Abstract
Mutations in ZFHX1B, encoding Smad-interacting protein 1 (SIP1), have been recently reported to cause a form of Hirschsprung disease (HSCR). Patients with ZFHX1B deficiency typically show mental retardation, delayed motor development, epilepsy, microcephaly, distinct facial features, and/or congenital heart disease, in addition to the cardinal form of HSCR. To investigate the breadth of clinical variation, we studied DNA samples from six patients with clinical profiles quite similar to those described elsewhere for ZFHX1B deficiency, except that they did not have HSCR. The results showed the previously reported R695X mutation to be present in three cases, with three novel mutations—a 2-bp insertion (760insCA resulting in 254fs262X), a single-base deletion (270delG resulting in 91fs107X), and a 2-bp deletion (2178delTT resulting in 727fs754X)—newly identified in the other three. All mutations occurred in one allele and were de novo events. These results demonstrate that ZFHX1B deficiency is an autosomal dominant complex developmental disorder and that individuals with functional null mutations present with mental retardation, delayed motor development, epilepsy, and a wide spectrum of clinically heterogeneous features suggestive of neurocristopathies at the cephalic, cardiac, and vagal levels.
Introduction
Hirschsprung disease (HSCR, or aganglionic megacolon [MIM 142623]) is the most common (1/5,000 live births) congenital malformation caused by defective development of embryonic neural crest cells at the vagal level. The patients show severe vomiting, abdominal distention, and constipation, because of the lack of submucosal and myenteric ganglion cells along variable lengths of the gastrointestinal tract, leading to intestinal obstruction after birth. HSCR is genetically heterogeneous, and six disease-causing genes have been recently identified: RET, encoding the RET proto-oncogene (Edery et al. 1994; Romeo et al. 1994); EDNRB, encoding the endothelin B receptor (Puffenberger et al. 1994); GDNF, encoding glial cell line–derived neurotrophic factor (Angrist et al. 1996; Salomon et al. 1996); EDN3, encoding endothelin 3 (Edery et al. 1996; Hofstra et al. 1996); Sox 10 (Pingault et al. 1998), and ECE-1, encoding endothelin-converting enzyme 1 (Hofstra et al. 1999). In spite of the discovery of this range of candidate genes for HSCR, there are some associated diseases with unknown genetic etiology.
Goldberg and Shprintzen (1981) first reported siblings with an HSCR-associated disease (MIM 235730) who presented with microcephaly, mental retardation, hypertelorism, submucous cleft palate, and short stature. Reports of patients with overlapping collections of these abnormalities in addition to HSCR have been reviewed in the literature (Fryer 1998; Mowat et al. 1998). Although they all exhibited HSCR, mental retardation, microcephaly, and mild facial dysmorphology, there is genetic and clinical heterogeneity among the patients. The presence of clinical features such as cleft palate, ptosis, and iris coloboma in some siblings suggests autosomal recessive disease (Goldberg and Shprintzen 1981; Hurst et al. 1988; Kumasaka and Clarren 1988; Yomo et al. 1991; Fryer 1998; Brooks et al. 1999), whereas other isolated patients with epilepsy or congenital heart disease are likely to have contiguous gene syndromes or dominant mutations (Hurst et al. 1988; Halal and Morel 1990; Tanaka et al. 1993; Lurie et al. 1994; Mowat et al. 1998). Recently, we reported that mutations in ZFHX1B (MIM 605802; GenBank accession number AB 056507), encoding Smad-interacting protein 1 (SIP1), are a cause of the dominant form of this disease (Wakamatsu et al. 2001).
