Abstract
Autism is characterized by impairments in reciprocal communication and social interaction and by repetitive and stereotyped patterns of activities and interests. Evidence for a strong underlying genetic predisposition comes from twin and family studies, although susceptibility genes have not yet been identified. A whole-genome screen for linkage, using 83 sib pairs with autism, has been completed, and 119 markers have been genotyped in 13 candidate regions in a further 69 sib pairs. The addition of new families and markers provides further support for previous reports of linkages on chromosomes 7q and 16p. Two new regions of linkage have also been identified on chromosomes 2q and 17q. The most significant finding was a multipoint maximum LOD score (MLS) of 3.74 at marker D2S2188 on chromosome 2; this MLS increased to 4.80 when only sib pairs fulfilling strict diagnostic criteria were included. The susceptibility region on chromosome 7 was the next most significant, generating a multipoint MLS of 3.20 at marker D7S477. Chromosome 16 generated a multipoint MLS of 2.93 at D16S3102, whereas chromosome 17 generated a multipoint MLS of 2.34 at HTTINT2. With the addition of new families, there was no increased allele sharing at a number of other loci originally showing some evidence of linkage. These results support the continuing collection of multiplex sib-pair families to identify autism-susceptibility genes.
Introduction
Autism (MIM 209850) is a neurodevelopmental disorder that affects at least 5 in 10,000 individuals (Fombonne 1999). Onset is in the first 3 years of life, and the core deficits comprise qualitative impairments in reciprocal communication and social interaction, as well as repetitive and stereotyped behaviors and interests. Autism is the prototypical pervasive developmental disorder (PDD); the PDDs are a group of related disorders that includes atypical autism, Asperger syndrome, and “pervasive developmental disorder not otherwise specified” (PDD NOS). The rate of autism amongst siblings of singleton probands is higher, at ∼3%, than that in the general population; the rate is further elevated when other PDDs are included (Bailey et al. 1998; Szatmari et al. 1998). In three same-sex twin studies, concordance for autism in MZ pairs varied between 36% and 91%, compared with 0% in DZ pairs (Folstein and Rutter 1977; Steffenburg 1991; Bailey et al. 1995), leading to an estimated heritability of >90% (Bailey et al. 1995). Two twin studies (Folstein and Rutter 1977; Bailey et al. 1995) have also found that the behavioral phenotype extends beyond autism and other PDDs to include related, but milder, abnormalities in social behavior and language, a finding replicated in family studies (Bailey et al. 1998; Szatmari et al. 1998; Pickles et al. 2000). The sibling recurrence risks and twin concordance findings are incompatible with a Mendelian mode of inheritance, and statistical modeling suggests that 3 or 4 loci are probably implicated, although as many as 15 loci may be involved (Pickles et al. 1995).
The high concordance rates in MZ twins for autism and the broader phenotype (Folstein and Rutter 1977; Bailey et al. 1995) implicate genetic influences in the vast majority of idiopathic cases. As yet, however, there is no consensus about the neuropathological and neurophysiological basis of autism, and, thus, there are no very strong candidate genes. Autism is associated with recognized medical disorders in a small minority of cases, and these associations might indicate genes that are potentially of wider significance. The strongest specific associations are with fragile-X syndrome and tuberous sclerosis. Hallmayer et al. (1994) found no evidence that the fragile-X locus is implicated in idiopathic cases. Similarly there is no evidence that previously undetected tuberous sclerosis accounts for a significant proportion of cases, although TSC1 and TSC2 cannot yet be excluded as possible susceptibility loci (Smalley 1998; Mbarek et al. 1999). A wide range of chromosomal abnormalities are found in up to 5% of cases of autism (Gillberg 1998), the most consistent association being with 15q11-q13 interstitial duplications or a supernumerary pseudodicentric chromosome 15 (inv-dup[15]) that overlaps the Prader-Willi Angelman critical region (PWACR) of proximal 15q (see Lamb et al. [2000] for a review).
Some 75% of individuals with autism are male, raising the possibility that genes on the X chromosome might also be implicated in susceptibility. Hallmayer et al. found modest evidence of linkage for autism on the X chromosome (Hallmayer et al. 1996), with a maximum LOD score (MLS) of 1.24 at DXS424 on Xq24. Philippe et al. (1999) also reported a two-point MLS of 0.89 at DXS996 on Xp22.3. Skuse has suggested that there may be a protective imprinted locus on the X chromosome (Skuse 2000), although a higher threshold for phenotypic expression in females could also explain the observed sex ratio. Reports of excess paternal transmissions on chromosome 7 and maternal/paternal differences in sharing on chromosome 15 have also led to speculation that there may be a parent-of-origin effect (Ashley-Koch et al. 1999, 2000; Sutcliffe and CLSA 2000).
The paucity of functional and positional candidate genes for autism has led most research groups to follow a strategy of a genomewide screen for linkage using affected relative–pair families, followed by a positional candidate-gene approach. The published genome screens have found preliminary convergent evidence for linkage in several genomic regions (for reviews, see Lamb et al. [2000] and Turner et al. [2000]), with a region on chromosome 7q showing increased allele sharing in all screens. Nevertheless, the largest genome screen to date (Risch et al. 1999), which analyzed 97 independent affected sib pairs with 519 microsatellite markers and an additional 50 sib pairs with a subset of 149 markers in candidate regions, failed to generate a LOD score with genomewide significance. This study did, however, provide modest support for loci on chromosomes 7 and 17, with a multipoint MLS of 0.93 at D7S1804 and 1.21 at D17S1584.
Some evidence for linkage has also been found in the Prader-Willi–Angelman critical region (PWACR) on chromosome 15 (Bass et al. 2000), with a multipoint MLS of 1.31 at D15S217. The γ aminobutyric acid receptor subunit β 3 (GABRB3) in this region has been studied by several groups, but the findings are not consistent (Cook et al. 1998; Maestrini et al. 1999; Salmon et al. 1999; Martin et al. 2000). Sequencing of E6-AP ubiquitin-protein ligase (UBE3A), which is also located in cytogenetic band 15q11-13, has not detected functional mutations (Veenstra-VanderWeele et al. 1999). Studies of the serotonin transporter as a candidate gene for autism have also proved inconclusive, with association found in different populations with both the long and short variant of an insertion/deletion polymorphism (Cook et al. 1997; Klauck et al. 1997; Persico et al. 2000; Vicente et al. 2000), as well as with neither variant (Maestrini et al. 1999; Zhong et al. 1999).
Elsewhere (IMGSAC 1998), we have reported analysis of 354 markers typed in 39 multiplex families and a subset of 175 markers typed in 99 families. Here, we present a complete genome screen for 83 sib pairs from the original 99 families (IMGSAC 1998), using 349 markers, and the results of genotyping at 13 putative susceptibility loci where linkage was indicated from the completed genome screen. A subset of 74 markers used in the original genome screen and 45 new markers have now been genotyped in a total of 152 sib pairs. The data have also been analyzed for evidence of linkage disequilibrium, including an examination of maternal and paternal sharing.
