyer PB, Gardner MJ (1980) Paget's disease of bone: the Lancashire focus. Br Med J 280:1105–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch MA, Taylor W, Fraser WD, Ralston SH, Hart CA, Gallagher JA (1994) Absence of paramyxovirus RNA in cultures of pagetic bone cells and in pagetic bone. J Bone Miner Res 9:11–16 [DOI] [PubMed] [Google Scholar]
- Boehnke M (1991) Allele frequency estimation from data on relatives. Am J Hum Genet 48:22–25 [PMC free article] [PubMed] [Google Scholar]
- Bouchard G, de Braekeleer M (1991) Histoire d'une génome: population et génétique dans l'est du Québec. Presse de l'Université Laval, Sillery, Québec [Google Scholar]
- Breanndan Moore S, Hoffman DL (1988) Absence of HLA linkage in a family with osteitis deformans (Paget's disease of bone). Tissue Antigens 31:69–70 [DOI] [PubMed] [Google Scholar]
- Cartwright EJ, Gordon MT, Freemont AJ, Anderson DC, Sharpe PT (1993) Paramyxoviruses and Paget's disease. J Med Virol 40:133–141 [DOI] [PubMed] [Google Scholar]
- Cody JD, Singer FR, Roodman GD, Otterund B, Lewis TB, Leppert M, Leach RJ (1997) Genetic linkage of Paget disease of the bone to chromosome 18q. Am J Hum Genet 61:1117–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detheridge FM, Barker DJ, Guyer PB (1983) Paget's disease of bone in Ireland. Br Med J (Clin Res Ed) 287:1345–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Fotino M, Haymovits A, Falk CT (1977) Evidence for linkage between HLA and Paget's disease. Transplant Proc 9:1867–1868 [PubMed] [Google Scholar]
- Freimer NB, Sandkuijl LA, Blower SM (1993) Incorrect specification of marker allele frequencies: effects on linkage analysis. Am J Hum Genet 52:1102–1110 [PMC free article] [PubMed] [Google Scholar]
- Gherardi G, Lo Cascio V, Bonucci E (1980) Fine structure of nuclei and cytoplasm of osteoclasts in Paget's disease of bone. Histopathology 4:63–74 [DOI] [PubMed] [Google Scholar]
- Good D, Busfield F, Duffy D, Lovelock PK, Kesting JB, Cameron DP, Shaw JT (2001) Familial Paget's disease of bone: nonlinkage to the PDB1 and PDB2 loci on chromosomes 6p and 18q in a large pedigree. J Bone Miner Res 16:33–38 [DOI] [PubMed] [Google Scholar]
- Gordon MT, Anderson DC, Sharpe PT (1991) Canine distemper virus localised in bone cells of patients with Paget's disease. Bone 12:195–201 [DOI] [PubMed] [Google Scholar]
- Guyer PB, Chamberlain AT (1980) Paget's disease of bone in two American cities. Br Med J 280:985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdy RC (1995) Clinical features and pharmacologic treatment of Paget's disease. Endocrinol Metab Clin North Am 24:421–436 [PubMed] [Google Scholar]
- Harvey L, Gray T, Beneton MN, Douglas DL, Kanis JA, Russell RG (1982) Ultrastructural features of the osteoclasts from Paget's disease of bone in relation to a viral aetiology. J Clin Pathol 35:771–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam SI, Van Hul W, Morales-Piga A, Balemans W, San-Millan JL, Nakatsuka K, Willems P, Haites NE, Ralston SH (1998) Paget's disease of bone: evidence for a susceptibility locus on chromosome 18q and for genetic heterogeneity. J Bone Miner Res 13:911–917 [DOI] [PubMed] [Google Scholar]
- Helfrich MH, Hobson RP, Grabowski PS, Zurbriggen A, Cosby SL, Dickson GR, Fraser WD, Ooi CG, Selby PL, Crisp AJ, Wallace RG, Kahn S, Ralston SH (2000) A negative search for a paramyxoviral etiology of Paget's disease of bone: molecular, immunological, and ultrastructural studies in UK patients. J Bone Miner Res 15:2315–2329 [DOI] [PubMed] [Google Scholar]
- Hocking L, Slee F, Haslam SI, Cundy T, Nicholson G, van Hul W, Ralston SH (2000) Familial Paget's disease of bone: patterns of inheritance and frequency of linkage to chromosome 18q. Bone 26:577–580 [DOI] [PubMed] [Google Scholar]
- Howatson AF, Fornasier VL (1982) Microfilaments associated with Paget's disease of bone: comparison with nucleocapsids of measles virus and respiratory syncytial virus. Intervirology 18:150–159 [DOI] [PubMed] [Google Scholar]
