Abstract
Inherited growth-hormone insensitivity (GHI) is a heterogeneous disorder that is often caused by mutations in the coding exons or flanking intronic sequences of the growth-hormone receptor gene (GHR). Here we describe a novel point mutation, in four children with GHI, that leads to activation of an intronic pseudoexon resulting in inclusion of an additional 108 nt between exons 6 and 7 in the majority of GHR transcripts. This mutation lies within the pseudoexon (A−1→G−1 at the 5′ pseudoexon splice site) and, under in vitro splicing conditions, results in inclusion of the mutant pseudoexon, whereas the wild-type pseudoexon is skipped. The presence of the pseudoexon results in inclusion of an additional 36–amino acid sequence in a region of the receptor known to be involved in homo-dimerization, which is essential for signal transduction.
Inclusion of intronic sequences by aberrant splicing is a common cause of genetic disease (Krawczak et al. 1992). This usually occurs through the creation or activation of a splice site within 100 nt of the normally used splice site (Nakai and Sakamoto 1994), as a result of a point mutation in the genomic DNA sequence. Such mutations lead to the inclusion of intronic sequences immediately flanking the exonic sequence. Intronic DNA frequently encodes potential exonic sequences (Dunham et al. 1999)—so-called “pseudoexons” (Sun and Chasin 2000)—that are not recognized by the splicing machinery.
Patients with growth-hormone insensitivity (GHI [MIM 600946 and MIM 245590]) express a variable phenotype. The most severe form, Laron syndrome (MIM 262500), is characterized by severe growth retardation and dysmorphic facial features, associated with elevated circulating growth hormone (GH) and subnormal levels of insulin-like growth factor 1 (IGF-1), IGF-binding protein 3 (IGFBP3), and acid-labile subunit (Savage et al. 1993). Levels of GH-binding protein (GHBP), the soluble extracellular domain of the GHR, are low or undetectable in 80% of cases (Buchanan et al. 1991; Woods et al. 1997), reflecting defective GHR expression or defective ligand binding as the underlying cause of the disease. In cases of partial or atypical GHI, GHBP levels and facial appearance are often normal, and growth retardation is less severe (Attie et al. 1995; Goddard et al. 1995; Carlsson 1996). Mutations within the GHR have frequently been implicated as the cause of GHI.
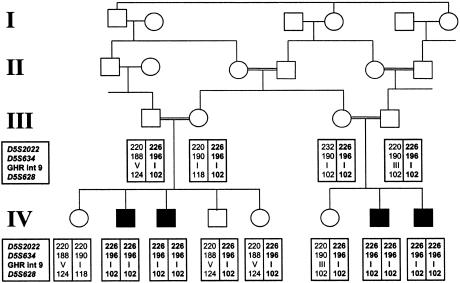
The present study concerns one highly consanguineous Pakistani kindred with four male individuals affected with atypical GHI (fig. 1). The four patients (two pairs of siblings) have marked short stature. The standard deviation score (SDS)—an indicator of height that is independent of age, is appropriate for the population under study, and reflects the divergence from the mean—was found to be −3.3 to −5.6 in these children. In addition, there were low levels of IGF-1 (20–29 ng/ml [normal level >50 ng/ml]), detectable levels of GHBP (28.1%–51.7% [normal range 14.1%–42.9%]), and a normal facial appearance. Homozygosity mapping of several polymorphic markers surrounding the GHR identified, in all four patients, a homozygous region that was absent in their unaffected siblings. This region (>3 cM) spanned three markers (D5S2022, D5S634, and D5S628; fig. 1). In addition, six nucleotide polymorphisms in intron 9 of the GHR (Amselem et al. 1989) were investigated by PCR and direct sequencing and were found to be homozygous in all four patients but not in other family members (GHR int 9; fig. 1). Sequencing of the coding exons flanking splice junctions and branch point sequences showed no mutations. We therefore hypothesized that the defect was either within one of the introns of the GHR or within another gene closely linked to it.
Figure 1.
Homozygosity mapping. The pedigree of the consanguineous Pakistani family is shown. Black and white symbols denote affected and unaffected individuals, respectively. D5S628, D5S634, and D5S2022 dinucleotide repeat markers were amplified by PCR, and resultant fragments were electrophoresed on a 5% PAGE gel on an ABI 377 automated sequencer; analysis was performed using Genescan software (Applied Biosystems). The complex single polymorphic region in intron 9 of GHR was sequenced using the ABI Prism Big Dye Sequencing kit and an ABI 377 automated DNA sequencer (Applied Biosystems), in accordance with the manufacturer’s instructions. Genotypes of this region are indicated by Roman numerals, as defined by Amselem et al. (1989).
