Abstract
Although the role of genetic factors in the origin of Parkinson disease has long been disputed, several genes involved in autosomal dominant and recessive forms of the disease have been localized. Mutations associated with early-onset autosomal recessive parkinsonism have been identified in the Parkin gene, and recently a second gene, PARK6, involved in early-onset recessive parkinsonism was localized on chromosome 1p35-36. We identified a family segregating early-onset parkinsonism with multiple consanguinity loops in a genetically isolated population. Homozygosity mapping resulted in significant evidence for linkage on chromosome 1p36. Multipoint linkage analysis using MAPMAKER-HOMOZ generated a maximum LOD-score of 4.3, with nine markers spanning a disease haplotype of 16 cM. On the basis of several recombination events, the region defining the disease haplotype can be clearly separated, by ⩾25 cM, from the more centromeric PARK6 locus on chromosome 1p35-36. Therefore, we conclude that we have identified on chromosome 1 a second locus, PARK7, involved in autosomal recessive, early-onset parkinsonism.
Parkinson disease (PD [MIM 168600]) is a neurodegenerative disorder characterized by bradykinesia, resting tremor, muscular rigidity, and postural instability. The cerebral pathology includes loss of dopaminergic neurons, in particular at the substantia nigra and cytoplasmatic eosinophilic inclusions (i.e., Lewy bodies). The role of genetic factors in the origin of PD has long been disputed. However, several families segregating the disease as a dominant or recessive trait have been identified. At present, four genes implicated in autosomal dominant forms of parkinsonism have been identified or localized, including the α-synuclein gene (Polymeropoulos et al. 1997), the ubiquitin carboxy-terminal hydrolase-L1 gene (Leroy et al. 1998), and two yet-unidentified genes—PARK3 on chromosome 2p13 (Gasser et al. 1998) and PARK4 on chromosome 4p14-16.3 (Farrer et al. 1999). Until now, most mutations have been found in the Parkin gene (Kitada et al. 1998) and have been found to lead to a recessive form of juvenile parkinsonism (ARJP [MIM 600116]). The onset age may vary from 20 to 65 years, but in most patients the onset is at age <40 years (Lücking et al. 2000). Parkin mutations are estimated to explain up to 50% of familial patients with ARJP (Lücking et al. 2000). Recently, a new locus, PARK6 (MIM 605909), involved in autosomal recessive early-onset parkinsonism was localized on chromosome 1p35-36 in a single Italian family (Valente et al. 2001). In the present report, we describe a family segregating early-onset parkinsonism with multiple consanguinity loops that we identified in a genetically isolated population. We report the findings of a genome search using homozygosity mapping in an effort to localize the gene involved in the disease.
Patients were from a genetically isolated community in the southwestern region of The Netherlands. The study is part of a larger research program named “Genetic Research In Isolated Populations” (GRIP). The isolated population was founded ∼1750, by ∼150 individuals. The scientific protocol of GRIP has been approved by the Medical Ethical Committee of the Erasmus Medical Center Rotterdam. From the population, initially three patients (VII-2, VII-3 and VII-7) diagnosed with early-onset parkinsonism were ascertained, including two affected siblings and one apparently sporadic patient (fig. 1). The patients and their first-degree relatives were personally examined by the research physician, a neurologist, and a clinical geneticist. Clinical diagnosis of parkinsonism required the presence of at least two of the three cardinal signs (i.e., resting tremor, bradykinesia, and muscular rigidity) and absence of atypical features and of signs of involvement of other neurological systems (pyramidal, cerebellar, or autonomic). The extrapyramidal signs were evaluated by Hoehn-Yahr staging (HY) (Hoehn and Yahr 1967) and the Unified Parkinson’s Disease Rating Scale (UPDRS; maximum of motor subscale = 108) (Fahn et al. 1987). Furthermore, structural and functional brain-imaging studies were performed. The clinical examinations of available relatives revealed a fourth affected individual (VII-6), who was not known to have the diagnosis of parkinsonism.
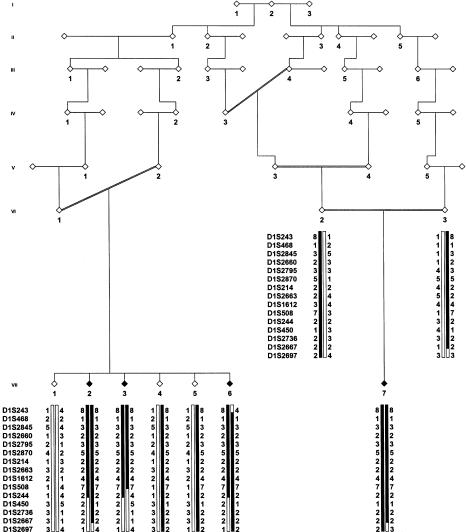
Figure 1.