Material and Methods
Phenotypic Assessment
The identification of families and the assessment methods used by the IMGSAC has been described in detail elsewhere (IMGSAC 1998). In brief, in families passing an initial screen, parents were given the Autism Diagnostic Interview-Revised (ADI-R) (Lord et al. 1994) and the Vineland Adaptive Behavior Scales (Sparrow et al. 1984). Potential cases, who were all ⩾4 years old, were also assessed using either the Autism Diagnostic Observation Schedule (ADOS) (Lord et al. 1994) or ADOS-Generic (ADOS-G) (Lord et al. 2000). When possible, psychometric evaluation was conducted using Raven's progressive matrices (Raven 1989) or the Mullen Scales of Early Learning (Mullen 1995), and the British Picture Vocabulary Scale (Dunn et al. 1982) or the Peabody Picture Vocabulary Test III (Dunn and Dunn 1997) or an appropriate translation; other tests were administered if clinically indicated. A history of language delay was defined as no single words before age 24 mo and/or no phrase speech before age 33 mo. Individuals who lost language that was not regained by these ages were also considered to have language delay. Individuals were assigned case status in a hierarchical fashion: they were designated as “case type 1” if they had a clinical diagnosis of autism, met ADI-R (Lord et al. 1994) and ADOS (Lord et al. 1994) or ADOS-G (Lord et al. 2000) algorithm criteria for autism, and had a history of language delay and a performance IQ of ⩾35. Individuals were designated as “case type 2” if they had a clinical diagnosis of autism, atypical autism, Asperger syndrome, or PDD NOS and met at least ADOS-G criteria for PDD; there was no requirement for a history of language delay, and individuals were allowed to fall one point below threshold on one behavioral domain of the ADI-R. Case type 2 was subdivided into case type 2a individuals, who had a performance IQ of ⩾35 (n=97), and case type 2b individuals, who had a performance IQ either measured at or inferred to be <35 (n=35). Individuals were designated as “case type 3” if they had a clinical diagnosis of autism or another PDD and either met ADI-R criteria for autism or fell one point below threshold in one behavioral domain but failed to meet ADOS-G criteria for PDD. The criteria for each of these case types are listed in table 1. These diagnostic criteria differ from those described in previous work published by IMGSAC (2001), since low-functioning individuals for whom an IQ could not be computed have now been classified as case type 2b rather than case type 3. Also, case type 3 individuals have been described elsewhere (IMGSAC 2001) as meeting ADI-R algorithm criteria in all domains, whereas, like case type 2 individuals, they can fall one point below threshold in one domain only. The mean performance IQ for case type 1 individuals was 79, for case type 2 individuals 82 (98 for individuals with case type 2a and 27 for individuals with case type 2b; IQ could not be computed for six individuals with case type 2b), and for case type 3 individuals 97. A detailed phenotypic description of the subjects included in this study is in preparation.
Table 1.
Criteria Used to Distinguish between Different Autism Case Types
| CaseType | Criteria |
| 1 | Clinical diagnosis of autism, meets ADI-R and ADOS or ADOS-G criteria for autism, history of language delay, and performance IQ of ⩾35 |
| 2: | Clinical diagnosis of autism, atypical autism, Asperger syndrome or PDDNOS; score on one ADI-R behavioral domain can be 1 point below threshold; meets at least ADOS-G criteria for PDD; no requirement of a history of language delay |
| 2a | Performance IQ of ⩾35 |
| 2b | No requirement of a performance IQ of ⩾35 |
| 3 | Clinical diagnosis of autism, atypical autism, Asperger syndrome or PDDNOS; score on one ADI-R behavioral domain can be one point below threshold; fails to meet ADOS-G criteria for PDD; performance IQ of ⩾35 |
Family Collection
Families have been collected and genotyped in four successive stages. Increased stringency of inclusion criteria has led to the current exclusion of nine families included in the original screen. Of these, eight families were excluded either because, so far, only the 1989 version of the ADOS had been administered to nonverbal individuals (for whom no algorithm exists) or because it has not been possible to compute valid scores on psychometric tests because of poor cooperation. The remaining family was excluded because of an abnormal karyotype result. Of the original 99 extended families, 11 are not included in this analysis, which examines only sib pairs. Eighty-three sib pairs from the 99 families published elsewhere are now included. Through subsequent family collection and assessment, 69 new sib pairs have been recruited. In total there are now 152 affected sib pairs, including three trios. Affected pairs were distributed in the following case type combinations: type 1/type 1, 43 pairs; type 1/type 2, 84 pairs; type 2/type 2, 16 pairs; type 1/type 3, 7 pairs; and type 2/type 3, 2 pairs. The sex ratio (male:female) of all affected relative pairs in the study sample was 4.8:1. There is no known overlap between the families included in this study and those ascertained by other research groups.
A physical examination was undertaken to exclude recognizable medical causes of autism, particularly tuberous sclerosis. A blood sample for DNA extraction was taken, when possible, from all cases and available first-degree relatives. In addition, lymphoblastoid cell lines were generated from peripheral blood leukocytes, to ensure a renewable source of DNA. In cases in which a blood sample could not be obtained, buccal swabs were taken (7.2% of DNA samples). When possible, karyotyping was performed—or previous results obtained—on all affected individuals, and molecular genetic testing for fragile-X syndrome was performed—or previous results obtained—on one affected individual from each family. Karyotype abnormalities were excluded in at least one affected individual in each family in 94% of families and in both affected individuals in 86% of families. Fragile-X syndrome was excluded in at least one affected individual in all 152 sib pairs. Written informed consent was given by all parents/guardians and, where possible, by affected individuals. The study has been reviewed by the relevant ethical committees.
Genotyping
Genomic DNA was extracted and primer extension preamplification performed as described elsewhere (IMGSAC 1998). Genotyping was performed using a semiautomated fluorescence-based method (Reed et al. 1994). PCR was performed in a 15-μl volume in 96-well plates, containing 5 μl of 40 ng genomic DNA, 10 mM Tris pH 8.3, 50 mM KCl, 1–3 mM MgCl2, 200 μM dNTPs, 0.2 μM of each fluorescent primer, and 0.25 U Taq or Taq Gold polymerase. The PCR consisted of 35 cycles of 30 s at 94°C, 30 s at 45°C–66°C, and 30 s at 72°C and was performed in MJ Research Thermocyclers. When Taq Gold was used, the program began with 15 min at 94°C and finished with 7 min at 72°C. PCR products were combined into pools and typed using ABI 373, 377, and 3700 sequencing machines and GENESCAN/GENOTYPER software (Applied Biosystems). Tamra and Rox were used as internal lane-size standards, and fluorescent markers were labeled with either HEX, TET, NED, or FAM. Allele designation was performed without knowledge of the individual’s status. A check for non-Mendelian inheritance of markers and conversion of allele sizes to whole numbers was performed using GAS (version 2). Global binning was performed for markers run on different types of sequencing machines, using allele sizes generated from two positive control DNA samples. Genotypic data was initially stored in Genbase (version 2.0.5) and later was transferred to Discovery Manager 2.3 (Genomica), along with phenotypic data. All data for statistical analysis were exported from Discovery Manager.
Genetic Markers
392 microsatellites and 2 single-nucleotide polymorphisms were typed in 83 sib pairs and a subset of 119 markers typed in a total of 152 sib pairs in the 13 candidate regions. The microsatellites include di-, tri-, and tetranucleotide repeats and were taken from a variety of sources, including Généthon (Dib et al. 1996), ABI PRISM Linkage Mapping Sets (Perkin-Elmer), The Co-operative Human Linkage Centre (Murray et al. 1994), the Genome Database, and Kwiatkowski et al. (1992). A 5-cM map with 25 markers is used throughout this article to refer to chromosome 7. Further genotyping and analyses have been performed on the region of interest on chromosome 7 (IMGSAC 2001) and are not repeated here. The average intermarker distance for the completed genome screen on 83 sib pairs was 9.92 cM. In the 13 regions where further genotyping was done on a total of 152 sib pairs, the average intermarker distance was reduced to one marker every 4.88 cM. Five microsatellites were deleted from those used in the original publication because of low informativeness.