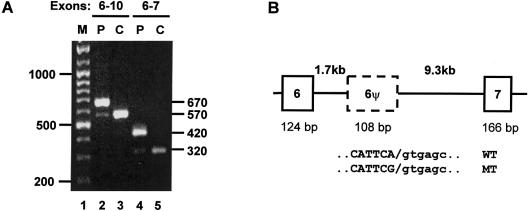
GHR mRNA transcripts in fibroblasts from patients and from control subjects were investigated by reverse-transcriptase PCR (RT-PCR). Two nested amplifications were performed. First-round primers were directed to exons 2 and 10, and second-round primers were directed either to exons 6 and 7 or to exons 6 and 10 of the GHR. In cDNA from control fibroblasts, the only products were of the expected size (fig. 2A, lanes 3 [574 bp] and 5 [320 bp]), whereas in cDNA from patient samples, a larger band was produced (fig. 2A, lanes 2 [682 bp] and 4 [428 bp]) in addition to the much less abundant band of the expected size. Direct sequencing of each product was performed, revealing that the fragments of the expected size for both patient and control samples corresponded to the predicted mRNA sequence. The larger fragments seen in patient cDNA contained a 108-bp insertion between exons 6 and 7.
Figure 2.
Pseudoexon inclusion demonstrated in cDNA from a patient, but not from a control. A, mRNA was extracted from patient and control skin fibroblasts, and cDNA was generated using MMLTV reverse transcriptase (Promega). PCR amplification was achieved by a nested PCR, using primers in exons 2 and 10 in first rounds and 6 and 10 (lanes 2 and 3) or 6 and 7 (lanes 4 and 5) in second rounds. Products obtained from the patient (P) or control (C) are indicated above the lanes. Sizes (in bp) of the markers (M) are given on the left of the gel, and sizes of the PCR products are given on the right. B, Scheme of the genomic structure between exons 6 and 7, derived from long PCR through use of the Extensor Long PCR system (Advanced Biotechnologies). Boxes represent exons, the dashed box represents the pseudoexon, and horizontal lines represent introns. Intron lengths (in kb) are given above the diagram in bold type, and exon lengths (in bp) are given below the diagram. The sequences of the wild-type (WT) and mutant (MT) pseudoexon (in capital letters) and the flanking intronic 5′ splice site (in lowercase letters) is shown below. The slash (/) indicates the cleavage site.
The novel 108-bp inserted sequence did not match the sequence flanking either exon 6 or 7, implying that the sequence was derived either from activation of a pseudoexon (6ψ) within intron 6 or from an exon introduced into the intron by a recombination event. We use the term “pseudoexon” to indicate a potential exon, containing adequate 5′ and 3′ splice sites, that is not normally spliced into mature mRNA by the cellular splicing machinery (Sun and Chasin 2000). To ascertain the position of the new sequence, long PCR was performed to span intron 6 in patient and control DNA samples. Intron 6 is 11 kb in length, and 6ψ is located 1.7 kb from exon 6 (fig. 2B). This sequence has not been reported previously and has now been deposited in GenBank (accession numbers AF344653 and AF344654). Analysis by PCR and sequencing of a 650-bp fragment spanning 6ψ from a control genomic DNA sample revealed that 6ψ was flanked by 5′ and 3′ splice sites. Patient DNA was found to have a homozygous base change (A→G) at the last nucleotide of the pseudoexon (fig. 2B). The parents and several unaffected siblings were heterozygous for the mutation but were of normal stature, as was a single sibling who was homozygous for the wild-type A allele. The mutation was not detected by minisequencing (SnaPshot, Applied Biosystems) in 100 unrelated control chromosomes of similar ethnic origin.