Pedigree of family with autosomal recessive early-onset parkinsonism, and haplotypes of marker loci spanning the linked region on chromosome 1p36. To protect patient confidentiality, a diamond symbol has been used to mask the sex of all family members. Individuals VI-2, VI-3, and VII-1–VII-7 were examined clinically. Black symbols indicate definitely affected individuals; black bars denote the disease-associated haplotype.
The clinical details of the four affected subjects are reported in table 1. Onset of symptoms was at age ⩽40 years in three subjects (VII-2, VII-3, and VII-7). The fourth affected individual (VII-6), who was age 40 years at the time of the clinical examination, was newly diagnosed with parkinsonism and could not indicate the age at onset of the disease. None of the patients exhibited atypical features or signs of involvement of additional neurological systems. The parkinsonian signs responded well to therapy with l-dopa or dopamine agonists in two patients (VII-3 and VII-7); at the time of clinical examination, the two other patients still had not been treated with antiparkinsonian drugs. One patient (VII-7) showed motor fluctuations of “wearing-off” type, l-dopa–induced dyskinesias in the “on” phase, and “off”-phase dystonia (i.e., laterocollis). Mild blepharospasm was present in patient VII-3, who receives dopamine-agonist therapy. Tendon reflexes in lower limbs were brisk in three of the patients (VII-3, VII-6, and VII-7). Babinski sign was absent. Two of the patients also showed neurotic signs, and one had suffered from psychotic episodes. In all patients, the progression of disease was slow. Structural brain images were unremarkable, in agreement with the diagnosis of idiopathic parkinsonism. Brain single-positron–emission computed tomography with dopamine-transporter tracer, performed in patients VII-2 and VII-3, showed severe abnormalities consistent with presynaptic dysfunction of nigrostriatal dopaminergic systems. Autopsy data are not available. At the time of clinical examination, the ages of unaffected individuals VII-1, VII-4 and VII-5 were 54, 50, and 46 years, respectively.
Table 1.
Clinical Phenotype of Four Patients with Parkinsonism
|
Age at(years) |
Symptoma |
|||||||||||
| Patient | Time of Study | Onsetb | RT | PT | B | R | P | AS | Therapy | HY | UPDRS | Other Characteristics |
| VII-2 | 50 | 40 | X | X | X | X | Untreated | II | 29 | Psychotic episodes | ||
| VII-3 | 48 | 31 | X | X | X | X | X | X | Pergolide | III | 57 | Neurotic signs, blepharospasm |
| VII-6 | 40 | NA | X | X | X | X | Untreated | III | 22 | Neurotic signs | ||
| VII-7 | 38 | 27 | X | X | X | X | X | l-dopa/IDD, entacapone | III | 42 | Wearing-off, dyskinesias, off dystonia | |
RT = resting tremor; PT = postural tremor; B = bradykinesia; R = muscular rigidity; P = loss of postural reflexes; AS = asymmetric onset of symptoms.
NA = not available.
The genealogical history of the isolate has been computerized as far back as 1750. Although the patients were not aware of consanguinity in their family, the pedigree of each of the two sibships showed multiple consanguinity loops (fig. 1). The pedigree is drawn on the basis of the two shortest consanguinity loops for both parent pairs (VI-1/V-2 and VI-2/VI-3), which links the two sibships to a common ancestor six generations ago. However, other connections between the sibships exist, including a second loop linking VI-1 and V-2 to a common ancestor, who is also related to VI-2.
The pedigree structure is consistent with an autosomal recessive mode of inheritance, and the kindred therefore was considered to be suitable for homozygosity mapping (Lander and Botstein 1987). For all patients and available first-degree relatives, genomic DNA was isolated from peripheral blood, according to the method described by Miller et al. (1988). For the systematic genome scan, short tandem-repeat polymorphisms (STRPs) from the ABI PRISM Linkage Mapping Set MD-10 (Applied Biosystems) were used. Additional markers for fine-mapping and the PARK6 region were obtained from the Généthon and Marshfield (Center for Medical Genetics, Marshfield Medical Research Foundation) genetic-marker sets. Marker order and distances were obtained from the Marshfield (Center for Medical Genetics, Marshfield Medical Research Foundation) integrated linkage map. Markers were amplified from genomic DNA, according to methods specified by the manufacturers. PCR products were pooled and loaded onto an ABI377 automated sequencer (filter set D; 5% denaturing FMC LongRanger acrylamide gel), and data were analyzed by ABI GENESCAN3.1 and ABI GENOTYPER2.1 software.