Genetic marker order and map distances were taken from Généthon (Dib et al. 1996) and other published maps (Murrell et al. 1995; Gilbert et al. 2000). These were confirmed by running sib_map, part of the ASPEX package.
Error Checking
Prior to statistical analysis, SIBMED (sib-pair mutation error detection) was run on all data from the autosomes to remove possible genotyping errors (Douglas et al. 2000). A prescribed false-positive rate of <.001 was set given a prior genotyping-error rate of .01. After the removal of identified genotyping errors, the data were analyzed using the ASPEX statistical package. It should be noted that SIBMED is most effective at eliminating errors in regions where marker density is >1 marker every 5 cM; this requirement was met for regions on chromosomes 2 and 16, where marker density is ∼1 marker every 2 cM. The number of errors identified by SIBMED was small compared to those in other published work (Lathrop et al. 1983). This was due, in part, to the high percentage of sib pairs with parental genotypes available, >90% of families. All the MLSs quoted were generated subsequent to data checking by SIBMED.
Statistical Analysis
The data were prepared using Mega2 (Mukhopadhyay et al. 1999) and were analyzed using the ASPEX package to compute multipoint LOD scores. Despite the high proportion of parental genotypes available, sib_phase was used, rather than sib_ibd, to maximize the amount of information available. Analyses were performed using case status definitions to divide the sample into three groups: type 1/type 1 pairs (N=43), type 1/type 2 pairs (N=84), and all strict criteria pairs (type 1/type 1 + type 1/type 2 pairs) (N=127). Case types 2a and 2b were not analyzed separately, because of small sample sizes. Paternal and maternal transmission was analyzed using sib_ibd with the sex_split option, to elucidate the number of transmissions of maternal and paternal alleles contributing to sharing for each marker.
The transmission/disequilibrium test (TDT) (Spielman et al. 1993) was also performed, using sib_tdt on markers beneath peaks of linkage. Empirical P values for χ2 statistics were calculated to test for association independent of linkage (multiple sibs were used per family during analysis of the data for linkage).
To assess data quality and support the multipoint analysis, SPLINK was used to calculate single-point MLSs, under “possible triangle” restrictions, by estimating the probability of affected sibs sharing 0, 1, or 2 alleles identical by descent (IBD) (Holmans 1993; Holmans and Clayton 1995). Both SPLINK and ASPEX use maximum-likelihood methods to estimate marker allele frequencies within the database. ASPEX multipoint analysis was performed under an additive model (no dominance variance), so that if zi is the probability of an affected sib pair sharing i alleles identical by descent, then z0=.25/λs, z1=.5, and z2=.50-z0, where λs is defined as the sibling recurrence risk.
Results
Multipoint Linkage Analysis
The results of the multipoint linkage analysis of the 83 sib pairs from the IMGSAC (1998) screen indicated 12 chromosomes with regions showing an MLS >0.82 (chromosomes 1, 2, 4, 7, 8, 9, 10, 14, 16, 17, 19, and 22). Chromosome 7 had the highest multipoint MLS in the initial screen (IMGSAC 1998), and further analysis using this enlarged data set has recently been reported (IMGSAC 2001). The remaining 11 chromosomal areas had multipoint MLSs between 0.82 and 2.05 and a supporting single-point MLS. Further genotyping was also performed on chromosome 15, because of previous reports of linkage and chromosomal abnormalities associated with autism (Gillberg 1998). Table 2 presents results for these 12 chromosomes in the enlarged sample of 152 sib pairs at loci where either the SPLINK single-point MLS or the ASPEX multipoint MLS were highest, as well as for chromosome 15. The results from the 83 sib pairs currently included from the initial screen (IMGSAC 1998) are also shown for comparison. The mean effective sample size for each marker across the genome was 59.2, and the estimated average marker heterozygosity was .74.
Table 2.
Single-Point MLS Determined by SPLINK and Multipoint MLS Generated by ASPEX, for the 13 Chromosomes Genotyped in 152 Sib Pairs[Note]
|
Single-Point MLS |
Multipoint MLS |
|||||||
| Marker | cMa | All | ESSb | Oldc (83) | All (152) | 1/1d (43) | 1/2e (84) | Strictf (127) |
| D1S218 | 225.21 | .64 | 81.22 | [.57] | .58 | .00 | .97 | .68 |
| D1S249 | 255.59 | .65 | 120.48 | .30 | .60 | .21 | .16 | .35 |
| D2S131 | 24.31 | .16 | 58.10 | .22 | .39 | 1.97 | .00 | .09 |
| D2S1351 | 111.43 | 2.52 | 87.33 | .71 | 1.60 | .68 | 1.17 | 1.22 |
| D2S2188 | 206.39 | 3.59 | 85.94 | [.44] | 3.74 | 1.24 | 3.66 | 4.80 |
| D4S2936 | .00 | 1.58 | 98.58 | 1.31 | .71 | .48 | .03 | .31 |
| D4S430 | 140.57 | .42 | 53.85 | .37 | .44 | .05 | .61 | .59 |
| D4S408 | 230.15 | .02 | 22.93 | .04 | .43 | 1.04 | .00 | .75 |
| D7S477 | 119.60 | 2.93 | 70.95 | [1.18] | 3.20 | .00 | 3.55 | 2.33 |
| D8S552 | 29.21 | .28 | 65.88 | .04 | .49 | .47 | .22 | .58 |
| D8S261 | 38.84 | .30 | 73.97 | .65 | 1.12 | .16 | .77 | .90 |
| D8S1786 | 48.37 | .67 | 97.73 | .23 | .19 | .00 | .24 | .02 |
| D9S157 | 18.93 | .79 | 85.41 | [.00] | .75 | .00 | 3.11 | 2.02 |
| D9S164 | 140.25 | .96 | 79.29 | .20 | 1.09 | .59 | 1.31 | 1.90 |
| D9S1826 | 141.87 | 1.83 | 38.13 | [.24] | 1.46 | .06 | 3.59 | 2.23 |
| D9S158 | 142.07 | 2.97 | 33.65 | .23 | 1.36 | .00 | 3.16 | 2.09 |
| D10S197 | 53.66 | .65 | 74.66 | .91 | 1.08 | .75 | .59 | 1.26 |
| D10S208 | 64.29 | 1.38 | 90.72 | [.72] | 1.43 | .11 | .98 | 1.00 |
| D10S201 | 116.63 | 1.37 | 105.99 | .60 | 1.22 | .47 | .17 | .56 |
| D14S80 | 19.70 | 1.13 | 92.94 | .64 | .33 | .02 | .37 | .34 |
| D14S1034 | 24.62 | .89 | 93.08 | .78 | .41 | .05 | .44 | .46 |
| D14S68 | 90.45 | .14 | 61.52 | .06 | .07 | .00 | .67 | .34 |
| D14S267 | 119.65 | .50 | 73.03 | .92 | .99 | .00 | 1.07 | .51 |
| D15S129 | 34.10 | .30 | 86.84 | .02 | .76 | 1.47 | .44 | 1.49 |
| CYP19g | 40.46 | .60 | 42.54 | .12 | .65 | 2.21 | .14 | 1.20 |
| D16S407 | 16.70 | 2.19 | 107.61 | 1.26 | 1.59 | .14 | 2.22 | 2.11 |
| D16S3114 | 21.80 | 2.52 | 114.61 | 1.11 | 2.12 | .36 | 2.05 | 1.81 |
| D16S3102 | 23.10 | .78 | 49.07 | .96 | 2.93 | .49 | 2.20 | 2.61 |
| D17S786 | 7.60 | .47 | 49.99 | .18 | .80 | .00 | 1.63 | .92 |
| HTTINT2h | 45.37 | 3.60 | 37.75 | .14 | 2.34 | .61 | 1.27 | 1.87 |
| D17S250 | 53.68 | 1.77 | 105.54 | .09 | 1.60 | .51 | .57 | 1.04 |
| D19S220 | 63.02 | 1.11 | 105.24 | .18 | .13 | .03 | .07 | .10 |
| CYP2Di | 41.43 | .82 | 41.17 | .39 | .33 | .00 | 1.22 | .25 |
Note.— Markers with the highest single-point and multipoint MLSs are shown, including markers where the highest multipoint MLS was generated when the data were divided by case type status. Underlined markers and figures indicate that an MLS >3 was generated. Numbers in parentheses indicate the number of sib-pairs analyzed.