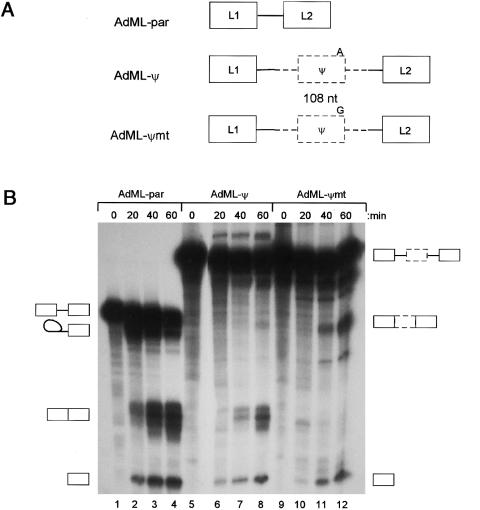
To ascertain whether this base change was necessary and sufficient for 6ψ inclusion in transcription, we inserted the pseudoexon and flanking intronic sequences into the intron of a well-characterized splicing reporter derived from the adenovirus major late first and second leader exons (AdMLpar) (Gozani et al. 1994). Two heterologous substrates were made, one containing the wild-type 6ψ pseudoexon (AdML-ψ; fig. 3A) and the other the mutated 6ψ exon (AdML-ψmt). Under splicing conditions, the pseudoexon was skipped in AdML-ψ but was efficiently included in AdML-ψmt (fig. 3B).
Figure 3.
Pseudoexon inclusion caused by the A→G mutation. A, Structure of the three pre-mRNA substrates used for in vitro splicing experiments. Solid boxes and lines indicate AdML sequences, and dashed boxes and lines are pseudoexon fragments and flanking intron. B, In vitro splicing. PCR fragments containing wild-type and mutant pseudoexons and surrounding introns were cloned into pCR Blunt (Invitrogen), sequenced, and used as further templates for PCR. AdML-ψ and AdML-ψmt were made by overlap-extension proof-reading PCR, using primers GHR-pe-AdMLpar2-S (5′-ccctcactaaagCAGGAGTATCATGCTGCT) and GHR-pe-AdMLpar2-A (5′-cttgactactgcTAATGACAAAATTGGCATCT), and primers AdMLpar-int51-S (5′-GCAGTAGTCAA) and AdMLpar-L2-A (5′-ATCCAAGAGTACTGGAA), and a second-round PCR with GHR-pe-AdMLpar2-S and AdMLpar-L2-A. The second-round PCR fragment was used as a megaprimer in a third-round PCR with a T7 primer, using AdMLpar as a template. Assembled AdML-ψ and AdML-ψmt PCR products were gel purified, cloned into pCR-blunt, and sequenced. Transcription templates were generated by a proof-reading PCR, with a T7 primer and AdMLpar-L2-A, and capped pre-mRNA transcribed with T7 RNA polymerase in the presence of [32P]-labeled GTP. Splicing reactions used 20 fmol of RNA, 32% Hela nuclear extract, 0.5 mM ATP, 3.2 mM MgCl2, 2.6% polyvinyl alcohol, and 60 mM KCl at 30°C (Chew et al. 2000). Reactions were deproteinized with phenol, precipitated with ethanol, and run on polyacrylamide gels ranging from 5.5% to 8%, before autoradiography. A representative in vitro splicing experiment is shown. Pre-mRNA substrates are indicated above the lanes, together with the time of the splicing reactions. The identity of the bands is indicated by the symbols to the sides of the gel and was confirmed by comparison with AdMLpar, by electrophoresis on higher-percentage gels to shift the lariat-intermediate, and by isolation of the mRNA from the gel, RT-PCR, and sequencing.
Pseudoexon inclusion has previously been reported only in association with the creation, by mutations in intronic sequence, of novel splice or branch sites (Highsmith et al. 1994; Wang et al. 1997; Chillon et al. 1995; De Klein et al. 1998; Vervoort et al. 1998; Dwi Pramono et al. 2000). By contrast, the wild-type pseudoexon in this case already possesses a legitimate branch site, 5′ and 3′ splice-site sequences, and the mutation is within the pseudoexonic sequence itself. The exon 6ψ mutation changes A−1 to G−1 at the 5′ splice site (consensus G−1/G1U2G3A4G5C6, where the slash indicates the cleavage site). The mutation brings the sequence closer to complementarity with U1 snRNP, but the mechanism by which this causes such a dramatic change in splice-site selection is unclear. First, an A−1 is found in ∼15% of natural exons (Long et al. 1998). Second, a comparison between G−1 and A−1 shows that A−1 does not reduce in vivo splicing efficiency in a constitutively spliced exon (Aebi et al. 1986) or selection of alternative 5′ splice sites (Lear et al. 1990). Previous work has shown that the A−1 change increases exon skipping in vitro by only a small amount (∼15%) (Aebi et al. 1986), whereas exon skipping is dramatically increased in the wild type 6ψ (A-1) when compared with the mutant (G−1) (fig. 3B). In mammals, binding of U1 snRNP to 5′ splice sites does not correlate well with splicing in vitro (Eperon et al. 1993); however, we hypothesize that the point mutation improves the interaction of the pre-mRNA with U1 snRNP, resulting in recognition, by the spliceosome, of the mutant 6ψ as an exon. This interaction is the subject of continuing investigation.
