Abstract
Improved molecular understanding of the pathogenesis of type 2 diabetes is essential if current therapeutic and preventative options are to be extended. To identify diabetes-susceptibility genes, we have completed a primary (418-marker, 9-cM) autosomal-genome scan of 743 sib pairs (573 pedigrees) with type 2 diabetes who are from the Diabetes UK Warren 2 repository. Nonparametric linkage analysis of the entire data set identified seven regions showing evidence for linkage, with allele-sharing LOD scores ⩾1.18 (P⩽.01). The strongest evidence was seen on chromosomes 8p21-22 (near D8S258 [LOD score 2.55]) and 10q23.3 (near D10S1765 [LOD score 1.99]), both coinciding with regions identified in previous scans in European subjects. This was also true of two lesser regions identified, on chromosomes 5q13 (D5S647 [LOD score 1.22] and 5q32 (D5S436 [LOD score 1.22]). Loci on 7p15.3 (LOD score 1.31) and 8q24.2 (LOD score 1.41) are novel. The final region showing evidence for linkage, on chromosome 1q24-25 (near D1S218 [LOD score 1.50]), colocalizes with evidence for linkage to diabetes found in Utah, French, and Pima families and in the GK rat. After dense-map genotyping (mean marker spacing 4.4 cM), evidence for linkage to this region increased to a LOD score of 1.98. Conditional analyses revealed nominally significant interactions between this locus and the regions on chromosomes 10q23.3 (P=.01) and 5q32 (P=.02). These data, derived from one of the largest genome scans undertaken in this condition, confirm that individual susceptibility-gene effects for type 2 diabetes are likely to be modest in size. Taken with genome scans in other populations, they provide both replication of previous evidence indicating the presence of a diabetes-susceptibility locus on chromosome 1q24-25 and support for the existence of additional loci on chromosomes 5, 8, and 10. These data should accelerate positional cloning efforts in these regions of interest.
Introduction
Type 2 diabetes (T2D [MIM 125853]) is a global disease of rapidly increasing prevalence, for which current preventative and therapeutic strategies remain suboptimal. It is estimated that ∼160 million people worldwide have T2D, a figure expected to rise by 40% by 2010 (McCarty and Zimmet 1994). Furthermore, T2D is one component of a broader constellation of phenotypes (including hyperlipidemia, central obesity, and hypertension—the “metabolic syndrome”) that contribute in a major way to the burden of cardiovascular morbidity and mortality and that therefore represent dominant causes of disease worldwide (Reaven 1988). Further advances in treating or preventing these disorders are predicated on an improved molecular description of their pathogenesis. To this end, recognition that individual susceptibility to T2D has a substantial genetic component provides a coherent framework for efforts to elucidate the fundamental disease processes.
The relative recurrence risk of T2D in first-degree relatives of affected subjects is 3.5 in European populations (Köbberling and Tillil 1982; Rich 1990). Candidate-gene studies have identified several loci with modest effects on T2D susceptibility (Kadowaki et al. 1994; Hani et al. 1998, 1999; Altshuler et al. 2000; Huxtable et al. 2000). In addition, the past 5 years have seen global (i.e., genomewide) scans for linkage that have been conducted in a wide range of populations, including African Americans (Ehm et al. 2000), Ashkenazim (Permutt et al. 2001), Finns (Mahtani et al. 1996; Ghosh et al. 2000; Watanabe et al. 2000), French (Vionnet et al. 2000), Mexican Americans (Hanis et al. 1996; Duggirala et al. 1999; Ehm et al. 2000), Native Americans (Hanson et al. 1998) and individuals in the United States who are of European descent (Elbein et al. 1999; Ehm et al. 2000). Although several regions showing evidence for linkage have emerged from these studies, and although at least one susceptibility gene (i.e., CAPN10, for calpain 10 [MIM 605286]; Horikawa et al. 2000) has subsequently been identified by linkage-disequilibrium mapping, these analyses have reinforced the concept of T2D as an etiologically complex, heterogeneous, and multifactorial condition, with individual susceptibility determined by the integrated effect of variation at multiple genetic loci and predisposing environmental exposures.
The reduction in power that is associated with susceptibility-gene–discovery efforts in the presence of complex etiology may be ameliorated by the analysis of large samples, by careful attention to population selection so as to minimize the possible effects of ethnic and clinical heterogeneity, and, arguably, by use of population isolates (Lernmark and Ott 1998; Ott 1999, p. 321). In certain circumstances, statistically controlled exploratory methods, such as stratification based on quantitative phenotypes (Ghosh et al. 1999) and conditional analyses (Cox et al. 1999), may yield increases in power (Leal and Ott 2000). Support for a putative susceptibility locus is enhanced considerably by statistically significant replication in additional populations: indeed, such replication is widely considered a vital component in the assessment of complex-trait linkage effects (Lander and Kruglyak 1995).
The Warren 2 Consortium was established by Diabetes UK (formerly the British Diabetic Association) to contribute to global efforts to identify T2D-susceptibility genes. The objectives have been to assemble a number of substantial patient and family cohorts of U.K. individuals of European descent and to exploit these to advance understanding of the genetic basis of T2D in northern-European populations. Here, we report the results of the autosomal genomewide scan for linkage, conducted in a total of 573 U.K. families that were multiplex for T2D, together with the fine-scale mapping data obtained from one of the chromosomal regions implicated by the primary analysis.
Subjects and Methods
Subjects
The Warren 2 Sib Pair Repository (see the Warren 2 Project Information [Wellcome Trust Centre for Human Genetics, Oxford] web site) currently comprises 843 sibship pedigrees ascertained, during 1995–98, through six U.K. research centers (two in London and one each in Exeter, Oxford, Cambridge/Norwich, and Newcastle), according to a unified ascertainment protocol. All ascertained families include, at minimum, a sib pair with T2D, together with parents and additional siblings when available. Validation of the diagnosis of diabetes in the index sib pair was based on either current prescribed treatment with sulfonyl ureas, biguanides, and/or insulin or, in the case of individuals treated with diet alone, historical or contemporary laboratory evidence of hyperglycemia (as defined by World Health Organization [1985] guidelines in place at the time of recruitment). Age at diagnosis (AAD), of both members of the index sib pair, was initially restricted to the age range 35–75 years and subsequently was narrowed to 35–70 years, with 97.6% families meeting the latter criterion. Other forms of diabetes (e.g., maturity-onset diabetes of the young, mitochondrial diabetes, and type 1 diabetes) were excluded by standard clinical criteria based personal and family history, including an absence of first-degree relatives with type 1 diabetes and an interval of ⩾1 year between diagnosis and institution of regular insulin therapy. In addition (see the following paragraph), evidence for autoimmunity to islet antigens was sought by measurement of titers of antibodies to glutamic acid decarboxylase (anti-GAD). All sibships were of European descent, with all four grandparents having exclusively British and/or Irish origin, both by self-reported ethnicity and by place of birth. Finally, pedigrees either reporting bilineal inheritance (both parents diabetic) or having a high proportion of affected individuals within large sibships were excluded from collection.
These 843 pedigrees included a total of 2,147 ascertained individuals, 2,112 siblings (of whom 1,820 were diabetic), and 35 parents (15 of whom were diabetic). The numbers of affected siblings in the sibships ranged from 2 (in 729 pedigrees) to 5 (in 5 pedigrees). All available pedigree members were assessed for standard anthropometric measures (weight, height, and waist and hip circumferences). Blood samples were taken for DNA extraction and the establishment of transformed cell lines (undertaken at the European Collection of Cell Cultures, Centre for Applied Microbiology & Research, Salisbury, England). Serum was reserved from all diabetic sibship members, for detection of latent islet autoimmunity (Tuomi et al. 1993). Anti-GAD titers were measured by radio-ligand assay, as described elsewhere (Petersen et al. 1994; Bingley et al. 1997). This assay achieved 100% sensitivity and specificity in the Third GAD Proficiency Program, in 1997, with an intra-assay variation of 7%. An anti-GAD titer >10 U (corresponding to ∼8 SD above the mean of 88 normal control subjects) in duplicate samples was considered positive. Informed consent for participation was obtained from all subjects and relatives, after approval had been granted by the relevant Research Ethics Committees.
Microsatellite Genotyping
Sufficient DNA was available from all ascertained members in a total of 687 pedigrees at the initiation of the genotyping analyses. The autosomal genome scan in these 1,721 individuals used a panel of 418 microsatellite markers based principally on the ABI Prism Linkage Mapping Set MD-10 (Applied Biosystems), supplemented (where necessary, for reasons of marker performance or heterozygosity) by additional markers optimized in-house. All markers were dinucleotide repeats of the type (CA)n, originally chosen from the Généthon/CEPH map (Dib et al. 1996). Unlabeled primers were “PIG-tailed” to facilitate automated allele calling (Brownstein et al. 1996). After the primary, 418-marker scan, a further 17 microsatellite markers were typed on chromosome 1q. Details of all markers used are available from the Warren 2 Project Information (Wellcome Trust Centre for Human Genetics, Oxford) web site.
Template DNA (25 ng/reaction) was amplified in simplex 15-μl PCR reactions using Taq Gold polymerase, according to the instructions supplied with the MD-10 marker set. For each individual, 10–20 PCR products were pooled into coelectrophoresis panels, on the basis of pooling ratios designed to normalize signal intensity across markers. Electrophoresis and signal recording were performed on ABI377 automated Sequencers (Applied Biosystems), by a standard protocol, with trace analysis by GENESCAN and GENOTYPER. Allele calling by the latter program incorporated several components: an initial round of fully automated typing with standardized analysis settings was followed, when required, by manual inspection and correction. Manually approved genotypes were then submitted to a pipeline of rigorous quality-control checks (e.g., absolute and relative peak intensities and outlier peaks), with a subsequent manual reevaluation (by a second operator) of selected genotypes. Database-aided quality-control procedures included confirmation of standard individual genotypes (CEPH standard 1347-02; Coriell Institute), plate identity and orientation, and allele size.