cM = approximate distance, in Haldane centimorgans, from pter to marker.
ESS = effective sample size, as calculated by SPLINK.
Old = previously published genotype data analyzed with 83 original sib pairs included. Numbers in square brackets indicate MLS at new marker position.
1/1 = case type 1/type 1 pairs.
1/2 = case type 1/type 2 pairs.
Strict = case type 1/type 1 pairs + type 1/type 2 pairs.
CYP19 is on chromosome 15.
HTTINT2 is on chromosome 17.
CYP2D is on chromosome 22.
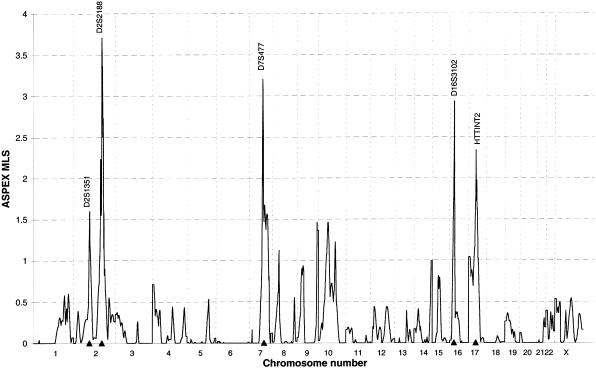
The results from ASPEX analysis for all 152 sib pairs after the removal of errors identified by SIBMED are shown in figure 1. SIBMED identified 232 genotypes for removal from a total of 308,450 genotypes, giving an error rate of .075%. None of the reported peak MLSs were increased by >0.28 by the removal of potential genotyping errors.
Figure 1.
Multipoint maps across all chromosomes, from pter to qter, generated by ASPEX under an additive model with no dominance variance. The position of markers attaining a maximum MLS >1.5 are shown as triangles and have been typed in 152 sib pairs.
In the total sample of 152 sib pairs, an ASPEX multipoint MLS >3 was generated on two chromosomes, the highest being an MLS of 3.74 on the long arm of chromosome 2 at D2S2188; this finding was supported by a SPLINK single-point MLS of 3.59. Chromosome 7 provided an MLS of 3.20 at D7S477, supported by a single-point MLS of 2.93. The third-highest multipoint MLS, 2.93, was on the short arm of chromosome 16 at D16S3102. A SPLINK single-point MLS of 2.97 and a multipoint MLS of 1.36 were generated at D9S158 on chromosome 9. However, the low effective sample size (33.73) at this locus and the telomeric location should be taken into account when considering the significance of this result. Four chromosomes (2, 7, 16, and 17) generated a multipoint MLS >1.5, whereas the previous evidence of linkage on chromosomes 4, 14, 19, and 22 diminished.
Allelic Association
When the ASPEX multipoint MLS was >1.5, multiallelic TDTs were performed to test for association. Thirty-three markers were tested, and no significant evidence for association (P>.05) was found in these regions. Some evidence for association has been reported elsewhere in the area of linkage on chromosome 7 (IMGSAC 2001).
Analysis of Data by Case Status
Grouping of affected individuals into specific case types, as described in table 1, provides an opportunity to analyze data hierarchically and, potentially, to minimize heterogeneity. Accordingly, multipoint analysis was performed using ASPEX for specific case type combinations in regions genotyped in 152 sib pairs. Regions where the analysis by case type status gave the highest multipoint MLS are presented in table 2.
In general, the 43 case type 1/type 1 affected sib pairs do not contribute strongly to multipoint MLS in the regions studied. The exceptions were on chromosome 15 at CYP19, where the highest multipoint for case type 1/type 1 pairs was achieved with a multipoint MLS of 2.21, and on chromosome 2, at D2S188, where they attained a multipoint MLS of 1.24.
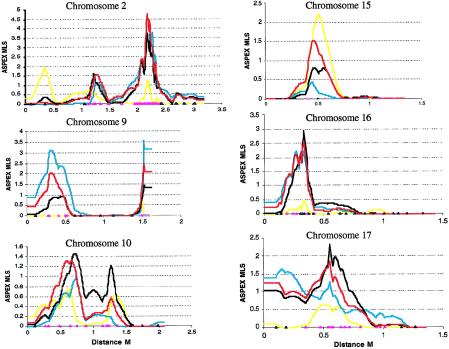
The larger group of 84 type 1/type 2 pairs showed considerably more sharing, and five chromosomes (2, 7, 9, 16, and 17) achieved a multipoint MLS >1.5. Combining type 1/type 1 and type 1/type 2 pairs gives 127 strictly defined sib pairs, which, together, generated an MLS >1.0 on seven chromosomes (2, 7, 9, 10, 15, 16, and 17); the highest MLS was 4.80 on chromosome 2 at D2S2188. Multipoint plots by case type are shown for chromosomes 2, 9, 10, 15, 16, and 17 in figure 2.
Figure 2.
Multipoint maps generated in ASPEX for sib pairs analyzed by case type. The yellow line represents the multipoint MLS for case type 1/type 1 pairs (43), the blue line represents type 1/type 2 pairs (84), and the red line represents strict criteria pairs (127). The black line represents the total multipoint MLS for 152 sib pairs. Blue triangles represent markers typed in 83 sib pairs, and pink triangles represent markers typed in 152 sib pairs. Distances are approximate, in haldane M, from pter to qter.
Paternal and Maternal Allele Transmission
Sex-specific analysis on chromosomes 2, 9, 10, 15, 16, and 17 was carried out using ASPEX to calculate separate LOD scores for paternal and maternal sharing. There was substantial variation in the contribution of paternal and maternal sharing to the multipoint MLS across these six chromosomes. On chromosomes 2 and 15, paternal and maternal sharing was approximately equal. At D2S2188, paternal- and maternal-specific multipoint LODs were 2.10 and 1.95, respectively, and, at D15S129, paternal- and maternal-specific multipoint LODs were 0.45 and 0.32, respectively. There was some evidence for an increased paternal contribution to the multipoint MLS for chromosomes 16 and 17 at D16S3102 and HTTINT2, with paternal multipoint LODs of 2.21 and 2.32, compared with maternal multipoint LODs of 0.96 and 0.16, respectively. Conversely, on chromosomes 9 and 10, at D9S157 and D10S208, there was evidence that all sharing at these loci was maternally derived, with paternal multipoint LODs of 0 and maternal multipoint LODs of 1.43 and 0.75, respectively.