The inclusion of 6ψ is predicted to lead to the insertion of 36 amino acid residues, between exons 6 and 7, that will be in-frame and that will lack a stop codon. This region of the GHR protein is critically involved in receptor dimerization (Cunningham et al. 1991). It is likely that disruption of this process—and, hence, failure of GH signalling—would result from this mutation, as has been demonstrated with another mutation in this region (Duquesnoy et al. 1994). Classical GHBP-negative GHI exhibits a mean height SDS of −6.45 (range −3.0 to −10.4), whereas the height SDS of these subjects was less severe and more typical of GHBP-positive GHI (mean −4.89; range −2.2 to −10.4) (Woods et al. 1997). This slightly less severe phenotype of the four affected boys may be due to a small amount of normally spliced GHR mRNA (fig. 2A), which produces an entirely normal GHR.
We believe that this is the first demonstration that a pseudoexonic point mutation is necessary and sufficient to activate a pseudoexon and cause disease. Mutations causing pseudoexonic inclusion may be more prevalent than the current literature suggests; pseudoexonic sequences are frequently found within introns (Dunham et al. 1999), and, in the human hprt gene, pseudoexons outnumber legitimate exons by as many as 10:1 (Sun and Chasin 2000). Detection of these events in disease is hampered since introns, because of their frequently large size, are often excluded from mutational analyses, and it is often impossible or impractical to perform cDNA analysis.
Acknowledgments
We thank David Curtis, for his helpful advice; Andrew Overall, for supplying the control chromosome DNA; and Ragnar Bjarnason and Peter Clayton, for maintenance of the fibroblast cell line. This work was supported by the Wellcome Trust. S.A.A. is supported by a Research Fellowship from the Joint Research Board, St. Bartholomew’s Hospital.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for accession numbers AF344653 and AF344654)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for GHI [MIM 600946, MIM 245590, and MIM 262500])
References
- Aebi M, Hornig H, Padgett RA, Reiser J, Weissmann C (1986) Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell 47:555–565 [DOI] [PubMed] [Google Scholar]
- Amselem S, Duquesnoy P, Attree O, Novelli G, Bousnina S, Postel-Vinay MC, Goossens M (1989) Laron dwarfism and mutations of the growth hormone-receptor gene. N Engl J Med 321:989–995 [DOI] [PubMed] [Google Scholar]
- Attie KM, Carlsson LMS, Rundle AC, Sherman B (1995) Evidence for partial growth hormone insensitivity among patients with idiopathic short stature. J Pediatr 127:244–250 [DOI] [PubMed] [Google Scholar]
- Buchanan CR, Maheshwari HG, Norman MR, Morrell DJ, Preece MA (1991) Laron-type dwarfism with apparently normal high affinity serum growth hormone-binding protein. Clin Endocrinol (Oxf) 35:179–185 [DOI] [PubMed] [Google Scholar]
- Carlsson LMS (1996) Partial growth hormone insensitivity in childhood. In: Ross RJM, Savage MO (eds) Clinical endocrinology and metabolism: growth hormone resistance. Ballière Tindall, Sydney, Philadelphia, Tokyo, Toronto, pp 389–400 [DOI] [PubMed] [Google Scholar]
- Chew SL, Baginsky L, Eperon IC (2000) An exonic splicing silencer in the testes-specific DNA ligase III beta exon. Nucleic Acids Res 28:402–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillon M, Dork T, Casals T, Gimenez J, Fonknechten N, Will K, Ramos D, Nunes V, Estivill X (1995) A novel donor splice site in intron 11 of the CFTR gene, created by mutation 1811=1.6kbA→G, produces a new exon: high frequency in Spanish cystic fibrosis chromosomes and association with severe phenotype. Am J Hum Genet 56:623–629 [PMC free article] [PubMed] [Google Scholar]
- Cunningham BC, Ultsch M, De Vos AM, Mulkerrin MG, Clauser KR, Wells JA (1991) Dimerization of the extracellular domain of the human growth hormone receptor by a single hormone molecule. Science 254:821–825 [DOI] [PubMed] [Google Scholar]