Inheritance Checking
Pedigrees containing three or more typed individuals were examined by PEDCHECK (O’Connell and Weeks 1998), to detect Mendelian inconsistencies. Reported family relationships in all 687 genotyped pedigrees were examined by identity-by-descent–based methods implemented in RELATIVE (Göring and Ott 1997) and RELPAIR (Boehnke and Cox 1997), with all available genotyped markers. A family was excluded if all methods failed to produce concordant results. Individuals (or samples) apparently unrelated to other “family” members were excluded. Instances of unsuspected half-sib relationships (present in 27 families) were also excluded, to avoid the possible influence of biological parents whose adherence to the ascertainment criteria could not be verified. A “final” pedigree data set (573 families) was obtained by excluding all families that contained one or more members positive for anti-GAD antibodies. All analyses reported herein are based on this 573-pedigree data set.
Linkage Analysis
Allele frequencies for the 418 autosomal markers were estimated on the basis of the entire data set, by RECODE (Division of Statistical Genetics, Department of Human Genetics, University of Pittsburgh). Marker order and intermarker distances were taken from published Généthon/CEPH maps (Dib et al. 1996) and were converted into Haldane centimorgans, for multipoint linkage analysis using ALLEGRO version 1.1b (Gudbjartsson et al. 2000). All distances quoted herein are relative to the most p-terminal marker in the Généthon/CEPH maps (Dib et al. 1996), unless otherwise stated. ALLEGRO implements both the nonparametric linkage (NPL) Z-score of Kruglyak et al. (1996) and the allele-sharing LOD score of Kong and Cox (1997), designed to accommodate the conservativeness of the NPL Z-score when inheritance information is incomplete, as is the case with missing genotype data. In addition, for the genome scan and subsequent simulations, ALLEGRO was preferred over GENEHUNTER (Kruglyak et al. 1996) and GENEHUNTER PLUS (Kong and Cox 1997), for reasons of computational efficiency. The Sall scoring statistic was used for calculation of the NPL Z-score (Kruglyak et al. 1996), and the exponential model was used for the allele-sharing LOD score (Kong and Cox 1997). Significance levels of LOD scores quoted herein are nominal, unless otherwise stated.
Genomewide Exclusion Mapping
As a complement to the search for evidence of linkage, we conducted exclusion mapping to determine which genomic regions could be excluded as candidates for major susceptibility effects, using GENEHUNTER version 2.0 (Kruglyak et al. 1996). Five different effect sizes were considered, under the assumption that the overall sibling relative risk (λS) for T2D is 3.5 (Köbberling and Tillil 1982; Rich 1990). Locus-specific relative risks of 3.50, 1.87, 1.52, 1.37, and 1.28 correspond, respectively, to one to five affection-trait loci (ATLs) of equal effect size acting multiplicatively (Risch 1990). Analyses were performed under the assumption of no dominance variance (hence λS=λO).
Genomewide Empirical-Significance Calculations and Power Estimates
We determined the genomewide empirical significance of regions of excess allele sharing that were identified by our genome scan. A complete genome with marker characteristics reproducing those observed in the final pedigree data set (573 families) was simulated under the null hypothesis of no linkage (by SIMULATE; see the Lab of Statistical Genetics [Rockefeller University] web site). Ten thousand replicates (incorporating the exact pattern of missing genotypes observed in our scan) were generated and analyzed by ALLEGRO, as above (see the “Linkage Analysis” subsection). In addition, simulations were performed to estimate our data set's power to detect T2D ATLs with different effect sizes. An “average” chromosome, comprising 20 markers and reproducing the mean number of alleles, heterozygosity, and marker spacing of the marker set used, was simulated for the final pedigree data set, under the assumption of linkage to a single ATL. As described above (see the “Genomewide Exclusion Mapping” subsection of the Subjects and Methods section), five gene-effect sizes were considered, and an (average) missing-genotype rate of 15% was optionally included. One thousand replicates under each set of conditions were analyzed by ALLEGRO.
Conditional Analyses
To identify possible interactions between the regions of excess allele sharing that were identified in the genome scan, we performed conditional analyses (Cox et al. 1999) using GENEHUNTER PLUS (Kong and Cox 1997). Reevaluating the evidence for linkage at one locus after having accounted for the evidence for linkage at a second, unlinked but phenotypically correlated locus may increase power and indicate the nature of any interaction between the two (Cox et al. 1999). To control the extent of multiple testing, we elected only to consider interactions in which both loci had multipoint LOD scores ⩾1.18 (corresponding to P⩽.01 [Ott 1999, p. 66]) and in which at least one of the loci had a multipoint LOD score ⩾1.50. Spearman's correlation coefficient, ρ, between the family NPL scores at each pair of loci was calculated by SAS, release 8.2 (SAS Institute). The evidence for linkage at a given (“conditioned”) locus was then reevaluated after each family had been assigned a weight according to the NPL score at the site of maximal linkage to the other (“conditioning”) locus. In the case of positive correlations (indicating possible epistasis), those weights were +1 for each family with a positive NPL score at the conditioning locus and 0 for families with 0 or negative NPL scores; for negative correlations (representing possible heterogeneity), weights were +1 when the NPL score at the conditioning locus was negative and were 0 when the NPL score was 0 or positive. When the value of the weighted LOD score exceeded that of the unweighted LOD score, the significance of the change was assessed by permutation: the family-specific weights were permuted, and the LOD score was recalculated on the basis of the GENEHUNTER PLUS probability files.
Ordered-Subset Analysis
Interfamilial differences in associated phenotypes such as age at onset and body-mass index (BMI) may reflect clinically and etiologically distinct subsets of T2D. To explore possible heterogeneity in our data set—and, thereby, to overcome any associated reduction in power to detect ATLs—we conducted ordered-subset analyses (Hauser et al. 1998) of the regions of interest that were highlighted by our primary genome scan, subsetting either on age- and sex-adjusted BMI or on AAD (as a measure of age at onset). Each family was ranked according to the mean value of the associated trait (BMI or AAD) in diabetic individuals within that family. The LOD score for linkage to T2D was then recalculated (under the linear model) at each chromosomal position in successive data sets formed by adding each family in rank order. The analysis was conducted in both rank orders, to ensure detection of subsets at either end of the phenotypic distribution. The significance of the maximized LOD score was determined by 10,000 replicates of rank permutation and reanalysis, as before.
Results
Characteristics of Analyzed Data Set
The autosomal genome scan was completed on 687 pedigrees. Examination of genotype data with PEDCHECK, RELATIVE and RELPAIR resulted in exclusion of 87 pedigrees, on the grounds of demonstrable non-paternity, half-paternity or inadequate genotype data for confirmation of presumed family relationships. A further 27 families were excluded on the basis that each contained at least one member positive for anti-GAD auto-antibodies and therefore suspected of harboring latent autoimmune diabetes (Tuomi et al. 1993). This generated a final pedigree data set for analysis of 573 pedigrees, corresponding to 743 affected sib pairs. Clinical characteristics of these families are shown in table 1.
Table 1.
Clinical and Family-Structure Characteristics of the Entire 573-Pedigree Data Set
|
Mean (SD) in |
||
| Males (n=659) | Females (n=564) | |
| Age at examination (years) | 63.7 (8.4) | 64.5 (8.4) |
| AAD (years) | 55.2 (8.6) | 56.1 (8.6) |
| BMI (kg/m2)a | 27.8 (4.3) | 29.8 (5.6) |
| Waist-to-hip ratioa | .95 (.07) | .86 (.07) |
| Affected sib pairs: | ||
| All possibleb | 743 | |
| Independentc | 650 | |
Significant difference (P<.0001) between males and females.
Calculated as s(s-1)/2, where s is the number of sibs.
Calculated as s-1, where s is the number of sibs (Suarez and Hodge 1979).
Genotyping Characteristics
Mean marker spacing for the 418 autosomal microsatellite markers in the primary genome scan was 9.3 cM (Haldane) with a mean marker heterozygosity of ∼78%. Overall, the success rate for completed genotypes in the 573 pedigree data set was ∼86%, reflecting the rigorous quality-control procedures necessary for small sibships when opportunities for inheritance checking are limited. The mean entropy-based information content (measuring the proportion of the total inheritance information extracted at each chromosomal position, given the available genotype data [Kruglyak et al. 1996]) of the 418 typed markers was 42% in the two-point analysis and 51% in the multipoint analysis.
Power Calculations
Power calculations for the 573-pedigree data set under different values of λS and missing genotype frequency are presented in table 2. Under the optimistic assumption of a single ATL for T2D (locus-specific λS=3.50), there was ∼100% power for detection with a LOD score of 3, when an “average Warren 2 marker map” was used, regardless of the missing genotype rate. Power remained good (84%) for detection of an ATL of modest effect (λS=1.52) with a LOD score of 3, even with 15% missing genotypes, and, under similar assumptions, was close to 100% at a LOD score of 1. Predictably, power to detect ATLs of smaller effect (λS=1.28) with a LOD score of 3 was poor (27% with 15% missing genotypes), although such loci should still be detectable at lower thresholds (∼56% power for a LOD score of 2.0 and ∼86% for a LOD score of 1.0).
Table 2.
Power Calculations for Detection of Linkage, with Entire 573-Pedigree Data Set
|
Powera(%) |
||||||
| No Genotypes Missing |
15% of Genotypes Missing |
|||||
| Locus-Specific λS | Threshold LOD Score = 1 | Threshold LOD Score = 2 | Threshold LOD Score = 3 | Threshold LOD Score = 1 | Threshold LOD Score = 2 | Threshold LOD Score = 3 |
| 3.50 | 100 | 100 | 100 | 100 | 100 | 100 |
| 1.87 | 100 | 100 | 100 | 100 | 99.8 | 99.5 |
| 1.52 | 99.8 | 98.1 | 88.8 | 99.3 | 95.2 | 84.3 |
| 1.37 | 98.1 | 84.9 | 58.7 | 96.2 | 76.1 | 51.0 |
| 1.28 | 90.4 | 59.8 | 30.8 | 86.3 | 55.5 | 26.8 |
Derived from simulations of 1,000 replicates of an “average Warren 2 chromosome,” under the assumption that one, two, three, four, or five ATLs (corresponding to locus-specific λS values of 3.50, 1.87, 1.52, 1.37, or 1.28) contribute to disease.