To evaluate the significance of the maternal and paternal sharing differences, single-point calculations were also performed; of 34 markers tested, only 2 supported the multipoint analysis (P<.05). The increased maternal sharing at D10S208 was supported with 62/92 maternal transmissions, compared with 40/82 paternal transmissions, giving a P value of .0196. Significantly increased paternal sharing was found at D17S798, 1.8 cM distal to HTTINT2, with 62/92 paternal transmissions, compared with 44/95 maternal transmissions, giving a P value of .0058. Single-point analysis of flanking markers at the above loci did not show significant differences in parental sharing, indicating that these results may have arisen by chance.
Discussion
The completion of a whole-genome screen for autism in 83 sib pairs indicated 12 regions with nominal evidence for linkage. These regions were further characterized by the addition of 69 new sib pairs who were genotyped using a subset of 74 of the original markers and 45 new markers. The effect of adding new sib pairs and markers has been to reduce “noise” across the genome. Loci on chromosomes 4, 19, and 22 previously reported as having an ASPEX multipoint MLS >1 (IMGSAC 1998) now all have a multipoint MLS <1. Among the 12 loci with an MLS >0.82, genomewide significance—as defined by Lander and Kruglyak (1995)—has been reached at one locus on chromosome 2, with a multipoint MLS of 3.74 at D2S2188. Suggestive linkages have been found on chromosomes 7, 16, and 17, with multipoint MLSs of 3.20, 2.93, and 2.34, respectively.
The chromosome 2 result is the highest multipoint MLS generated to date in an autism genome screen; it is supported by a reported nominal linkage of 0.64 by Philippe et al. (1999) between markers D2S382 and D2S364, which flank D2S2188, and by Buxbaum et al. (2001), who found a maximum multipoint nonparametric linkage score of 2.39 between D2S335 and D2S364, also flanking D2S2188. When the subset of 127 sib-pair families identified using strict criteria was considered, the multipoint MLS at D2S2188 increased to 4.80. As described by Lamb et al. (2000), the chromosome 7 linkage has also been replicated, although location estimates vary. Our findings on chromosome 16 directly overlap those of Philippe et al. (1999), who found a multipoint MLS of 0.74 between 23.3 and 39.0 cM. The modest MLS of 1.43 at D10S208 on chromosome 10 should be viewed with caution, following reports of transmission ratio distortion in CEPH pedigrees in this region (Paterson and Petronis 1999). Increasing the number of markers and sib pairs genotyped in this region will help guard against any artificial inflation of LOD score (Greenwood and Morgan 2000).
It is unclear why case type 1/type 1 pairs contribute so little to the strongest linkage findings. This group may be more genetically heterogeneous than other combinations of case types (although it includes no severely handicapped cases), or, perhaps, the presence of language delay in both affected individuals identifies a small specific subgroup of families. It may be relevant that the 43 case type 1/type 1 pairs generated a multipoint MLS of 2.21 on chromosome 15 at CYP19. The linkage on chromosome 15 extends to D15S129, which had a multipoint MLS of 1.49 and is ∼25 cM distal from the position of chromosomal abnormalities associated with autism. It is possible to interpret this finding as supporting a susceptibility locus that is potentially specific to severe autism with language delay.
Despite the width of the peaks of linkage and despite evidence from simulation studies that there is large variation in peak location relative to actual disease-gene location (Roberts et al. 1999), a number of potential candidate genes are worth noting briefly. On chromosome 2, two distalless homeobox genes (DLX1 and DLX2) are located in 2q32 (Zerucha et al. 2000), and their homologues (DLX5 and DLX6) are located in 7q22, beneath the second highest peak of linkage. Another gene of interest on chromosome 2 is alpha-1-chimaerin (α1CHIM) and the alternatively spliced α2CHIM, a GTPase-activating protein. The rat homologue of α1CHIM and α2CHIM (n-chimaerin) has mRNA expression restricted to neurons and is increased during cellular differentiation and synaptogenesis (Lim et al. 1992). Other interesting genes in the region include t brain 1 (TBR1), neuropilin (NRP2), and cAMP response element-binding protein 1 and 2 (CREB1 and CREB2).
On chromosome 7q32, potential candidate genes include plexin (PLXNA4), which forms a complex with neuropilin and mediates repulsion by the axonal guidance signal semaphorin 3A (Rohm et al. 2000). The imprinted genes PEG1/MEST (Hayashida et al. 2000) and γ2-COP (Blagitko et al. 1999) are also within the candidate region on chromosome 7q32 and may be relevant, given the possibility of a parent-of-origin–specific effect in autism. Ashley-Koch has reported increased paternal transmission in this region, and there was some evidence to support this in our families (Ashley-Koch et al. 2000; IMGSAC 2001). It may also be useful to consider candidate genes in the 5.6-cM SPCH1 region on 7q31 (Lai et al. 2000). Several recent studies suggest Reelin (RELN) on 7q22 as a candidate gene for autism. TDT analysis of a polymorphic trinucleotide repeat in the 5′ UTR of Reelin has been reported to show preferential transmission of a particular allele/alleles in autistic probands (Zhang et al. 2000; Persico et al. 2001).
The locus with the third-highest MLS, generating a multipoint MLS of 2.93, was on 16p13. An n-methyl-d-aspartate receptor (NMDA) gene is located at 16p13.3, and this receptor plays an important role in synaptic plasticity. The NMDA channel is formed by two members of the glutamate receptor channel (GluR)–subunit family and has been found to cause a deficit in spatial learning in mutant mice when knocked out (Sakimura et al. 1995). Other important loci in the chromosome 16 region include tuberous sclerosis 2 (TSC2) (Burn et al. 1996); a gene for one of the netrins (NTN2L), a family of chemotrophic factors that play a central role in axon guidance (Van Raay et al. 1997); and the CREB-binding protein (CREBBP).
The significance of the reported parent-of-origin effect in autism is unclear from the analysis of our data. We found no evidence to support claims for a significant skew in IBD parental transmission of alleles on chromosomes 7 and 15 (Ashley-Koch et al. 1999, 2000; Sutcliffe and CLSA 2000) or for the peak on chromosome 2. There was, however, some evidence from the multipoint MLS to support increased paternal-allelic transmission on chromosomes 16 and 17, with the locus on chromosome 17 supported by single-point analysis. Maternal allele transmission was also increased in both multipoint MLS and single-point analysis on chromosome 10. The lack of support for these results from flanking marker single-point analysis indicates that they may simply reflect chance observations.
These findings demonstrate the utility of enlarging collections of affected relative–pair families, both in terms of strengthening putative true linkages and indicating which preliminary findings are likely to have been false positives. Indeed, as yet, no group has attained a sample size of 200 affected relative pairs, the generally accepted minimum sample size for a whole genome scan of complex diseases (Weeks and Lathrop 1995). The replication of findings on chromosomes 2, 7, and 16 across different groups—whose families are drawn from somewhat different populations—is very promising for the general applicability of the findings. Efforts to establish a meta-analysis should further advance this process.