In the initial screens, only three definitely affected individuals (VII-2, VII-3, and VII-7) were genotyped. We first tested for linkage of our family to known PD loci (Polymeropoulos et al. 1997; Gasser et al. 1998; Kitada et al. 1998; Leroy et al. 1998; Farrer et al. 1999), using STRPs from the ABI PRISM Linkage Mapping Set MD-10. At chromosome 4q22, the markers D4S414 and D4S1572 were assessed; at chromosome 4p14, markers D4S419 and D4S405; at chromosome 4p14-16.3, markers D4S419 and D4S1592; at chromosome 6q25.2-q27, markers D6S1581 and D6S264; and, at chromosome 2p13, markers D2S337 and D2S2368. The analysis of the markers flanking the known PD genes did not show evidence for homozygosity, indicating that the disease in the family cannot be explained by one of these known genes (data not shown). We then performed a systematic genome screen. LOD scores were generated by the program MAPMAKER-HOMOZ (Kruglyak et al. 1995), under the assumptions of equal recombination for males and females, autosomal recessive inheritance, and gene frequency .004. In the initial genome screen, the family structure used in the analysis is depicted in figure 1, and allele frequencies were estimated on the basis of the DNA of 51 spouses of patients participating in the various studies in GRIP. The three individuals included in the initial screen (VII-2, VII-3, and VII-7) were homozygous for six markers, located on chromosomes 1, 5, 11, 17, and 21. Only for the chromosome 1 region, two adjacent markers (D1S468 and D1S214) were found to be homozygous in all patients, leading to a maximum LOD score of 2.8.
The newly diagnosed patient (VII-6) was identified after this initial screen and was typed, together with unaffected first-degree relatives (VI-2, VI-3, VII-1, VII-4, and VII-5), only for the five regions at which the first three patients were homozygous.
Homozygosity remained only for markers on chromosomes 1, 5, and 17. On chromosomes 5 and 17, homozygosity was found with very-frequent (34% and 42%, respectively) alleles of the markers D5S1981 and D17S944. For closely linked (i.e., <10 cM distant) markers, all four patients were heterozygous, indicating that the observed alleles for these markers could be identical by state (IBS). In contrast, the population frequency of the homozygous allele at D1S468 was 9%. The chances that the homozygous allele in our distantly related patients is IBS are very small. In addition, all four patients were homozygous for the flanking marker D1S214, indicating that the alleles were most likely identical by descent. We therefore saturated the region surrounding D1S468 and D1S214 with 13 additional markers and found that all patients were homozygous for 9 of the markers studied, whereas the unaffected parents and two unaffected siblings were all either heterozygous or noncarriers of the disease haplotype (fig. 1). When LOD scores for the disease haplotype were calculated, equal allele frequencies for each marker were assumed, because population allele frequencies were not available. Penetrance was assumed to be 100% by age 40 years. The most conservative analysis—that is, under the assumption that there is a second-degree relationship between the parents of the sibships—resulted in a maximum multipoint LOD score of 4.3. Subsequent haplotype analysis showed homozygosity of all four patients, for a region of ∼16 cM on the sex-averaged linkage map. The first recombination events were observed for marker D1S243 on the telomeric side in individual VII-6 and for marker D1S244 on the centromeric side in individual VII-3.
Because of Valente et al.'s (2001) recent report of linkage of an early-onset autosomal recessive form of parkinsonism to chromosome 1p35-36, we investigated whether we could exclude linkage to the PARK6 region in the family that we studied; we tested nine markers from the PARK6 region (table 2). The patients in this family showed no homozygosity at any of these markers. The PARK6 critical region, between markers D1S483 and D1S247, is localized ⩾25 cM centromeric of our critical region. Considering the evidence that the critical region in the family that we studied does not overlap with the region encompassing the PARK6 gene identified by Valente et al. (2001) and taking into account the recommendations that Lander and Kruglyak (1995) have made with regard to the reporting of linkage findings, we report a significant linkage finding for the presence, on chromosome 1p, of a second locus for autosomal recessive early-onset parkinsonism; and we propose to name it “PARK7.”
Table 2.