Linkage Analysis
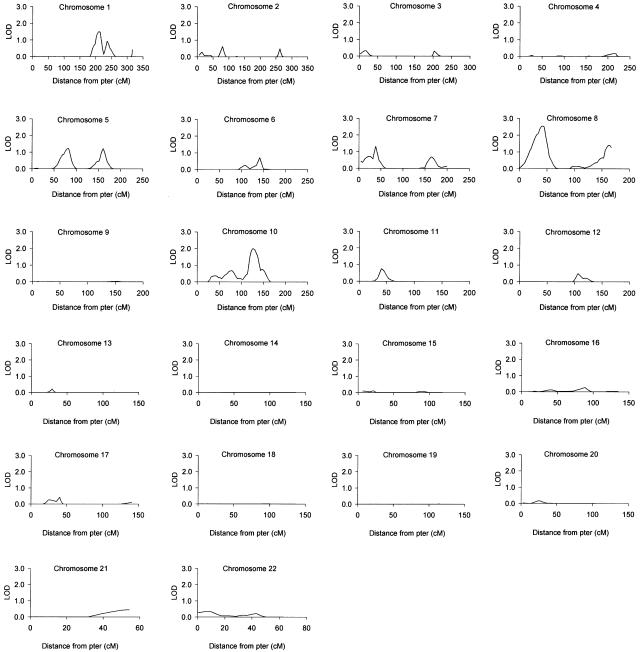
Multipoint linkage maps for the autosomes are shown in figure 1. In all, seven regions showing evidence for linkage, with multipoint LOD scores ⩾1.18 (P⩽.01), were detected: 1q24-25 (LOD score 1.50; 209.8 cM), 5q13 (LOD score 1.22; 82.1 cM), 5q32 (LOD score 1.22; 160.8 cM), 7p15.3 (LOD score 1.31; 38.0 cM), 8p21.3-22 (LOD score 2.55; 42.2 cM), 8q24.21-24.23 (LOD score 1.41; 162.6 cM), and 10q23.31-23.33 (LOD score 1.99; 125.2 cM) (table 3). The widths, at a LOD score ⩾0.59 (P⩽.05), of the three regions showing the best evidence for linkage were estimated: the locus on 1q24-25 covered an ∼29-cM region between D1S484 and D1S238; that on 8p21.3-22, an ∼41-cM region between D8S277 and D8S505; and that on 10q23, an ∼39-cM region between D10S1730 and D10S1693. There were three additional regions—on chromosomes 11, 12 and 17—that generated two-point, but not multipoint, LOD scores ⩾1.18 (table 3), and a further six regions—on chromosomes 1, 2, 6, 7, 10, and 11—showed nominal multipoint evidence for linkage (LOD score ⩾0.59) (table 4). As expected on the basis of the pointwise LOD scores (Lander and Kruglyak 1995), empirical-significance calculations (10,000 replicates) indicated that none of the regions reached genomewide significance at P<.05; for example, the 8p21.3-22 locus was associated with a genomewide empirical-significance level of .098.
Figure 1.
Multipoint analyses for the primary 418-marker autosomal genome scan of 573 sib-pair pedigrees. Analyses were performed on the entire 573-pedigree data set, by ALLEGRO, as described in the text. Allele-sharing LOD scores calculated under the exponential model are shown.
Table 3.
Summary of Regions Displaying Multipoint or Two-Point Evidence for Excess Allele-Sharing (LOD Score ⩾1.18)[Note]
|
Multipoint Datab |
Two-Point Datab |
||||||
| Marker | Positiona(cM) | Marker Heterozygosity(%) | LOD Score | Nominal P | Entropy | LOD Score | Nominal P |
| Chromosome 1: | |||||||
| D1S196 | 203.1 | 75.6 | 1.28 | .0076 | .53 | (.99) | (.016) |
| Interval | 209.8 | … | 1.50 | .0044 | .44 | … | … |
| D1S218 | 214.2 | 79.6 | 1.46 | .0048 | .55 | 1.64 | .0030 |
| D1S2836 | 316.2 | 81.9 | (.41) | (.084) | (.41) | 3.63 | .000022 |
| Chromosome 2: | |||||||
| D2S396 | 262.2 | 80.1 | (.48) | (.068) | (.53) | 1.26 | .0079 |
| Chromosome 5: | |||||||
| D5S647 | 82.1 | 81.9 | 1.22 | .0088 | .54 | 1.35 | .0064 |
| D5S436 | 160.8 | 79.0 | 1.22 | .0090 | .54 | 1.46 | .0048 |
| Chromosome 6: | |||||||
| D6S262 | 141.8 | 88.8 | (.70) | (.036) | (.57) | 1.97 | .0013 |
| Chromosome 7: | |||||||
| D7S493 | 38.0 | 88.1 | 1.31 | .0071 | .56 | 2.26 | .00063 |
| Chromosome 8: | |||||||
| D8S520 | 21.1 | 79.2 | 1.22 | .0087 | .51 | (.54) | (.056) |
| D8S549 | 33.8 | 51.8 | 2.11 | .00090 | .39 | (.68) | (.039) |
| Interval | 42.2 | … | 2.55 | .00031 | .37 | … | … |
| D8S258 | 44.3 | 71.3 | 2.49 | .00036 | .44 | 2.11 | .00091 |
| D8S284 | 156.1 | 83.5 | 1.11 | .012 | .55 | 2.50 | .00035 |
| Interval | 162.6 | … | 1.41 | .0054 | .39 | … | … |
| D8S272 | 166.9 | 75.9 | 1.28 | .0076 | .46 | (.53) | (.058) |
| Chromosome 10: | |||||||
| D10S1686 | 119.9 | 86.0 | 1.61 | .0033 | .54 | 1.09 | .012 |
| D10S1765 | 125.2 | 86.9 | 1.99 | .0012 | .58 | 1.37 | .0060 |
| Interval | 127.2 | … | 1.99 | .0012 | .52 | … | … |
| D10S185 | 135.0 | 76.0 | 1.65 | .0030 | .53 | (.25) | (.14) |
| Chromosome 11: | |||||||
| D11S904 | 40.1 | 80.9 | (.74) | (.032) | (.52) | 1.65 | .0030 |
| Chromosome 12: | |||||||
| D12S351 | 105.6 | 75.3 | (.49) | (.066) | (.47) | 1.70 | .0026 |
| Chromosome 17: | |||||||
| D17S921 | 40.2 | 77.4 | (.41) | (.084) | (.46) | 3.74 | .000017 |
Table 4.
Summary of Additional Regions Displaying Nominal Multipoint Evidence for Linkage (LOD Score ⩾0.59)[Note]
|
Multipoint Data |
Two-Point Data |
||||||
| Marker | Position(cM) | Marker Heterozygosity(%) | LOD Score | Nominal P | Entropy | LOD Score | Nominal P |
| D1S413 (chromosome 1) | 236.2 | 73.3 | .92 | .020 | .51 | .88 | .022 |
| D2S391 (chromosome 2) | 80.1 | 76.1 | .62 | .046 | .52 | 1.15 | .010 |
| D6S262 (chromosome 6) | 141.8 | 88.8 | .70 | .036 | .57 | 1.97 | .0013 |
| D7S684 (chromosome 7) | 163.9 | 78.9 | .68 | .038 | .50 | .44 | .078 |
| D10S196 (chromosome 10) | 79.7 | 73.9 | .66 | .041 | .45 | .58 | .051 |
| D11S904 (chromosome 11) | 40.1 | 80.9 | .74 | .032 | .52 | 1.65 | .0030 |
Note.— Data are as defined in the footnotes to table 3.
Dense Mapping on Chromosome 1q
The linkage results from the primary (∼9 cM) genome scan highlighted regions on 1q24-25, 8p21-22, and 10q23.3 as being the strongest candidates for dense-map genotyping. In each case, the inheritance information extracted by the primary scan was ∼50%. Here, we present dense-mapping data for the 1q24-25 region (table 5 and fig. 2). Work on the other regions is in progress and will be reported elsewhere.
Table 5.
Linkage Results in Chromosome 1q24 Region after Inclusion of Dense-Map Markers[Note]
|
Multipoint Data |
Two-Point Data |
||||||
| Chromosome 1 Marker | Position(cM) | Marker Heterozygosity(%) | LOD Score | Nominal P | Entropy | LOD Score | Nominal P |
| D1S2681 | 197.9 | 78.7 | 1.19 | .0095 | .64 | (.09) | (.26) |
| D1S196 | 201.6 | 75.6 | 1.91 | .0015 | .65 | (.99) | (.016) |
| D1S2799 | 203.8 | 85.2 | 1.94 | .0014 | .67 | 2.36 | .0005 |
| Interval | 206.0 | … | 1.98 | .0013 | .60 | … | … |
| D1S452 | 209.4 | 73.3 | 1.80 | .0020 | .64 | 1.48 | .0046 |
| D1S218 | 212.1 | 79.5 | 1.59 | .0034 | .65 | 1.74 | .0023 |
Note.— Data are as defined in the footnotes to table 3.
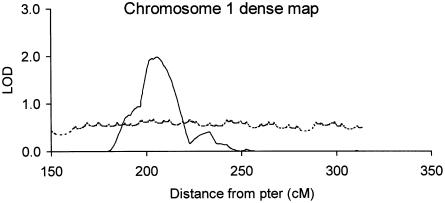
Figure 2.
Multipoint analysis of dense-marker map of chromosome 1q: reanalysis of the entire 573-pedigree data set after typing of an additional 17 markers on chromosome 1q. Allele-sharing LOD scores are denoted by the unbroken line, and entropy-based information content is denoted by the broken line.