- Hughes AE, Ralston SH, Marken J, Bell C, MacPherson H, Wallace RG, Van Hul W, Whyte MP, Nakatsuka K, Hovy L, Anderson DM (2000) Mutations in TNFRSF11A, affecting the signal peptide of RANK, cause familial expansile osteolysis. Nat Genet 24:45–48 [DOI] [PubMed] [Google Scholar]
- Hughes AE, Shearman AM, Weber JL, Barr RJ, Wallace RG, Osterberg PH, Nevin NC, Mollan RA (1994) Genetic linkage of familial expansile osteolysis to chromosome 18q. Hum Mol Genet 3:359–361 [DOI] [PubMed] [Google Scholar]
- Janssen B, Halley D, Sandkuijl L (1997) Linkage analysis under locus heterogeneity: behaviour of the A-test in complex analyses. Hum Hered 47:223–233 [DOI] [PubMed] [Google Scholar]
- Jeanpierre M (1987) A rapid method for the purification of DNA from blood. Nucleic Acids Res 15:9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanis JA (1998) Pathophysiology and treatment of Paget's disease of bone, 2d ed. Martin Dunitz, London [Google Scholar]
- Klein RM, Norman A (1995) Diagnostic procedures for Paget's disease. Radiologic, pathologic, and laboratory testing. Endocrinol Metab Clin North Am 24:437–450 [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Leach RJ, Singer FR, Roodman GD (2001) The genetics of Paget's disease of the bone. J Clin Endocrinol Metab 86:24–28 [DOI] [PubMed] [Google Scholar]
- McKusick VA (1972) Paget's disease of the bone. In: Heritable disorders of connective tissue, 4th ed. CV Mosby, St Louis, pp 718–723 [Google Scholar]
- Mee AP, Dixon JA, Hoyland JA, Davies M, Selby PL, Mawer EB (1998) Detection of canine distemper virus in 100% of Paget's disease samples by in situ–reverse transcriptase–polymerase chain reaction. Bone 23:171–175 [DOI] [PubMed] [Google Scholar]
- Meunier PJ, Salson C, Mathieu L, Chapuy MC, Delmas P, Alexandre C, Charhon S (1987) Skeletal distribution and biochemical parameters of Paget's disease. Clin Orthop (April): 37–44 [PubMed] [Google Scholar]
- Mills BG, Singer FR (1976) Nuclear inclusions in Paget's disease of bone. Science 194:201–202 [DOI] [PubMed] [Google Scholar]
- Mills BG, Singer FR, Weiner LP, Suffin SC, Stabile E, Holst P (1984) Evidence for both respiratory syncytial virus and measles virus antigens in the osteoclasts of patients with Paget's disease of bone. Clin Orthop (March): 303–311 [PubMed] [Google Scholar]
- Morales-Piga AA, Rey-Rey JS, Corres-Gonzalez J, Garcia-Sagredo JM, Lopez-Abente G (1995) Frequency and characteristics of familial aggregation of Paget's disease of bone. J Bone Miner Res 10:663–670 [DOI] [PubMed] [Google Scholar]
- Morissette J (1992) L'informatisation de l'information en génétique humaine. In: Knoppers B-M, Cadiet L, Laberge CM (eds) La génétique humaine de l'information à l'informatisation. Thémis & Litec, Montréal and Paris, pp 89–99 [Google Scholar]
- Morissette J, Clépet C, Moisan S, Dubois S, Winstall E, Vermeeren D, Nguyen TD, Polansky JR, Côté G, Anctil JL, Amyot M, Plante M, Falardeau P, Raymond V (1998) Homozygotes carrying an autosomal dominant TIGR mutation do not manifest glaucoma. Nat Genet 19:319–321 [DOI] [PubMed] [Google Scholar]
- Morissette J, Côté G, Anctil JL, Plante M, Amyot M, Héon E, Trope GE, Weissenbach J, Raymond V (1995) A common gene for juvenile and adult-onset primary open-angle glaucomas confined on chromosome 1q. Am J Hum Genet 56:1431–1442 [PMC free article] [PubMed] [Google Scholar]
- Nance MA, Nuttall FQ, Econs MJ, Lyles KW, Viles KD, Vance JM, Pericak-Vance MA, Speer MC (2000) Heterogeneity in Paget disease of the bone. Am J Med Genet 92:303–307 [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1995) The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet 11:402–408 [DOI] [PubMed] [Google Scholar]
- ——— (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi CG, Walsh CA, Gallagher JA, Fraser WD (2000) Absence of measles virus and canine distemper virus transcripts in long- term bone marrow cultures from patients with Paget's disease of bone. Bone 27:417–421 [DOI] [PubMed] [Google Scholar]