With regard to future work, there is a need to utilize all the available genetic and phenotypic information in nuclear families. This requires accurate description of the affected status of relatives who do not have a PDD. Eventually it may be possible, by means of QTL approaches, to combine data from affected individuals and their relatives. Future work will also involve conditional analysis to examine locus interaction (epistasis) and the extent of heterogeneity in autism (Cox et al. 1999). Careful consideration of potential candidate genes will also be necessary to provide a biological basis for testing the presence of interaction—for example, between the DLX genes on chromosomes 2 and 7.
Acknowledgments
This work would not have been possible without the cooperation of the individuals with autism, their families, and the many professionals who referred them. We also thank the computer, bioinformatics, and secretarial support staff at the Wellcome Trust Centre and at the Centre for Social, Genetic, and Developmental Psychiatry for their assistance. This work has been funded by the U.K. Medical Research Council, The Wellcome Trust, BIOMED 2 (grant CT-97-2759), EC Fifth Framework (grant QLG2-CT-1999-0094), Telethon-Italy (grant E.1007), the Janus Korczak Foundation, Deutsche Forschungsgemeinschaft, Fondation France Télécom, Conseil Régional Midi-Pyrénées, Danish Medical Research Council, Sofiefonden, the Beatrice Surovell Haskells Fund for Child Mental Health Research of Copenhagen, the Danish Natural Science Research Council (grant 9802210), the National Institute of Child Health and Development (grant 5-P01-HD-35482), and the National Institutes of Health (grants MO1 RR06022 GCRC NIH, NIH K05 MH01196, and K02 MH01389), particularly the Collaborative Programs for Excellence in Autism Research. A.P.M. is a Wellcome Trust Principal Research Fellow. IMGSAC members, United Kingdom: Centre for Social, Genetic and Developmental Psychiatry and Department of Child and Adolescent Psychiatry, Institute of Psychiatry, London—Sarah Palferman, Nicola Matthews, Martha Turner, Janette Moore, Amaia Hervas, Anne Aubin, Simon Wallace, Janine Michelotti, Catherine Wainhouse, Alina Paul, Elaine Thompson, Ramyani Gupta, Claire Garner, Marianne Murin, Christine Freitag, Nicola Ryder, Emily Cottington, Jeremy Parr, Andrew Pickles, Michael Rutter, and Anthony Bailey; Wellcome Trust Centre for Human Genetics, University of Oxford—Gabrielle Barnby, Janine A. Lamb, Angela Marlow, Pat Scudder, and Anthony P. Monaco; Newcomen Centre, Guy’s Hospital, London—Gillian Baird and Anthony Cox; Regional Genetics Centre, Division of Medical and Molecular Genetics, Guy’s Hospital, London—Zoe Docherty, Pamela Warburton, Elizabeth P. Green, and Stephen J. Abbs; Flemming Nuffield Unit, Newcastle—Ann Le Couteur, Helen R. McConachie, and Tom Berney; Neuropsychology Department, Newcastle General Hospital—Thomas P. Kelly; Developmental Psychiatry Section, University of Cambridge Clinical School—Petrus J. De Vries and Patrick F. Bolton; Booth Hall Children’s Hospital, Manchester—Jonathan Green, Anne Gilchrist, and Jane Whittacker; European Centre for Collection of Animal Cell Cultures, Salisbury—Bryan Bolton and Ros Packer. Italy: Dipartimento di Biologia, Universita’ di Bologna—Elena Maestrini and Francesca Blasi. The Netherlands: AZU, Department of Child and Adolescent Psychiatry, Utrecht—Herman Van Engeland, Maretha V. De Jonge, and Chantal Kemner. Germany: Deutsches Krebsforschungszentrum, Molecular Genome Analysis—Sabine M. Klauck, Kim S. Beyer, Sabine Epp, and Annemarie Poustka; Deutsches Krebsforschungszentrum, Biostatistics—Axel Benner; J. W. Goethe-Universität, Department of Child and Adolescent Psychiatry, Frankfurt—Fritz Poustka, Dorothea Rühl, Gabriele Schmötzer, Sven Bölte, and Sabine Feineis-Matthews. France: Unite de Diagnostic et Evaluation de l’autisme, Hopital la Grave, Toulouse—Eric Fombonne, Bernadette Rogé, Jeanne Fremolle-Kruck, Catherine Pienkowski, and Marie-Thérèse Tauber. Denmark: National Centre for Autism—Lennart Pedersen, Karen-Brondum-Nielsen, Gunna Eriksen, and Demetrious Haracopos; Biophysics, Danish Technical University—Rodney M. J. Cotterill. Greece: Department of Psychological Paediatrics, Agia Sophia Children’s Hospital, Athens—John Tsiantis and Katerina Papanikolaou. U.S.A.: Department of Psychiatry, University of Chicago—Catherine Lord, Christina Corsello, Stephen Guter, Bennett Leventhal, and Edwin Cook; UCLA Center for Neurobehavioral Genetics—Susan L. Smalley, Julia Bailey, James McGough, and Jennifer Levitt; Child Study Center, Sterling Hall of Medicine, Yale University—David Pauls and Fred Volkmar; Department of Human Genetics, University of Pittsburgh—Daniel E. Weeks.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Applied Biosystems, http://www.appliedbiosystems.com/
- ASPEX program, ftp://lahmed.stanford.edu/pub/aspex
- Genome Database, http://www.gdb.org/
- IMGSAC, http://www.well.ox.ac.uk/~maestrin/iat.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for autism [MIM 209850] and Rubinstein-Taybi syndrome [MIM 180849])
- SIBMED program, http://www.sph.umich.edu/group/statgen/software
References
- Ashley-Koch A, Menold M, Joyer K, Mason S, Poole C, Donnelly S, Wolpert C, Raven S, Abramson R, DeLong G, Cuccaro M, Von Wendt L, McClain C, Wright H, Vance JM, Gilbert JG, Pericak-Vance MA (2000) Examination of epigentic factors and gene-gene interaction influencing genetic susceptibility at chromosome 7 and 15 for autistic disorder. Am J Hum Genet Suppl 67:A46 [Google Scholar]
- Ashley-Koch A, Wolpert CM, Menold MM, Zaeem L, Basu S, Donnelly SL, Ravan SA, Powell CM, Qumsiyeh MB, Aylsworth AS, Vance JM, Gilbert JR, Wright HH, Abramson RK, DeLong GR, Cuccaro ML, Pericak-Vance MA (1999) Genetic studies of autistic disorder and chromosome 7. Genomics 61:227–236 [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M (1995) Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25:63–77 [DOI] [PubMed] [Google Scholar]
- Bailey A, Palferman S, Heavey L, Le Couteur A (1998) Autism: the phenotype in relatives. J Autism Dev Disord 28:369–392 [DOI] [PubMed] [Google Scholar]
- Bass MP, Menold MM, Wolpert CM, Donnelly SL, Ravan SA, Hauser ER, Maddox LO, Powell CM, Qumsiyeh MB, Aylsworth AS, Vance JM, Gilbert JR, Wright HH, Abramson RK, DeLong GR, Cuccaro ML, Pericak-Vance MA (2000) Genetic studies in autistic disorder and chromosome 15. Neurogenetics 2:219–226 [DOI] [PubMed] [Google Scholar]
- Blagitko N, Schulz U, Schinzel AA, Ropers HH, Kalscheuer VM (1999) gamma2-COP, a novel imprinted gene on chromosome 7q32, defines a new imprinting cluster in the human genome. Hum Mol Genet 8:2387–2396 [DOI] [PubMed] [Google Scholar]