All pedigrees were typed for 17 additional 1q markers, distributed from D1S498 (174.9 cM) to D1S2682 (313.6 cM) on 1q, with the majority mapping under the broad 1q24-25 peak (fig. 1) and with others in the 1qter region (around D1S2836, which, on the primary scan, had provided evidence for linkage, with a two-point LOD score of 3.63). This resulted in a dense map of 33 markers, with a mean spacing of 4.4 cM across this region, and elevated the mean information extraction (entropy) to ∼60% (table 5).
Evidence for linkage in this region increased from a LOD score of 1.50 in the primary map to a LOD score of 1.98 (P=.0013) in the dense map, between D1S2799 and D1S452 (206 cM) on 1q24.2. The width of this peak of linkage, at a LOD score of 0.59 (P=.05), increased slightly, to ∼32 cM; at a LOD score of 1.18 (P=.01), the width was ∼17 cM. At the same time, the excess allele sharing that, in the primary scan, was observed close to D1S413 (LOD score 0.92 at 236.2 cM; table 4) was reduced considerably (LOD score 0.41), further localizing the locus for this chromosomal arm, into the region around 206 cM. The region that, in the primary scan, showed excess allele sharing at 1qter (around D1S2836) disappeared completely when the additional markers were included.
Genomewide Exclusion Mapping
Results from the exclusion mapping of the genome are shown in table 6. Segregation of an ATL of very large effect (i.e., λS=3.50) could be excluded across the entire autosomal genome, with a LOD score <−7. Exclusion of an ATL with λS=1.87 was possible for 99% of the genome (with LOD score <−3), the sole exception being the region around the peak of allele sharing, on 8p21-22. At λS=1.52, 92% of the genome was excluded (with LOD score <−2); only the regions described above (see the “Linkage Analysis” subsection)—on 1q24.2, 5q13, 5q32, 7p15.3, 8p21-22, 8q24.2, and 10q23.3 (LOD score ⩾−0.99)—and a region from 44 cM to qter, on chromosome 21 (LOD score −1.19), were not. Modest effect sizes (locus-specific λS values of 1.37 and 1.28) could be excluded from 78% and 57% of the genome, respectively, with the same eight regions being the only ones with positive LOD scores.
Table 6.
Exclusion-Mapping Data for Warren 2 Genome Scan
|
Proportion of Chromosome Excluded at LOD Score <−2.0 When Locus-Specific λS =(%) |
|||||
| Chromosome | 1.28 | 1.37 | 1.52 | 1.87 | 3.50 |
| 1: | |||||
| Primary map | 68 | 74 | 86 | 100 | 100 |
| Dense map | 76 | 85 | 92 | 100 | 100 |
| 2 | 63 | 86 | 100 | 100 | 100 |
| 3 | 70 | 83 | 100 | 100 | 100 |
| 4 | 51 | 93 | 100 | 100 | 100 |
| 5 | 35 | 58 | 77 | 100 | 100 |
| 6 | 60 | 80 | 100 | 100 | 100 |
| 7 | 34 | 49 | 73 | 100 | 100 |
| 8 | 14 | 35 | 56 | 84 | 100 |
| 9 | 85 | 100 | 100 | 100 | 100 |
| 10 | 20 | 31 | 61 | 100 | 100 |
| 11 | 67 | 88 | 100 | 100 | 100 |
| 12 | 73 | 87 | 100 | 100 | 100 |
| 13 | 86 | 94 | 100 | 100 | 100 |
| 14 | 78 | 100 | 100 | 100 | 100 |
| 15 | 64 | 100 | 100 | 100 | 100 |
| 16 | 16 | 88 | 100 | 100 | 100 |
| 17 | 63 | 78 | 100 | 100 | 100 |
| 18 | 87 | 100 | 100 | 100 | 100 |
| 19 | 81 | 97 | 100 | 100 | 100 |
| 20 | 62 | 100 | 100 | 100 | 100 |
| 21 | 56 | 63 | 82 | 100 | 100 |
| 22 | 9 | 75 | 100 | 100 | 100 |
| Entire genome | 57 | 78 | 92 | 99 | 100 |
Conditional Analyses
As described above, Spearman correlation coefficients, ρ, between thefamily NPL scores obtained at the regions showing maximum evidence for linkage, on 1q24.2, 8p21-22, and 10q23.3, were calculated. Further correlations, between each of these three regions and the peaks on 5q13, 5q32, 7p15.3, and 8q24.2, were calculated.
A significant positive correlation was detected between the 1q24.2 and 10q23.3 loci (ρ=.099; P=.018), suggesting the presence of epistasis. We detected an increase in the evidence for linkage at 1q24.2 when dense-map markers were included, after weighting for the linkage at 10q23.3 under an epistatic model. This LOD-score increase, from 1.98 to 3.07, was nominally significant by permutation ( P=.017). In the reciprocal analysis, we found a similarly significant increase in the LOD score at 10q23.3, from 1.99 to 2.78, after weighting for linkage at 1q24.2 (P=.032). There were no significant correlations between either of the two linkages on chromosome 8 and any other region of interest.
There was a nominally significant, negative, correlation between 1q24.2 and 5q32 (ρ=-.099; P=.018), suggesting heterogeneity. The evidence for linkage to 1q24.2, when conditioned for linkage to the 5q32 locus, increased from a LOD score of 1.98 to a LOD score of 3.01 (P=.023). Again, the reciprocal analysis was also nominally significant (LOD score increase, at 5q32, to 2.45; P=.010).
These exploratory conditional analyses involve 15 tests, independent under the null hypothesis of no interaction. Full adjustment for this aspect of the multiple testing would render these conditioning results nonsignificant, at P<.05.
Ordered-Subset Analysis
Ordered-subset analysis was conducted on the chromosome 1q24.2 dense map, with age-at-diagnosis (AAD) and age- and sex-adjusted BMI being used as the stratifying variables. No analysis resulted in a significant increase in the maximized LOD (MOD) score. The baseline LOD score (under a linear model) of 1.89 increased to 2.57 (P=.20) for the bottom 84% (n=481) of AAD-ranked families; the reverse ranking gave a MOD score of 2.43 (P=.28). When BMI was stratified with a low-to-high ranking, there was no increase in the LOD score; the reverse ranking yielded a MOD score of 2.65 (P=.29).
Discussion
The Warren 2 sib pair repository (see the Warren 2 Project Information [Wellcome Trust Centre for Human Genetics, Oxford] web site) represents one of the largest available collections of families segregating T2D. The genomewide scan for linkage reported herein is expected, therefore, to contribute significantly to the global picture emerging from similar studies in other populations, including African American (Ehm et al. 2000), Ashkenazim (Permutt et al. 2001), Finnish (Mahtani et al. 1996; Ghosh et al. 2000; Watanabe et al. 2000), French (Vionnet et al. 2000), Mexican American (Hanis et al. 1996; Duggirala et al. 1999; Ehm et al. 2000), Native American (Hanson et al. 1998), and U.S. individuals of European descent (Elbein et al. 1999; Ehm et al. 2000). In particular, synthesis of data from these different genome scans, most of which, individually, have failed to generate highly significant linkages in primary analyses, should improve the prospects for successful identification of those regions with the strongest claims for genuine susceptibility effects, thereby targeting positional cloning efforts toward the most rewarding loci.
Our study has involved a primary (∼9 cM) genome scan, followed by nonparametric linkage analysis and exclusion mapping, in a sample of 573 U.K. affected sibships of European descent. Loci with allele-sharing LOD scores ⩾1.50 were detected on 1q24.2, 8p21-22, and 10q23.3; other regions, with LOD scores ⩾1.18 (P⩽.01), were detected on 5q13, 5q32, 7p15.3, and 8q24.2. As outlined below (also see table 7), several of these regions—specifically, 1q24.2, 5q13, 5q32, 8p21-22, and 10q23.3—coincide with (and, in one case, clearly replicate) regions of excess allele sharing that have been identified in other scans. In addition, more-detailed analyses of the 1q24.2 region of interest have demonstrated increased evidence for linkage, by use of dense-map genotyping, and nominally significant evidence for an interaction with other regions of interest, both of which are consistent with an underlying susceptibility gene. Finally, by exclusion mapping, we have been able to exclude, from 92% of the genome, the presence of a locus with an effect size (i.e., locus-specific λS) of 1.52, with the remaining 8% representing the regions of interest on chromosomes 1, 5, 7, 8, and 10, together with an additional region on 21qter.
Table 7.
Correspondence between Regions of Interest from Warren 2 Genome Scan and Results from Other T2D Genome Scans
|
Linkage Evidence in Regions of Interest in the Present Studya |
|||||
| Group (Reference) | 1q24.2 | 5q13 | 5q32 | 8p21-22 | 10q23.3 |
| French European, 148 pedigrees (Vionnet et al. 2000) | APOA2–D1S484 on 1q21-24 | … | D5S410–D5S4 36 on 5q31-33 | … | D10S1655 and D10S212 on 10q26.3 |
| Finnish: | |||||
| 26 multiplex pedigrees from Botnia (Mahtani et al. 1996) | … | … | … | … | … |
| 478 pedigrees, FUSION (Ghosh et al. 2000) | … | … | … | … | D10S185–D10S1267 on 10q23.33-24.32 |
| Israeli Ashkenazim, 267 multiplex pedigrees (Permutt et al. 2001) | … | … | … | … | … |
| American (U.S.), European descent: | |||||
| 42 multigenerational Utah Mormon pedigrees (Elbein et al. 1999) | CRP-APOA2 on 1q21-23 | … | … | D8S136 on 8p21.3, D8S87–D8S532 on 8p12 | … |
| 77 pedigrees (Ehm et al. 2000) | … | D5S1404 on 5q13 | … | … | … |
| African American, 65 pedigrees (Ehm et al. 2000) | … | … | … | … | … |
| Mexican American: | |||||
| 53 pedigrees (Ehm et al. 2000) | … | … | … | … | … |
| 170 sibships (Hanis et al. 1996) | … | … | … | … | … |
| 27 extended pedigrees (Duggirala et al. 1999) | … | … | … | … | D10S587 on 10q26.12 |
| Native American–Pima, 264 nuclear pedigrees (Hanson et al. 1998) | D1S2127 on 1q25.3 | … | … | … | … |
Results are for other studies' genome scans (of ⩾20 pedigrees) for either T2D or, in the case of the studies by Ehm et al. (2000), T2D and impaired glucose homeostatis, for five regions of interest emerging from the Warren 2 genome scan used in the present study. The two regions of interest not represented here (i.e., 7p15 and 8q24) have not been reported in other studies and therefore are not listed. For additional details, see the text.