- De Klein, A Riegman PH, Bijlsma EK, Heldoorn A, Muijtjens M, den Bakker MA, Avezaat CJ, Zwarthoff EC (1998) A G→A transition creates a branch point sequence and activation of a cryptic exon, resulting in the hereditary disorder neurofibromatosis 2. Hum Mol Genet 7:393–398 [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, et al (1999) The DNA sequence of human chromosome 22. Nature 402:489–495 [DOI] [PubMed] [Google Scholar]
- Duquesnoy P, Sobrier ML, Duriez B, Dastot F, Buchanan CR, Savage MO, Preece MA, Craescu CT, Blouquit Y, Goossens M, Amselem S (1994) A single amino acid substitution in the exoplasmic domain of the human growth hormone (GH) receptor confers familial GH resistance (Laron syndrome) with positive GH-binding activity by abolishing receptor homodimerization. EMBO J 13:1386–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwi Pramono ZA, Takeshima Y, Surono A, Ishida T, Matsuo M (2000) A novel cryptic exon in intron 2 of the human dystrophin gene evolved from an intron by acquiring consensus sequences for splicing at different stages of anthropoid evolution. Biochem Biophys Res Commun 267:321–328 [DOI] [PubMed] [Google Scholar]
- Eperon IC, Ireland DC, Smith RA, Mayeda A, Krainer AR (1993) Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J 12:3607–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard AD, Covello R, Luoh SM, Clackson T, Attie KM, Gesundheit N, Rundle AC, Wells JA, Carlsson LM (1995) Growth hormone receptor mutations in idiopathic short stature. N Engl J Med 333:1093–1098 [DOI] [PubMed] [Google Scholar]
- Gozani O, Patton JG, Reed R (1994) A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J 13:3356–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highsmith WE, Burch LH, Zhou Z, Olsen JC, Boat TE, Spock A, Gorvoy JD, Quittel L, Friedman KJ, Silverman LM, Boucher RC, Knowles MR (1994) A mutation in the cystic fibrosis gene in patients with pulmonary disease but normal sweat chloride concentrations. N Engl J Med 331:974–980 [DOI] [PubMed] [Google Scholar]
- Krawczak M, Reiss J, Cooper DN (1992) The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet 90:41–54 [DOI] [PubMed] [Google Scholar]
- Lear AL, Eperon LP, Wheatley IM, Eperon IC (1990) Hierarchy for 5′ splice site preference determined in vivo. J Mol Biol 211:103–115 [DOI] [PubMed] [Google Scholar]
- Long M, de Souza SJ, Rosenberg C, Gilbert W (1998) Relationship between “proto-splice sites” and intron phases: evidence from dicodon analysis. Proc Natl Acad Sci USA 95:219–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Sakamoto H (1994) Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene 141:171–177 [DOI] [PubMed] [Google Scholar]
- Savage MO, Blum WF, Ranke MB, Postel-Vinay MC, Cotterill AM, Hall K, Chatelain PG, Preece MA, Rosenfeld RG (1993) Clinical features and endocrine status in patients with growth hormone insensitivity (Laron syndrome). J Clin Endocrinol Metab 77:1465–1471 [DOI] [PubMed] [Google Scholar]
- Sun H, Chasin LA (2000) Multiple splicing defects in an intronic false exon. Mol Cell Biol 20:6414–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoort R, Gitzelmann R, Lissens W, Liebaers I (1998) A mutation (IVS8+0.6kbdelTC) creating a new donor splice site activates a cryptic exon in an Alu-element in intron 8 of the human beta-glucuronidase gene. Hum Genet 103:686–693 [DOI] [PubMed] [Google Scholar]
- Wang M, Dotzlaw H, Fuqua SA, Murphy LC (1997) A point mutation in the human estrogen receptor gene is associated with the expression of an abnormal estrogen receptor mRNA containing a 69 novel nucleotide insertion. Breast Cancer Res Treat 44:145–151 [DOI] [PubMed] [Google Scholar]
- Woods KA, Dastot F, Preece MA, Clark AJL, Postel-Vinay M-C, Chatelain PG, Ranke MB, Rosenfeld RG, Amselem S, Savage MO (1997) Phenotype:genotype relationship in growth hormone insensitivity syndrome. J Clin Endocrinol Metab 82:3529–3535 [DOI] [PubMed] [Google Scholar]