Genotypes of Affected Individuals, for Markers of the PARK6 Region[Note]
|
Genotype ofAffected Individual |
||||
| Markera | VII-2 | VII-3 | VII-6 | VII-7 |
| D1S199 | 2 5 | 3 5 | 2 5 | 3 4 |
| D1S483 | 1 1 | 1 2 | 1 1 | 2 2 |
| D1S478 | 3 5 | 1 3 | 3 5 | 2 3 |
| D1S2828 | 4 1 | 4 5 | 4 1 | 4 3 |
| D1S2732 | 2 4 | 2 3 | 2 4 | 1 4 |
| D1S2702 | 2 4 | 2 3 | 2 4 | 4 1 |
| D1S2734 | 5 4 | 5 5 | 5 4 | 4 1 |
| D1S2885 | 4 1 | 4 2 | 4 1 | 1 3 |
| D1S247 | 4 2 | 1 6 | 4 2 | 3 5 |
Source.— Valente et al. (2001).
Order is according to the Marshfield (Center for Medical Genetics, Marshfield Medical Research Foundation) integrated linkage map.
Both PARK6 (Valente et al. 2001) and PARK7 were identified by homozygosity mapping. Although this is a very powerful approach in consanguineous pedigrees (Lander and Botstein 1987), some possible methodological problems recently have been raised in articles published in the Journal (Miano et al. 2000). Miano et al. (2000) have shown that patients may be homozygous for large regions by chance—in particular if there are multiple loops in the pedigree, such as occur in the pedigree that we studied. However, our most conservative analysis, based on an analysis assuming a second-degree relationship, still yielded a LOD score >4. In addition, the fact that the affected individuals from the two sibships share an identical homozygous haplotype, whereas none of the unaffected first-degree relatives was homozygous for this haplotype, makes a false-positive finding less likely.
Using homozygosity mapping, we have identified a third locus for autosomal recessive early-onset parkinsonism: PARK7. Similar to what has been reported in other studies of early-onset parkinsonism and Parkin mutations (Lücking et al. 2000), the disease in the patients of the family that we studied is characterized by a slow progression. Dystonic features, including blepharospasm and laterocollis, and brisk tendon reflexes have also been found in patients with Parkin mutations (Abbas et al. 1999; Lücking et al. 2000; Tassin et al. 2000). PARK7 is the second locus, in the chromosome 1p36 region, involved in autosomal recessive, early-onset parkinsonism. The region defining the disease haplotype can be clearly separated from that of the more centromeric PARK6 locus. Although the genomic sequence of the critical region is far from complete and many gaps remain, a first analysis of the Project Ensembl and Human Genome Project Working Draft at UCSC databases showed that inside the critical region ⩾25 genes are located, including 7 genes with an unknown function. Possible candidate genes are the gene encoding the vesicle-associated membrane protein 3 (VAMP3) and the gene encoding protein kinase C, zeta (PRKCZ). Telomeric of D1S243, a ubiquitin-conjugating enzyme has been localized. Although this would be an obvious candidate, its location is outside the candidate region. The observation, on chromosome 1p, of two closely linked loci for early-onset parkinsonism raises the question whether there might have been a duplication of this chromosomal region during evolution; however, comparison of the gene contents from the PARK6 and PARK7 critical regions and the syntenic region in the mouse does not support this idea.
Recessive mutations in the Parkin gene have been shown to be a common cause of early-onset parkinsonism (Lücking et al. 2000). Our findings and those of Valente et al. (2001) strongly suggest there may be other recessive mutations, in at least two unidentified genes involved in the disease. Analysis of additional families with early-onset parkinsonism lacking Parkin mutations will reveal the importance of these two new loci for early-onset parkinsonism.
The frequency of recessive mutations in early-onset parkinsonism may even have masked the genetic origin of PD for a long time, since familial aggregation and the identification of consanguinity require extensive genealogical research. Therefore, it remains to be determined whether these common recessive mutations are involved in late-onset forms of the disease as well. Studies of inbred and/or isolated populations such as GRIP may therefore help to further dissect the pathogenesis of PD.