Methodological Issues in T2D Genome Scans
Any study aiming to detect complex trait linkages needs to take explicit account of the reduction in power that is associated with the anticipated etiological complexity (as reflected by clinical, genetic, and ethnic heterogeneity; oligogenicity; and gene-environmental interactions) (Suarez et al. 1994; Risch and Merikangas 1996). The Warren 2 study was designed to mitigate these factors, in a number of ways. First, the final data set of 573 pedigrees (generated from 687 typed families after poor sample amplification, suspected nonpaternity, and presumed etiological admixture had been taken into account) represents one of the largest resources studied for linkage to T2D. As the power studies indicate, this study size improves the prospects for detection of loci with modest effects (particularly those with locus-specific λS values of 1.25–1.50, which remain within the reach of adequately powered linkage studies [Risch and Merikangas 1996]). Simulation-based power calculations demonstrate that our data set had excellent power to detect either a locus with λS=1.52 at a LOD score of 2 (95% power) or a locus with λS=1.28 at a LOD score of 1 (86% power). Second, we have reduced the risk of etiological heterogeneity by using a wide range of clinical and laboratory-based measurements, to minimize contamination by other known subtypes of diabetes; for example, the relatively low prevalence of anti-GAD antibodies (∼2.5% of all T2D sibs, compared to >7% in unselected T2D groups [Turner et al. 1997]) suggests that the clinical criteria employed were effective at reducing the admixture of autoimmune diabetes. The clinical characteristics of the T2D subjects recruited (table 1) are, save for an earlier AAD, typical of T2D seen in clinical practice in the United Kingdom. The advanced AAD is partly implicit in the ascertainment criteria and suggests successful enrichment for a relatively early-onset, familial T2D phenotype. Third, we have attempted to reduce the extent of any genetic heterogeneity arising from ethnic heterogeneity, by recruiting only subjects of European descent who have at least a three-generation history of British and/or Irish origin.
The question of how statistical significance should be attributed to the results arising from a genome scan such as this remains controversial, but published guidelines recommend that evidence for linkage should not be declared to be significant unless it has a genomewide significance level <.05, equivalent to an allele-sharing LOD score of ⩾3.63 (Lander and Kruglyak 1995; Nyholt 2000). In our own study, none of the regions showing evidence for linkage reached this level of significance (the closest was the 8p21.3-22 locus, with P=.098). By these significance standards, relatively few genome scans for T2D have detected loci of genomewide significance in primary (10-cM, nonstratified) analyses (Hanis et al. 1996; Elbein et al. 1999; Ehm et al. 2000).
There are likely to be several factors contributing to this paucity of strong linkage signals for multifactorial traits, a phenomenon certainly not restricted to analyses of T2D. Foremost is the anticipated etiological complexity of most such traits (see above), which is such that the effect size attributable to any individual susceptibility locus will often be modest. It is clear, for example, from the genome scans conducted for T2D, that, although there is some evidence for replication between data sets (discussed below, in the “Comparison of U.K. Findings with Results of Other Genome Scans for T2D” subsection), there is, for T2D, no major gene that has a status equivalent to that of HLA in type 1 diabetes (Davies et al. 1994). Other contributory factors include limitations of current technical and analytical methods. Thus, the typical 10-cM microsatellite scan fails to capture a considerable proportion of the inheritance information, particularly in small sibships without parents. Although a two-stage strategy (with dense mapping in regions of interest that have arisen from the primary scan) may allow some of this “missing” information to be recovered, this approach may still miss regions if the evidence for linkage has, by chance, been underestimated in the primary scan such that thresholds for dense mapping have not been attained. In addition, undiscovered residual genotyping errors (extremely difficult to eliminate entirely in typical sib-pair data sets when opportunities for Mendelian checking are limited) (Ewen et al. 2000) may have a disproportionate effect on the power to detect linkage, especially linkage to loci of modest effect (Douglas et al. 2000; Abecasis et al. 2001). Finally, since complex-trait susceptibility reflects the interactions of multiple genes, analytical methods capable of modeling such mechanisms may yield improved power over current “locus-by-locus” approaches (Goddard et al. 2001).
It is important to emphasize, with reference to assessment of the statistical significance of our findings, that the primary analyses reported herein (and the related empirical significance calculations) include the entire data set of 573 pedigrees, without any additional stratification or analytical manipulation. In a sib-pair data set such as ours, in which the information extraction from a primary 9-cM scan is relatively modest, additional genotyping planned in these and other families available to us may increase the evidence for linkage for some of these regions. However, given the considerable investment in obtaining the genotype data, as well as the expectation that the power to detect ATLs may be enhanced by further data exploration (albeit at the cost of an increase in type 1 error rates [Lernmark and Ott 1998; Leal and Ott 2000; Cardon and Bell 2001]), we have also initiated stratification and conditional analyses of our data.
Stratification is designed to address latent etiological heterogeneity within the data set (Leal and Ott 2000). Ordered-subset analysis (Hauser et al. 1998) obviates possible arbitrariness implicit in a priori stratification and has previously been applied both to T2D, as part of the FUSION study (Ghosh et al. 1999, 2000), and to alcoholism, in the COGA study (Watanabe et al. 1999a). Conditional analysis, which, in essence, is pedigree stratification by mean allele sharing, is designed to derive additional power by taking explicit account of the oligogenic basis of complex-trait phenotypes (Cox et al. 1999). This approach has contributed to the identification of CAPN10 as a susceptibility gene for T2D in Mexican Americans (Horikawa et al. 2000), by demonstrating significant epistatic interaction between two regions of linkage that were identified in the original genome scan (Hanis et al. 1996; Cox et al. 1999). Some of the most persuasive linkage evidence observed in other recent T2D genome scans has arisen through such exploratory analyses (Mahtani et al. 1996; Hanson et al. 1998; Vionnet et al. 2000).
Comparison of U.K. Findings with Results of Other Genome Scans for T2D
In the absence of highly significant evidence for linkage in most populations studied, the importance of comparing the data from multiple genome scans is well established (Lander and Kruglyak 1995; Hanis et al. 1996; Lernmark and Ott 1998). Replications of linkage results from additional populations, whether as extension/follow-up studies in the same population or as independent genome scans in different populations, provide vital confirmation of the original findings, and guidelines regarding the level of significance necessary in order to declare replication have been suggested (Lander and Kruglyak 1995). The importance of linkage peak location, in addition to peak height, in the context of replication has received recent attention (Hauser and Boehnke 1997; Roberts et al. 1999). Simulation studies have demonstrated that very considerable variation can exist in the location estimate (i.e., the position of the maximum LOD score relative to the true location of the susceptibility gene). Not surprisingly, larger sample sizes, larger gene effects, and denser maps all yield better localization (Hauser and Boehnke 1997; Roberts et al. 1999).
Table 7 summarizes, in the context of our own findings, the results of the major T2D genome scans performed thus far. The clearest evidence for correspondence between multiple data sets is obtained for the 1q region. Our linkage results here concord strongly with results from three published scans (Hanson et al. 1998; Elbein et al. 1999; Vionnet et al. 2000) and with further data from analysis of Amish pedigrees (St. Jean et al. 2000). Elbein et al. (1999) identified a T2D locus between CRP and APOA2 on 1q21-23, in a population of 42 multigenerational pedigrees resident in Utah that are of northern-European ancestry, with an allele-sharing (Kong and Cox 1997) LOD score of 2.260, and a parametric LOD score of 4.295, under a recessive model with an age-dependent penetrance function. In their study of Pima Indians, Hanson et al. (1998) observed evidence for linkage of T2D to both D1S2127 on 1q25.3, with a LOD score of 4.1, in a subset of 55 sib pairs (with age at onset <25 years) taken from their study population of 264 nuclear families, and to D1S1677 on 1q23.3, with a LOD score of 2.5 in a Haseman-Elston analysis comparing sib pairs concordant for T2D versus those discordant for the disease. Most recently, Vionnet et al. (2000) also found evidence for T2D linkage, between D1S498 and D1S484 on 1q21-24, with a maximum-likelihood binomial (MLB)-LOD score of 2.47, in a subset of 57 pedigrees with T2D and BMI <27 kg/m−2, that were selected, by stratification, from their study population of 148 French families of European descent with either T2D or glucose intolerance. Fine mapping of this region with an additional 16 markers increased their evidence for linkage at D1S484/APOA2, with an MLB-LOD score of 2.99.
Further support for the importance of this region comes from analyses in the Goto-Kakizaki (GK) rat, a nonobese animal model of T2D. These analyses have identified a locus on rat chromosome 2 (Galli et al. 1996; Gauguier et al. 1996), in a region syntenic with human 1q21 (Watanabe et al. 1999b); in one GK-rat genome scan (Galli et al. 1996) the locus, Niddm2, was implicated in glucose tolerance (LOD score 4.82), whereas in the second (Gauguier et al. 1996) scan it was most clearly linked to fasting insulin levels.
In our data set of U.K. families, the strongest evidence for linkage in this region was obtained between D1S2799 and D1S452, in our dense map, with a LOD score of 1.98. This locus, together with those detected in the Utah, Pima and French studies, fall within an ∼30-cM interval on 1q. This variation in the localization of the 1q locus is modest compared with both the width of the linkage peaks in these studies (∼32 cM, at P<.05, in the U.K. study) and the imprecision of location estimates as indicated by theoretical analyses (Hauser and Boehnke 1997; Roberts et al. 1999). These observations are therefore entirely consistent with detection of a common susceptibility locus in the different studies. The statistical interactions between this locus and other regions of interest in the U.K. genome scan provide additional support.