- Burn TC, Connors TD, Van Raay TJ, Dackowski WR, Millholland JM, Klinger KW, Landes GM (1996) Generation of a transcriptional map for a 700-kb region surrounding the polycystic kidney disease type 1 (PKD1) and tuberous sclerosis type 2 (TSC2) disease genes on human chromosome 16p3.3. Genome Res 6:525–537 [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Kilifarski M, Reichert J, Hollander E, Lawlor BA, Fizgerald M, Greenberg DA, Davis KL (2001) Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am J Hum Genet 68:1514–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH Jr, Courchesne RY, Cox NJ, Lord C, Gonen D, Guter SJ, Lincoln A, Nix K, Haas R, Leventhal BL, Courchesne E (1998) Linkage-disequilibrium mapping of autistic disorder, with 15q11-13 markers. Am J Hum Genet 62:1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH Jr, Courchesne R, Lord C, Cox NJ, Yan S, Lincoln A, Haas R, Courchesne E, Leventhal BL (1997) Evidence of linkage between the serotonin transporter and autistic disorder. Mol Psychiatry 2:247–250 [DOI] [PubMed] [Google Scholar]
- Cox NJ, Frigge M, Nicolae DL, Concannon P, Hanis CL, Bell GI, Kong A (1999) Loci on chromosomes 2 (NIDDM1) and 15 interact to increase susceptibility to diabetes in Mexican Americans. Nat Genet 21:213–215 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites [see comments]. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Douglas JA, Boehnke M, Lange K (2000) A multipoint method for detecting genotyping errors and mutations in sibling-pair linkage data. Am J Hum Genet 66:1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn LM, Dunn LM (1997) Peabody picture vocabulary test. American Guidance Service, Circle Pines, MN [Google Scholar]
- Dunn LM, Dunn LM, Whetton C, Pintilie D (1982) British picture vocabulary scale. NFER-Nelson, Windsor, United Kingdom [Google Scholar]
- Folstein S, Rutter M (1977) Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry 18:297–321 [DOI] [PubMed] [Google Scholar]
- Fombonne E (1999) The epidemiology of autism: a review. Psychol Med 29:769–786 [DOI] [PubMed] [Google Scholar]
- Gilbert JR, Kumar A, Newey S, Rao N, Ioannou P, Qiu H, Lin D, Xu P, Pettenati MJ, Pericak-Vance MA (2000) Physical and cDNA mapping in the DBH region of human chromosome 9q34. Hum Hered 50:151–157 [DOI] [PubMed] [Google Scholar]
- Gillberg C (1998) Chromosomal disorders and autism. J Autism Dev Disord 28:415–425 [DOI] [PubMed] [Google Scholar]
- Greenwood CM, Morgan K (2000) The impact of transmission-ratio distortion on allele sharing in affected sibling pairs. Am J Hum Genet 66:2001–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmayer J, Hebert JM, Spiker D, Lotspeich L, McMahon WM, Petersen PB, Nicholas P, Pingree C, Lin AA, Cavalli-Sforza LL, Risch N, Ciaranello RD (1996) Autism and the X chromosome: multipoint sib-pair analysis. Arch Gen Psychiatry 53:985–989 [DOI] [PubMed] [Google Scholar]
- Hallmayer J, Pintado E, Lotspeich L, Spiker D, McMahon W, Petersen PB, Nicholas P, Pingree C, Kraemer HC, Wong DL, Ritvo E, Lin A, Herbert J, Cavalli-Sforza LL, Ciaranella RD (1994) Molecular analysis and test of linkage between the FMR-1 gene and infantile autism in multiplex families. Am J Hum Genet 55:951–959 [PMC free article] [PubMed] [Google Scholar]
- Hayashida S, Yamasaki K, Asada Y, Soeda E, Niikawa N, Kishino T (2000) Construction of a physical and transcript map flanking the imprinted MEST/PEG1 region at 7q32. Genomics 66:221–225 [DOI] [PubMed] [Google Scholar]
- Holmans P (1993) Asymptotic properties of affected-sib-pair linkage analysis. Am J Hum Genet 52:362–374 [PMC free article] [PubMed] [Google Scholar]
- Holmans P, Clayton D (1995) Efficiency of typing unaffected relatives in an affected-sib-pair linkage study with single-locus and multiple tightly linked markers. Am J Hum Genet 57:1221–1232 [PMC free article] [PubMed] [Google Scholar]
- IMGSAC (International Molecular Genetic Study of Autism Consortium) (1998) A full genome screen for autism with evidence for linkage to a region on chromosome 7q. Hum Mol Genet 7:571–578 [DOI] [PubMed] [Google Scholar]
- IMGSAC (International Molecular Genetic Study of Autism Consortium) (2001) Further characterization of the autism susceptibility locus AUTS1 on chromosome 7q. Hum Mol Genet 10:973–982 [DOI] [PubMed] [Google Scholar]
- Klauck SM, Poustka F, Benner A, Lesch KP, Poustka A (1997) Serotonin transporter (5-HTT) gene variants associated with autism? Hum Mol Genet 6:2233–2238 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Henske EP, Weimer K, Ozelius L, Gusella JF, Haines J (1992) Construction of a GT polymorphism map of human 9q. Genomics 12:229–240 [DOI] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Levy ER, Hodgson S, Fox M, Jeremiah S, Povey S, Jamison DC, Green ED, Vargha-Khadem F, Monaco AP (2000) The SPCH1 region on human 7q31: genomic characterization of the critical interval and localization of translocations associated with speech and language disorder. Am J Hum Genet 67:357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JA, Moore J, Bailey A, Monaco AP (2000) Autism: recent molecular genetic advances. Hum Mol Genet 9:861–868 (erratum Hum Mol Genet 9:1461) [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Hooper AB, Huntsman JW, Ward RH (1983) Evaluating pedigree data. I. The estimation of pedigree error in the presence of marker mistyping. Am J Hum Genet 35:241–262 [PMC free article] [PubMed] [Google Scholar]
- Lim HH, Michael GJ, Smith P, Lim L, Hall C (1992) Developmental regulation and neuronal expression of the mRNA of rat n-chimaerin, a p21rac GAP:cDNA sequence. Biochem J 287:415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, Cook EH Jr, Leventhal BL, DiLavore PC, Pickles A, Rutter M (2000) The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord 30:205–223 [PubMed] [Google Scholar]
- Lord C, Rutter M, Goode S, Heemsbergen J, Jordan H, Mawhood L, Schopler E (1989) Autism diagnostic observation schedule: a standardized observation of communicative and social behavior. J Autism Dev Disord 19:185–212 [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A (1994) Autism diagnostic interview-revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 24:659–685 [DOI] [PubMed] [Google Scholar]
- Maestrini E, Lai C, Marlow A, Matthews N, Wallace S, Bailey A, Cook EH, Weeks DE, Monaco AP (1999) Serotonin transporter (5-HTT) and gamma-aminobutyric acid receptor subunit beta3 (GABRB3) gene polymorphisms are not associated with autism in the IMGSAC families. Am J Med Genet 88:492–496 [DOI] [PubMed] [Google Scholar]
- Martin ER, Menold MM, Wolpert CM, Bass MP, Donnelly SL, Ravan SA, Zimmerman A, Gilbert JR, Vance JM, Maddox LO, Wright HH, Abramson RK, DeLong GR, Cuccaro ML, Pericak-Vance MA (2000) Analysis of linkage disequilibrium in gamma-aminobutyric acid receptor subunit genes in autistic disorder. Am J Med Genet 96:43–48 [DOI] [PubMed] [Google Scholar]