Acknowledgments
This study was supported by a grant from the Netherlands Organisation for Scientific research (NWO). Ed Boere, Petra Veraart, and Hilda Kornman are acknowledged for their contribution to the genealogical research; Angela Jakobs and Erwin Wauters are acknowledged for their help in genotyping. All patients and their relatives, as well as general practitioners, neurologists, and nursing-home physicians, are thanked for making this study possible.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for information on polymorphic markers and localization of polymorphic markers)
- Généthon, http://www.genethon.fr/ (for information on polymorphic markers and localization of polymorphic markers)
- Human Genome Project Working Draft at UCSC, http://genome.cse.ucsc.edu/ (for identification of candidate genes VAMP3 and PRKCZ)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PD [MIM 168600], ARJP [MIM 600116], and PARK6 [MIM 605909])
- Project Ensembl, http://www.ensembl.org/ (for identification of candidate genes VAMP3 and PRKCZ)
References
- Abbas N, Lücking CB, Ricard S, Durr A, Bonifati V, De Michele G, Bouley S, Vaughan JR, Gasser T, Marconi R, Broussolle E, Brefel-Courbon C, Harhangi BS, Oostra BA, Fabrizio E, Bohme GA, Pradier L, Wood NW, Filla A, Meco G, Denefle P, Agid Y, Brice A (1999) A wide variety of mutations in the Parkin gene are responsible for autosomal recessive parkinsonism in Europe: French Parkinson's Disease Genetics Study Group and the European Consortium on Genetic Susceptibility in Parkinson's Disease. Hum Mol Genet 8:567–574 [DOI] [PubMed] [Google Scholar]
- Fahn S, Elton RL, UPDRS Development Committee (1987) Unified Parkinson’s disease rating scale. In: Fahn S, Marsden CD, Calne DB (eds) Recent developments in Parkinson’s disease. Vol 2. MacMillan Healthcare Information, Florham Park, NJ, pp 153–163 [Google Scholar]
- Farrer MJ, Gwinn-Hardy K, Muenter M, DeVrieze FW, Crook R, Perez-Tur J, Lincoln S, Maraganore D, Adler C, Newman S, MacElwee K, McCarthy P, Miller C, Waters C, Hardy J (1999) A chromosome 4p haplotype segregating with Parkinson’s disease and postural tremor. Hum Mol Genet 8:81–85 [DOI] [PubMed] [Google Scholar]
- Gasser T, Müller-Myhsok B, Wszolek ZK, Oehlmann R, Calne DB, Bonifati V, Bereznai B, Fabrizio E, Vieregge P, Horstmann RD (1998) A susceptibility locus for Parkinson’s disease maps to chromosome 2p13. Nat Genet 18:262–265 [DOI] [PubMed] [Google Scholar]
- Hoehn MM, Yahr MD (1967) Parkinsonism: onset, progression and mortality. Neurology 17:427–442 [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N (1998) Mutations in the Parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392:605-608 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Lander ES (1995) Rapid multipoint linkage analysis of recessive traits in nuclear families, including homozygosity mapping. Am J Hum Genet 56:519–527 [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D (1987) Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236:1567–1570 [DOI] [PubMed] [Google Scholar]
- Lander ES, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, Dehejia A, Lavedan C, Gasser T, Steinbach PJ, Wilkinson KD, Polymeropoulos MH (1998) The ubiquitin pathway in Parkinson's disease. Nature 395:451–452 [DOI] [PubMed] [Google Scholar]
- Lücking CB, Durr A, Bonifati V, Vaughan J, De Michele G, Gasser T, Harhangi BS, Meco G, Denefle P, Wood NW, Agid Y, Brice A (2000) Association between early-onset Parkinson’s disease and mutations in the Parkin gene: French Parkinson’s Disease Genetics Study Group. N Engl J Med 342:1560–1567 [DOI] [PubMed] [Google Scholar]
- Miano MG, Jacobson SG, Carothers A, Hanson I, Teague P, Lovell J, Cideciyan AV, Haider N, Stone EM, Sheffield VC, Wright AF (2000) Pitfalls in homozygosity mapping. Am J Hum Genet 67:1348–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL (1997) Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 276:2045–2047 [DOI] [PubMed] [Google Scholar]
- Tassin J, Durr A, Bonnet AM, Gil R, Vidailhet M, Lücking CB, Goas JY, Durif F, Abada M, Echenne B, Motte J, Lagueny A, Lacomblez L, Jedynak P, Bartholome B, Agid Y, Brice A (2000) Levodopa-responsive dystonia. GTP cyclohydrolase I or Parkin mutations? Brain 123:1112–1121 [DOI] [PubMed] [Google Scholar]
- Valente EM, Bentivoglio AR, Dixon PH, Ferraris A, Ialongo T, Frontali M, Albanese A, Wood NW (2001) Localization of a novel locus for autosomal recessive early-onset parkinsonism, PARK6, on human chromosome 1p35-p36. Am J Hum Genet 68:895–900 [DOI] [PMC free article] [PubMed] [Google Scholar]