In accordance with published guidelines (Lander and Kruglyak 1995), our findings from the dense-map genotyping of chromosome 1q in the U.K. families, in common with the findings from the French study (Vionnet et al. 2000), constitute replication of the linkage on 1q, which previously had been observed in analyses of Utah families (Elbein et al. 1999) and in Pima Indians (Hanson et al. 1998). The observations from the GK-rat scans add further compelling evidence that the 1q locus contains a T2D-susceptibility gene of global importance.
Examination of available human genome sequence (International Human Genome Sequencing Consortium 2001) for the region (from clones with GenBank accession numbers AP002532–AL022310; ∼20 Mb), by publicly available (ENSEMBL [see the Project Ensembl web site]) and in-house (GANESH [see the Genome Integrated Force in Type 2 Diabetes web site]) genome-annotation tools, identifies 133 known genes, including several strong candidates, such as the gamma subunit of the retinoid X receptor (RXRG [MIM 180247]), lamin A (LMNA [MIM 150330]) and the potassium inwardly rectifying channel subfamily J member 9 (KCNJ9 [LocusLink accession number 3765]). The region also contains a further 145 putative transcripts with homology to known genes, in addition to as many as 393 further “potential expressed” sequences based on in silico prediction, mouse genome comparison, or expressed-sequence-tag similarity alone. We are engaged in further fine-scale mapping and positional candidate analysis of this region.
Several other regions highlighted in the U.K. scan correspond to regions identified in other T2D studies. Evidence for linkage on chromosome 5q has been demonstrated previously in populations of European descent that are from the United States (Ehm et al. 2000) and France (Vionnet et al. 2000). In their study population of 77 nuclear families containing 198 sib pairs affected with either T2D or impaired glucose homeostasis, Ehm et al. (2000) found evidence for linkage to D5S1404 on 5q13, with a LOD score of 3.26; however, they were unable to reproduce this result in a second sample of U.S. individuals of European descent. D5S1404 falls ∼4 cM telomeric to D5S647, the 5q13 marker that, in our study, shows maximum evidence for linkage. The genome scan of 148 French families (Vionnet et al. 2000) detected maximum evidence for linkage, in their entire data set, on 5q31-33, between D5S410 and D5S436 (MLB-LOD score 1.52), in the same location as in the U.K. study. In addition, a genome scan of 158 French families, for genes predisposing to obesity (Hager et al. 1998), detected evidence for linkage between BMI and markers D5S647 on 5q13 (maximum LOD score 1.57), and D5S436 on 5q32 (maximum LOD score 1.19), the same markers that in our study showed evidence for linkage. Taken together, these results suggest the possibility of loci on chromosome 5q that are responsible for a broader metabolic dysfunction, underlying both T2D and obesity.
The study by Elbein et al. (1999), of Utah families of European descent, found modest evidence for linkage to two regions on 8p. The first region, on 8p12 (D8S87–D8S532; LOD score 1.365), falls ∼25 cM(K) (K = Kosambi distances, according to the Marshfield map (see the web site of the Center for Medical Genetics, Marshfield Medical Research Foundation) from the locus that we found on 8p21-22; however, the second region, on 8p21.3 (D8S136; LOD score 1.348), falls within 5 cM(K). Given the importance of disordered lipid metabolism as a feature of T2D and the metabolic syndrome, the gene for lipoprotein lipase (LPL [MIM 238600]) represents one plausible regional candidate. This region on 8p21-22 is currently being subjected to fine-scale mapping using the family resources described in the present study.
Three previous T2D studies in humans have obtained evidence for linkage to 10q. In their study population of 27 extended Mexican American families, Duggirala et al. (1999) observed evidence for linkage between a locus close to D10S587 and both T2D (LOD score 2.88) and age at onset of T2D (LOD score 3.75). This region on 10q26.12 is ∼45 cM telomeric to the region on 10q23.3, where the present study has found maximum evidence for linkage (LOD score 1.99). The French genome scan (Vionnet et al. 2000) has reported evidence for linkage to D10S1655 (MLB-LOD score 1.59) and D10S212 (MLB-LOD score 1.44) on 10q26.3 (∼50 cM(K) [Center for Medical Genetics, Marshfield Medical Research Foundation] telomeric to the peak in our study), in subsets of 147 and 65 families, respectively. The possibility that these observations regarding 10q26 and 10q23 constitute evidence for the same T2D-susceptibility locus cannot be discounted entirely, given the substantial variation possible in location estimates (Hauser and Boehnke 1997; Roberts et al. 1999), but this seems unlikely. The FUSION study (Ghosh et al. 2000) detected only nominal evidence for linkage to the 10q region in its primary analysis of a complete data set of 478 Finnish pedigrees (containing 719 affected sib pairs); however, an ordered-subset analysis, with pedigrees ranked on the basis of the mean value of the fasting insulin:fasting glucose ratio (IR1), obtained more-substantial evidence for linkage, between D10S185 and D10S1267 on 10q23.33-24.32 (maximum LOD score 3.12), in the 40 families with the lowest IR1 value. This region coincides with the locus on 10q23.3, where the present study has found evidence for linkage.
Studies in the GK rat provide circumstantial support for the locus on 10q23.3, in that both genome scans with this model (Galli et al. 1996; Gauguier et al. 1996) map a locus for poststimulation glycemia (either Niddm1 or Nidd/gk1) to a rat chromosome 1 region syntenic with human 10q23-26 (Galli et al. 1999; Watanabe et al. 1999b; Kaisaki et al. 2000). Subsequent dissection of the Niddm1 region in congenic strains has identified two contributing loci, Niddm1b and Niddm1i, each containing at least one gene (Galli et al. 1999). Further analyses have mapped Niddm1b to a 1-cM region containing the gene for insulin-degrading enzyme (IDE [MIM 146680]) (Fakhrai-Rad et al. 2000). Two amino acid substitutions in IDE, specific to the GK-rat Niddm1b allele, were found in the congenic strains and, when present together, conferred postprandial hyperglycemia, reduced insulin degradation in isolated muscle cells, and other diabetes-related phenotypes (Fakhrai-Rad et al. 2000). In previous studies, human IDE had been mapped to 10q23-25 (Affholter et al. 1990; Espinosa et al. 1991) and, recently, has been more precisely localized, to 10q23.33, in the draft human genome sequence (International Human Genome Sequencing Consortium 2001). The evidence for linkage, in the neighborhood of the human IDE locus, to T2D and related traits in Mexican American and French populations has been noted elsewhere (Galli et al. 1999; Fakhrai-Rad et al. 2000), although (see above) IDE lies ∼45 cM centromeric. IDE falls within the interval between D10S1765 and D10S185, which lies at the peak of linkage in our study. The observations from the FUSION ordered subset, together with the recent studies in Niddm1b-congenic rat strains and the synteny between rat and human genomes in this region, substantially strengthen the candidacy of IDE for the 10q23.3 locus. We are currently pursuing fine-scale marker typing and candidate-gene studies, to further characterize this locus on 10q23.3.
To the best of our knowledge, the remaining two regions of interest in our genome scan (i.e., 7p15.3 and 8q24.2) are novel and have not been highlighted by previous studies, although two groups (Ehm et al. 2000; Vionnet et al. 2000) observed modest evidence for linkage on 8p22, ⩾50 cM from the peak of linkage in the present study. Further studies will be necessary in order to determine whether these are false-positive results or represent genuine evidence for linkage to loci of small effect.
Finally, we note that, in four chromosomal locations strongly implicated in other genome scans, the present study's U.K. genome scan has failed to detect even nominal evidence for linkage to T2D. We detected no positive allele sharing in that region of chromosome 2qter (particularly D2S125) which shows significant evidence for linkage in Mexican Americans (Hanis et al. 1996; Horikawa et al. 2000) and were able to exclude this region for λS⩾1.28. We did detect positive, but nonsignificant, allele sharing ∼30 cM centromeric to D2S125 (LOD score 0.48; P=.068), which could be excluded for λS⩾1.52 only. We found no positive allele sharing at the 12q locus detected in Finns (Mahtani et al. 1996) and U.S. individuals of European descent (Bowden et al. 1997) and were able to exclude this region for λS⩾1.28. A second locus on 12q, observed in U.S. individuals of European descent (Bektas et al. 1999; Ehm et al. 2000) and ∼50 cM centromeric to that in the Finnish study, showed positive, but nonsignificant, allele sharing in our study (LOD score 0.49; P=.066) and could be excluded for λS⩾1.52. The locus detected on 20q in U.S. individuals of European descent (Bowden et al. 1997; Ji et al. 1997) and in Finns (Ghosh et al. 1999, 2000) showed no evidence of positive allele sharing our study and could be excluded for λS⩾1.28. These data for chromosomes 12q and 20q extend to the full cohort the findings previously reported for these regions in a subset of the same pedigrees (Frayling et al. 2000).
Concluding Remarks
The results of this genome scan of 573 pedigrees provide further confirmation that no single locus plays a major role in T2D susceptibility. Given the modest degree of familial aggregation (λS∼3.5) associated with T2D, linkage-based gene-discovery studies (particularly if limited in size) are often operating close to the limits of detection for the locus-specific effect sizes anticipated. Nevertheless, two recent developments in particular provide substantial encouragement. The first of these two developments is the availability of published genome-scan data from multiple ethnic groups and the increasing trend for submission of genotypes to central data repositories, allowing combined analyses of very large data sets (examples include the European Genome Integrated Force in Type 2 Diabetes [GIFT] consortium and the International Type 2 Diabetes Linkage Analysis Consortium; also see Boehnke and The International Type 2 Diabetes Linkage Analysis Consortium 1998). The second development is the availability of novel analytical approaches, such as conditional analysis and ordered-subset analysis, which aim to improve, through detailed exploration of available data sets, the power to obtain evidence of linkage. Confirmation of hypotheses arising from such exploratory approaches in additional data sets (facilitated by access to large combined data repositories) should make it possible to address concerns about the type 1–error inflation associated with the unrestricted proliferation of exploratory analyses.