- Ott J (1986) The number of families required to detect or exclude linkage heterogeneity. Am J Hum Genet 39:159–165 [PMC free article] [PubMed] [Google Scholar]
- ——— (1991) Analysis of human genetic linkage. The Johns Hopkins University Press, Baltimore and London [Google Scholar]
- Ralston SH, Digiovine FS, Gallacher SJ, Boyle IT, Duff GW (1991) Failure to detect paramyxovirus sequences in Paget's disease of bone using the polymerase chain reaction. J Bone Miner Res 6:1243–1248 [DOI] [PubMed] [Google Scholar]
- Reasbeck JC, Goulding A, Campbell DR, Beale LR, Stewart RD (1983) Radiological prevalence of Paget's disease in Dunedin, New Zealand. Br Med J (Clin Res Ed) 286:1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebel A, Basle M, Pouplard A, Kouyoumdjian S, Filmon R, Lepatezour A (1980a) Viral antigens in osteoclasts from Paget's disease of bone. Lancet 2:344–346 [DOI] [PubMed] [Google Scholar]
- Rebel A, Basle M, Pouplard A, Malkani K, Filmon R, Lepatezour A (1980b) Bone tissue in Paget's disease of bone: ultrastructure and immunocytology. Arthritis Rheum 23:1104–1114 [DOI] [PubMed] [Google Scholar]
- Reddy SV, Menaa C, Singer FR, Cundy T, Cornish J, Whyte MP, Roodman GD (1999) Measles virus nucleocapsid transcript expression is not restricted to the osteoclast lineage in patients with Paget's disease of bone. Exp Hematol 27:1528–1532 [DOI] [PubMed] [Google Scholar]
- Reddy SV, Singer FR, Mallette L, Roodman GD (1996) Detection of measles virus nucleocapsid transcripts in circulating blood cells from patients with Paget disease. J Bone Miner Res 11:1602–1607 [DOI] [PubMed] [Google Scholar]
- Reddy SV, Singer FR, Roodman GD (1995) Bone marrow mononuclear cells from patients with Paget's disease contain measles virus nucleocapsid messenger ribonucleic acid that has mutations in a specific region of the sequence. J Clin Endocrinol Metab 80:2108–2111 [DOI] [PubMed] [Google Scholar]
- Rénier JC, Fanello S, Rodriguez N, Audran M (1995) Current prevalence of Paget's disease of bone in a region of France (Anjou). Rev Rhum Engl Ed 62:571–575 [PubMed] [Google Scholar]
- Rosenbaum HD, Hanson DJ (1969) Geographic variation in the prevalence of Paget's disease of bone. Radiology 92:959–963 [DOI] [PubMed] [Google Scholar]
- Schäffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- Siris ES, Canfield RE (1990) Paget's disease of bone. In: Becker KL (ed) Principles and practice of endocrinology and metabolism. JB Lippincott, Philadelphia, pp 504–512 [Google Scholar]
- Siris ES, Ottman R, Flaster E, Kelsey JL (1991) Familial aggregation of Paget's disease of bone. J Bone Miner Res 6:495–500 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Sofaer JA, Holloway SM, Emery AE (1983) A family study of Paget's disease of bone. J Epidemiol Community Health 37:226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks AB, Peterson SN, Bell C, Loftus BJ, Hocking L, Cahill DP, Frassica FJ, Streeten EA, Levine MA, Fraser CM, Adams MD, Broder S, Venter JC, Kinzler KW, Vogelstein B, Ralston SH (2001) Mutation screening of the TNFRSF11A gene encoding receptor activator of NF kappa B (RANK) in familial and sporadic Paget's disease of bone and osteosarcoma. Calcif Tissue Int 68:151–155 [DOI] [PubMed] [Google Scholar]
- Tilyard MW, Gardner RJ, Milligan L, Cleary TA, Stewart RD (1982) A probable linkage between familial Paget's disease and the HLA loci. Aust NZ J Med 12:498–500 [DOI] [PubMed] [Google Scholar]
- Vignal A, Gyapay G, Hazan J, Nguyen S, Dupraz C, Cheron N, Becuwe N, Tranchant M, Weissenbach J (1993) Nonradioactive multiplex procedure for genotyping of microsatellite markers. In: Adolph KW (ed) Methods in molecular genetics. Vol 1: Gene and chromosome analysis. Academic Press, San Diego, pp 211–221 [Google Scholar]