- Mbarek O, Marouillat S, Martineau J, Barthelemy C, Muh JP, Andres C (1999) Association study of the NF1 gene and autistic disorder. Am J Med Genet 88:729–732 [PubMed] [Google Scholar]
- Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE (1999) Mega2, a data-handling program for facilitating genetic linkage and association analyses. Am J Hum Genet Suppl 65:A436 [DOI] [PubMed] [Google Scholar]
- Mullen EM (1995) Mullens scales of early learning. American Guidance Service, Circle Pines, MN [Google Scholar]
- Murray JC, Buetow KH, Weber JL, Ludwigsen S, Scherpbier Haddema T, Manion F, Quillen J, Sheffield VC, Sunden S, Duyk GM, Weissenbach J, Gyapay G, Dib C, Morrissette J (1994) A comprehensive human linkage map with centimorgan density. Science 265:2049–2054 [DOI] [PubMed] [Google Scholar]
- Murrell J, Trofatter J, Rutter M, Cutone S, Stotler C, Rutter J, Long K, Turner A, Deaven L, Buckler A, McCormick MK (1995) A 500-kilobase region containing the tuberous sclerosis locus (TSC1) in a 1.7-megabase YAC and cosmid contig. Genomics 25:59–65 [DOI] [PubMed] [Google Scholar]
- Paterson AD, Petronis A (1999) Transmission ratio distortion in females on chromosome 10p11-p15. Am J Med Genet 88:657–661 [PubMed] [Google Scholar]
- Persico A, D'Agruma L, Maiorano N, Totaro A, Militerni R, Bravaccio C, Wassink T (2001) Reelin gene alleles and haplotypes as a factor predisposing to autistic disorder. Mol Psychiatr 6:150–159 [DOI] [PubMed] [Google Scholar]
- Persico AM, Militerni R, Bravaccio C, Schneider C, Melmed R, Conciatori M, Damiani V, Baldi A, Keller F (2000) Lack of association between serotonin transporter gene promoter variants and autistic disorder in two ethnically distinct samples. Am J Med Genet 96:123–127 [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, Penet C, Feingold J, Brice A, Leboyer M, van Malldergerme L, Paris Autism Research International Sibpair Study (1999) Genome-wide scan for autism susceptibility genes. Hum Mol Genet 8:805–812 [erratum Hum Mol Genet 8:1353] [DOI] [PubMed] [Google Scholar]
- Pickles A, Bolton P, Macdonald H, Bailey A, Le Couteur A, Sim CH, Rutter M (1995) Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism. Am J Hum Genet 57:717–726 [PMC free article] [PubMed] [Google Scholar]
- Pickles A, Starr E, Kazak S, Bolton P, Papanikolaou K, Bailey A, Goodman R, Rutter M (2000) Variable expression of the autism broader phenotype: findings from extended pedigrees. J Child Psychol Psychiatry 41:491–502 [PubMed] [Google Scholar]
- Raven J (1989) Standard progressive matrices. Australian Council for Educational Research, Victoria [Google Scholar]
- Reed PW, Davies JL, Copeman JB, Bennett ST, Palmer SM, Pritchard LE, Gough SC, Kawaguchi Y, Cordell HJ, Balfour KM, Jenkins SC, Powell EE, Vignal A, Todd JA (1994) Chromosome-specific microsatellite sets for fluorescence-based, semi-automated genome mapping [see comments]. Nat Genet 7:390–395 [DOI] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, et al (1999) A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet 65:493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS (1999) Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet 65:876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohm B, Ottemeyer A, Lohrum M, Puschel AW (2000) Plexin/neuropilin complexes mediate repulsion by the axonal guidance signal semaphorin 3A. Mech Dev 93:95–104 [DOI] [PubMed] [Google Scholar]
- Sakimura K, Kutsuwada T, Ito I, Manabe T, Takayama C, Kushiya E, Yagi T, Aizawa S, Inoue Y, Sugiyama H, Mishina M (1995) Reduced hippocampal LTP and spatial learning in mice lacking NMDA receptor epsilon 1 subunit. Nature 373:151–155 [DOI] [PubMed] [Google Scholar]
- Salmon B, Hallmayer J, Rogers T, Kalaydjieva L, Petersen PB, Nicholas P, Pingree C, McMahon W, Spiker D, Lotspeich L, Kraemer H, McCague P, Dimiceli S, Nouri N, Pitts T, Yang J, Hinds D, Myers RM, Risch N (1999) Absence of linkage and linkage disequilibrium to chromosome 15q11-q13 markers in 139 multiplex families with autism. Am J Med Genet 88:551–556 [PubMed] [Google Scholar]
- Skuse DH (2000) Imprinting, the X-chromosome, and the male brain: explaining sex differences in the liability to autism. Pediatr Res 47:9–16 [DOI] [PubMed] [Google Scholar]
- Smalley SL (1998) Autism and tuberous sclerosis. J Autism Dev Disord 28:407–414 [DOI] [PubMed] [Google Scholar]
- Sparrow S, Balla D, Cicchetti D (1984) Vineland adaptive behaviour scales. American Guidance Service, Circle Pines, MN [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–516 [PMC free article] [PubMed] [Google Scholar]
- Steffenburg S (1991) Neuropsychiatric assessment of children with autism: a population-based study. Dev Med Child Neurol 33:495–511 [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, CLSA (Collaborative Linkage Study of Autism) (2000) Linkage disequilibrium in autism families to markers in the 15q-q13 autism candidate region. Am J Hum Genet Suppl 67:46 [Google Scholar]
- Szatmari P, Jones MB, Zwaigenbaum L, MacLean JE (1998) Genetics of autism: overview and new directions. J Autism Dev Disord 28:351–368 [DOI] [PubMed] [Google Scholar]
- Turner M, Barnby G, Bailey A (2000) Genetic clues to the biological basis of autism. Mol Med Today 6:238–244 [DOI] [PubMed] [Google Scholar]
- Van Raay TJ, Foskett SM, Connors TD, Klinger KW, Landes GM, Burn TC (1997) The NTN2L gene encoding a novel human netrin maps to the autosomal dominant polycystic kidney disease region on chromosome 16p13.3. Genomics 41:279–282 [DOI] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Gonen D, Leventhal BL, Cook EH Jr (1999) Mutation screening of the UBE3A/E6-AP gene in autistic disorder. Mol Psychiatry 4:64–67 [DOI] [PubMed] [Google Scholar]
- Vicente AM, Coutinho A, Mota-Vieira L, Marques C, Olivera G (2000) Genetic variation of serotonin system genes in a sample of autistic families from Portugal. Am J Hum Genet Suppl 67:303 [Google Scholar]
- Weeks DE, Lathrop GM (1995) Polygenic disease: methods for mapping complex disease traits. Trends Genet 11:513–519 [DOI] [PubMed] [Google Scholar]
- Zerucha T, Stuhmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JL, Ekker M (2000) A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci 20:709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang C, Robitaille S, Grayson DR, Guidotti AR, Macciardi F, Leggo J, Holden JJA (2000) The Reln gene as a candidate locus for autism spectrum disorders. Am J Hum Genet Suppl 67:359 [Google Scholar]
- Zhong N, Ye L, Ju W, Brown WT, Tsiouris J, Cohen I (1999) 5-HTTLPR variants not associated with autistic spectrum disorders. Neurogenetics 2:129–131 [DOI] [PubMed] [Google Scholar]