The U.K. genome-scan data presented here contribute to the evolution of our understanding of T2D etiology, by providing replication (according to published guidelines) of a linkage previously reported for chromosome 1q. In addition, regions of interest identified, in other European data sets, on chromosomes 5q, 8p, and 10q receive strong support from our data. We expect these data to accelerate efforts to positionally clone the susceptibility genes mapping to these regions and, thereby, to advance the understanding and treatment of this major worldwide health concern.
Acknowledgments
The work described herein was funded by Diabetes UK (formerly the British Diabetic Association), primarily via the Warren Bequest; by the National Lotteries Charities Board; and by the European Union (Framework Programme 5–GIFT Consortium grant QLG2-CT-1999-00546). We also acknowledge infrastructure support by the Wellcome Trust to the Wellcome Trust Centre for Human Genetics, Oxford. Ascertainment of the clinical resources would not have been possible without the efforts of many U.K. diabetes physicians, research nurses, and general practitioners and the contributions of the families themselves. We wish to highlight the contribution of the late Prof. Robert Turner, a founding member of the Warren 2 consortium. We also thank Alicia Vaux, Paul Beerling, and Prof. Tony Monaco (all of the Wellcome Trust Centre for Human Genetics, Oxford), Dr. Moira Murphy (Diabetes UK), Drs. Ros Packer and Bryan Bolton (both of the European Collection of Cell Cultures, Salisbury, U.K.), and Prof. Gianfranco Bottazzo (Royal London Hospital, London) for their contributions. We thank Goncalo Abecasis (Wellcome Trust Centre for Human Genetics, Oxford), for simulation code; Mr. William Duren (Department of Biostatistics, School of Public Health, University of Michigan, Ann Arbor), for the software used to perform ordered-subset analysis; and DECODE GENETICS (Reykjavik), for access to ALLEGRO. L.R.C. was supported in part by National Institutes of Health grant EY-12562. S.M. is a Diabetes (UK) R.D. Lawrence Fellow.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics (for genetic linkage maps)
- Division of Statistical Genetics (Department of Human Genetics, University of Pittsburgh) web site, ftp://watson.hgen.pitt.edu/pub/ (for RECODE)
- Généthon, http://www.genethon.fr/ (for genetic linkage maps)
- Genome Integrated Force in Type 2 Diabetes, http://www.gift.med.ic.ac.uk/ (for GANESH and GIFT Consortium)
- International Type 2 Diabetes Linkage Analysis Consortium, http://www.sfbr.org/external/diabetes/ [DOI] [PMC free article] [PubMed]
- Lab of Statistical Genetics (Rockefeller University) web site, ftp://linkage.rockefeller.edu/software/simulate/ (for SIMULATE)
- LocusLink, http://www.ncbi.nlm.nih.gov/LocusLink/ (for KCNJ9 [accession number 3765])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for T2D [MIM 125853], CAPN10 [MIM 605286], RXRG [MIM 180247], LMNA [MIM 150330], LPL [MIM 238600], and IDE [MIM 146680])
- Project Ensembl, http://www.ensembl.org/ (for ENSEMBL)
- Warren 2 Project Information (Wellcome Trust Centre for Human Genetics, Oxford), http://www.well.ox.ac.uk/warren2/ (for Warren 2–scan marker information)
References
- Abecasis GR, Cherny SS, Cardon LR (2001) The impact of genotyping error on family-based analysis of quantitative traits. Eur J Hum Genet 9:130–134 [DOI] [PubMed] [Google Scholar]
- Affholter JA, Hsieh CL, Francke U, Roth RA (1990) Insulin-degrading enzyme: stable expression of the human complementary DNA, characterization of its protein product, and chromosomal mapping of the human and mouse genes. Mol Endocrinol 4:1125–1135 [DOI] [PubMed] [Google Scholar]
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES (2000) The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26:76–80 [DOI] [PubMed] [Google Scholar]
- Bektas A, Suprenant ME, Wogan LT, Plengvidhya N, Rich SS, Warram JH, Krolewski AS, Doria A (1999) Evidence of a novel type 2 diabetes locus 50 cM centromeric to NIDDM2 on chromosome 12q. Diabetes 48:2246–2251 [DOI] [PubMed] [Google Scholar]
- Bingley PJ, Bonifacio E, Williams AJK, Genovese S, Bottazzo GF, Gale EAM (1997) Prediction of IDDM in the general population: strategies based on combinations of autoantibody markers. Diabetes 46:1701–1710 [DOI] [PubMed] [Google Scholar]
- Boehnke M, Cox NJ (1997) Accurate inference of relationships in sib-pair linkage studies. Am J Hum Genet 61:423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehnke M, The International Type 2 Diabetes Linkage Analysis Consortium (1998) Lessons learned in a combined linkage analysis (24 datasets, >2000 families) of type 2 diabetes on chromosome 20. Am J Hum Genet Suppl 63:A282 [Google Scholar]
- Bowden DW, Sale M, Howard TD, Qadri A, Spray BJ, Rothschild CB, Akots G, Rich SS, Freedman BI (1997) Linkage of genetic markers on human chromosomes 20 and 12 to NIDDM in Caucasian sib pairs with a history of diabetic nephropathy. Diabetes 46:882–886 [DOI] [PubMed] [Google Scholar]
- Brownstein MJ, Carpten JD, Smith JR (1996) Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. BioTechniques 20:1004–1010 [DOI] [PubMed] [Google Scholar]
- Cardon LR, Bell JI (2001) Association study designs for complex diseases. Nat Rev Genet 2:91–99 [DOI] [PubMed] [Google Scholar]
- Cox NJ, Frigge M, Nicolae DL, Concannon P, Hanis CL, Bell GI, Kong A (1999) Loci on chromosomes 2 (NIDDM1) and 15 interact to increase susceptibility to diabetes in Mexican Americans. Nat Genet 21:213–215 [DOI] [PubMed] [Google Scholar]
- Davies JL, Kawaguchi Y, Bennett ST, Copeman JB, Cordell HJ, Pritchard LE, Reed PW, Gough SCL, Jenkins SC, Palmer SM, Balfour KM, Rowe BR, Farrall M, Barnett AH, Bain SC, Todd JA (1994) A genome-wide search for human type 1 diabetes susceptibility genes. Nature 371:130–136 [DOI] [PubMed] [Google Scholar]
- Dib C, Fauré S, Fizames C, Samson D, Druout N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Douglas JA, Boehnke M, Lange K (2000) A multipoint method for detecting genotyping errors and mutations in sibling-pair linkage data. Am J Hum Genet 66:1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, O’Connell P, Stern MP (1999) Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet 64:1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehm MG, Karnoub MC, Sakul H, Gottschalk K, Holt DC, Weber JL, Vaske D, Briley D, Briley L, Kopf J, McMillen P, Nguyen Q, Reisman M, Lai EH, Joslyn G, Shepherd NS, Bell C, Wagner MJ, Burns DK, American Diabetes Association GENNID Study Group (2000) Genomewide search for type 2 diabetes susceptibility genes in four American populations. Am J Hum Genet 66:1871–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein SC, Hoffman MD, Teng K, Leppert MF, Hasstedt SJ (1999) A genome-wide search for type 2 diabetes susceptibility genes in Utah Caucasians. Diabetes 48:1175–1182 [DOI] [PubMed] [Google Scholar]
- Espinosa R III, Lemons RS, Perlman RK, Kuo WL, Rosner MR, Le Beau MM (1991) Localization of the gene encoding insulin-degrading enzyme to human chromosome 10, bands q23-q25. Cytogenet Cell Genet 57:184–186 [DOI] [PubMed] [Google Scholar]
- Ewen KR, Bahlo M, Treloar SA, Levinson DF, Mowry B, Barlow JW, Foote SJ (2000) Identification and analysis of error types in high-throughput genotyping. Am J Hum Genet 67:727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhrai-Rad H, Nikoshkov A, Kamel A, Fernström M, Zierath JR, Norgren S, Luthman H, Galli J (2000) Insulin-degrading enzyme identified as a candidate diabetes susceptibility gene in GK rats. Hum Mol Genet 9:2149–2158 [DOI] [PubMed] [Google Scholar]
- Frayling TM, McCarthy MI, Walker M, Levy JC, O’Rahilly S, Hitman GA, Subba Rao PV, Bennett AJ, Jones EC, Menzel S, Ellard S, Hattersley AT (2000) No evidence for linkage at candidate type 2 diabetes susceptibility loci on chromosomes 12 and 20 in UK Caucasians. J Clin Endocrinol Metab 85:853–857 [DOI] [PubMed] [Google Scholar]
- Galli J, Fakhrai-Rad H, Kamel A, Marcus C, Norgren S, Luthman H (1999) Pathophysiological and genetic characterization of the major diabetes locus in GK rats. Diabetes 48:2463–2470 [DOI] [PubMed] [Google Scholar]
- Galli J, Li LS, Glaser A, Östenson CG, Jiao H, Fakhrai-Rad H, Jacob HJ, Lander ES, Luthman H (1996) Genetic analysis of non-insulin dependent diabetes mellitus in the GK rat. Nat Genet 12:31–37 [DOI] [PubMed] [Google Scholar]
- Gauguier D, Froguel P, Parent V, Bernard C, Bihoreau MT, Portha B, James MR, Penicaud L, Lathrop M, Ktorza A (1996) Chromosomal mapping of genetic loci associated with non-insulin dependent diabetes in the GK rat. Nat Genet 12:38–43 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Hauser ER, Valle T, Magnuson VL, Erdos MR, Langefeld CD, et al (1999) Type 2 diabetes: evidence for linkage on chromosome 20 in 716 Finnish affected sib pairs. Proc Natl Acad Sci USA 96:2198–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Valle TT, Hauser ER, Magnuson VL, Langefeld CD, Ally DS, et al (2000) The Finland–United States Investigation of non–insulin-dependent diabetes mellitus genetics (FUSION) study. I. An autosomal genome scan for genes that predispose to type 2 diabetes. Am J Hum Genet 67:1174–1185 [PMC free article] [PubMed] [Google Scholar]
- Goddard KAB, Witte JS, Suarez BK, Catalona WJ, Olson JM (2001) Model-free linkage analysis with covariates confirms linkage of prostate cancer to chromosomes 1 and 4. Am J Hum Genet 68:1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göring HHH, Ott J (1997) Relationship estimation in affected sib pair analysis of late-onset diseases. Eur J Hum Genet 5:69–77 [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong CA (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 [DOI] [PubMed] [Google Scholar]
- Hager J, Dina C, Francke S, Dubois S, Houari M, Vatin V, Vaillant E, Lorentz N, Basdevant, Clement K, Guy-Grand B, Froguel P (1998) A genome-wide scan for human obesity genes reveals a major susceptibility locus on chromosome 10. Nat Genet 20:304–308 [DOI] [PubMed] [Google Scholar]
- Hani EH, Boutin P, Durand E, Inoue H, Permutt MA, Velho G, Froguel P (1998) Missense mutations in the pancreatic islet beta cell inwardly rectifying K+ channel gene (KIR6.2/BIR): a meta-analysis suggests a role in the polygenic basis of type II diabetes mellitus in Caucasians. Diabetologia 41:1511–1515 [DOI] [PubMed] [Google Scholar]
- Hani EH, Stoffers DA, Chèvre J-C, Durand E, Stanojevic V, Dina C, Habener JF, Froguel P (1999) Defective mutations in the insulin promoter factor-1 (IPF-1) gene in late-onset type 2 diabetes mellitus. J Clin Invest 104:R41–R48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanis CL, Boerwinkle E, Chakraborty R, Ellsworth DL, Concannon P, Stirling B, Morrison VA, et al (1996) A genome-wide search for human non-insulin-dependent (type 2) diabetes genes reveals a major susceptibility locus on chromosome 2. Nat Genet 13:161–171 [DOI] [PubMed] [Google Scholar]
- Hanson RL, Ehm MG, Pettitt DJ, Prochazka M, Thompson DB, Timberlake D, Foroud T, Kobes S, Baier L, Burns DK, Almasy L, Blangero J, Garvey WT, Bennett PH, Knowler WC (1998) An autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in Pima Indians. Am J Hum Genet 63:1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser ER, Boehnke M (1997) Confirmation of linkage results in affected-sib-pair linkage analysis for complex genetic traits. Am J Hum Genet Suppl 61:A278 [Google Scholar]
- Hauser ER, Watanabe RM, Duren WL, Boehnke M, The Fusion Study Group (1998) Stratified linkage analysis of complex genetic traits using related covariates. Am J Hum Genet Suppl 63:A45 [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, et al (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26:163–175 [DOI] [PubMed] [Google Scholar]
- Huxtable SJ, Saker PJ, Haddad L, Walker M, Frayling TM, Levy JC, Hitman GA, O'Rahilly S, Hattersley AT, McCarthy MI (2000) Analysis of parent-offspring trios provides evidence for linkage and association between the insulin gene and type 2 diabetes mediated exclusively through paternally transmitted class III variable number tandem repeat alleles. Diabetes 49:126–130 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Ji L, Malecki M, Warram JH, Yang Y, Rich SS, Krolewski AS (1997) New susceptibility locus for NIDDM is localized to human chromosome 20q. Diabetes 46:876–881 [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Kadowaki H, Mori Y, Tobe K, Sakuta R, Suzuki Y, Tanabe Y, Sakura H, Awata T, Goto Y, Hayakawa T, Matsuoka K, Kawamori R, Kamada T, Horai S, Nonaka I, Hagura R, Akanuma Y, Yazaki Y (1994) A subtype of diabetes mellitus associated with a mutation of mitochondrial DNA. N Engl J Med 330:962–968 [DOI] [PubMed] [Google Scholar]
- Kaisaki PJ, Rouard M, Danoy PA, Wallis RH, Collins SC, Rice M, Levy ER, Lathrop M, Bihoreau MT, Gauguier D (2000) Detailed comparative gene map of rat chromosome 1 with mouse and human genomes and physical mapping of an evolutionary chromosomal breakpoint. Genomics 64:32–43 [DOI] [PubMed] [Google Scholar]
- Köbberling J, Tillil H (1982). Empirical risk figures for first degree relatives of non-insulin-dependent diabetics. In: Köbberling J, Tattersall R (eds) The genetics of diabetes mellitus. Academic Press, London, pp 201–209 [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guideline for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Leal SM, Ott J (2000) Effects of stratification in the analysis of affected-sib-pair data: benefits and costs. Am J Hum Genet 66:567–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lernmark Å, Ott J (1998) Sometimes it’s hot, sometimes it’s not. Nat Genet 19:213–214 [DOI] [PubMed] [Google Scholar]
- Mahtani MM, Widén E, Lehto M, Thomas J, McCarthy M, Brayer J, Bryant B, Chan G, Daly M, Forsblom C, Kanninen T, Kirby A, Kruglyak L, Munnelly K, Parkkonen M, Reeve-Daly MP, Weaver A, Brettin T, Duyk G, Lander ES, Groop LC (1996) Mapping of a gene for NIDDM associated with an insulin secretion defect by a genome scan in Finnish families. Nat Genet 14:90–95 [DOI] [PubMed] [Google Scholar]
- McCarty D, Zimmet P (1994) Diabetes 1994 to 2010: global estimates and projections. Bayer AG, Leverkusen, Germany; and International Diabetes Institute, Melbourne [Google Scholar]
- Nyholt DR (2000) All LODS are not created equal. Am J Hum Genet 67:282–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1999) Analysis of human genetic linkage. The Johns Hopkins University Press, Baltimore [Google Scholar]
- Permutt MA, Wasson JC, Suarez BK, Lin J, Thomas J, Meyer J, Lewitzky S, Rennich JS, Parker A, DuPrat L, Maruti S, Chayen S, Glaser B (2001) A genome scan for type 2 diabetes susceptibility loci in a genetically isolated population. Diabetes 50:681–685 [DOI] [PubMed] [Google Scholar]
- Petersen JS, Hejnaes KR, Moody A, Karlsen AE, Marshall MO, Hoier-Madsen M, Boel E, Michelsen BK, Dyrberg T (1994) Detection of GAD65 antibodies in diabetes and other autoimmune diseases using a simple radioligand assay. Diabetes 43:459–467 [DOI] [PubMed] [Google Scholar]
- Reaven G (1988) Role of insulin resistance in human disease. Diabetes 37:1595–1607 [DOI] [PubMed] [Google Scholar]
- Rich SS (1990) Mapping genes in diabetes: genetic epidemiological perspective. Diabetes 39:1315–1319 [DOI] [PubMed] [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. I. Multilocus models. Am J Hum Genet 46:222–228 [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS (1999) Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet 65:876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Jean P, Hsueh W-C, Mitchell B, Ehm M, Wagner M, Burns D, Shuldiner AR (2000) Association between diabetes, obesity, glucose and insulin levels in the Old Order Amish and SNPs on 1q21-23. Am J Hum Genet 67 Suppl 2:332 [Google Scholar]
- Suarez BK, Hampe CL, van Eerdewegh P (1994) Problems of replicating linkage claims in psychiatry. In: Gershon ES, Cloninger CR (eds) Genetic approaches to mental disorders. American Psychiatric Press, Washington, DC, pp 23–46 [Google Scholar]
- Suarez BK, Hodge SE (1979) A simple method to detect linkage for rare recessive diseases: an application to clinical diabetes. Clin Genet 15:126–136 [DOI] [PubMed] [Google Scholar]
- Tuomi T, Groop LC, Zimmet PZ, Rowley MJ, Knowles W, Mackay IR (1993) Antibodies to glutamic acid decarboxylase reveal latent autoimmune diabetes mellitus in adults with a non-insulin-dependent onset of disease. Diabetes 42:359–362 [DOI] [PubMed] [Google Scholar]
- Turner R, Stratton I, Horton V, Manley S, Zimmet P, Mackay IR, Shattock M, Bottazzo GF, Holman R (1997) UKPDS 25: auto-antibodies to islet-cell cytoplasm and glutamic acid decarboxylase for prediction of insulin requirement in type 2 diabetes: UK Prospective Diabetes Study Group. Lancet 350:1288–1293 [DOI] [PubMed] [Google Scholar]
- Vionnet N, Hani EH, Dupont S, Gallina S, Francke S, Dotte S, De Matos F, Durand E, Lepêtre F, Lecoeur C, Gallina P, Zekiri L, Dina C, Froguel P (2000) Genomewide search for type 2 diabetes–susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2–diabetes locus on chromosome 1q21-q24. Am J Hum Genet 67:1470–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe RM, Ghosh S, Birznieks G, Duren WL, Mitchell BD (1999a) Application of an ordered subset analysis approach to the genetics of alcoholism. Genet Epidemiol 17 Suppl 1:S385–S390 [DOI] [PubMed] [Google Scholar]
- Watanabe RM, Ghosh S, Langefeld CD, Valle TT, Hauser ER, Magnuson VL, Mohlke KL, et al (2000) The Finland–United States investigation of non–insulin-dependent diabetes mellitus genetics (FUSION) study. II. An autosomal genome scan for diabetes-related quantitative-trait loci. Am J Hum Genet 67:1186–1200 [PMC free article] [PubMed] [Google Scholar]
- Watanabe TK, Bihoreau M-T, McCarthy LC, Kiguwa SL, Hishigaki H, Tsuji A, Brown J, et al (1999b) A radiation hybrid map of the rat genome containing 5,255 markers. Nat Genet 22:27–36 [DOI] [PubMed] [Google Scholar]
- World Health Organisation Study Group (1985) Diabetes Mellitus. WHO Tech Rep ser no 727. World Health Organization, Geneva [PubMed] [Google Scholar]