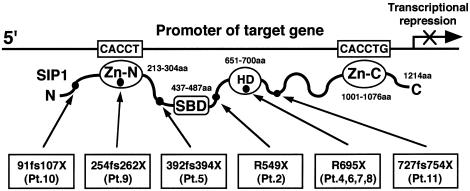
Zfx1b (GenBank accession number AF 033116), recently isolated from the mouse embryo through use of the yeast two-hybrid system as a Smad1-interacting protein, named “Sip1,” is a new member of the δEF1/Zfh-1 family of two-handed zinc-finger/homeodomain proteins. It contains a Smad-binding domain (SBD), a homeobox-like domain (HD), and two separated clusters of zinc fingers (ZF) at the amino (N) and carboxy (C) terminals (Verschueren et al. 1999). Each ZF cluster binds independently, as a DNA-binding transcriptional repressor, to the promoter regions of candidate target genes containing one CACCT and one CACCTG or its inverse sequence (Remacle et al. 1999; Postigo and Dean 2000). Zfx1b has also been shown to interact with receptor-mediated activated full-length Smads in vitro and is likely to be involved in the Smads-mediated signaling cascade (Verschueren et al. 1999). ZFXH1B spans ∼70 kb, consists of 10 exons and 9 introns, and encodes human SIP1. SIP1 consists of 1,214 amino acids (aa) and shows 97% similarity to the mouse form (Sip1) at the amino acid level, with exactly the same functional domains: SBD (437–487 aa), HD (651–700 aa), and ZF at the N and C terminals (213–304 aa and 1001–1076 aa, respectively) (fig. 1).
Figure 1.
Schematic representation of the structural organization of SIP1 and of the positions of the identified mutations. Three functional domains (SBD, HD, and N- and C-ZF) and six identified mutations are given, with patient numbers.
Because patients who have HSCR with ZFHX1B deficiency have complex developmental disorders caused by the disturbance of embryonic neural and neural-crest development at the cephalic, cardiac, and vagal levels (Wakamatsu et al. 2001), we speculated that they might demonstrate a great variety of clinical features. We therefore studied DNA samples from patients who had clinical features quite similar to those reported for ZFHX1B deficiency but did not have HSCR or congenital heart disease, and we identified mutations in six subjects.
Subjects and Methods
Subjects
Eleven patients (1–11) in our center were included in the present study; mutations of ZFHX1B were identified in 10 of them. All patients demonstrated severe mental retardation and delayed motor development, nine of whom (patients 1–9) had epilepsy with a variety of clinical presentations suggestive of neural crest impairment. The results of mutation analysis of ZFHX1B in five patients (1–5) who had HSCR with short-segment aganglionosis have been described elsewhere (Wakamatsu et al. 2001). In brief, patient 1 has a complete deletion of ZFHX1B at a de novo t(2;13)(q22;q22) translocation site on chromosome 2. Brain magnetic resonance imaging (MRI) revealed moderate cerebral atrophy without agenesis of the corpus callosum. At age 6 years, she still cannot stand without support. The other four patients (2–5) also showed delayed motor functions, and they began to walk without support at ages 3.5–8 years. Patients 2, 4, and 5 have nonsense or frameshift mutations of ZFHX1B (fig. 1), and the cases in three patients (1, 2, and 5) are also complicated by congenital heart disease (patent ductus arteriosus [PDA]). A brain MRI of patient 2 revealed agenesis of the corpus callosum.
Patient 6 is a 26-year-old male. He was born at 42 wk gestation, weighing 3,570 g. At age 1 year 2 mo, he developed epilepsy, and bilateral diffuse 2–4-Hz slow wave bursts and spikes were evident on electroencephalograms (EEG); seizures could be well controlled with anticonvulsants. He was admitted to our hospital at age 1 year 3 mo and was found to have microcephaly (head circumference 45.3 cm, 2 SD below the mean) and distinct facial features (hypertelorism, external strabismus, prognathism, and a low nasal root). He showed delayed motor development, beginning to walk at age 6 years. His mental development is also severely delayed, and he has limited speech. He has had severe constipation from birth; he has never developed ileus but still needs medication for a regular stool.
Patient 7 is a 28-year-old male. He was born at 40 wk gestation, weighing 3,950 g (height 55.0 cm). He was found to have hypertelorism and microcephaly (head circumference 33.0 cm, 2 SD below the mean) soon after birth. At age 6 mo, he developed epilepsy. He showed delayed motor milestones, developing neck control at age 5 mo and walking without support at age 4 years. He also has severe mental retardation with little linguistic ability.
Patient 8 is a 23-year-old male. He was born at 40 wk gestation, weighing 3,410 g (height 50.0 cm). He was also found to have hypertelorism and microcephaly (head circumference 33.0 cm, 2 SD below the mean) soon after birth. He had a very limited appetite in his first few years of life and developed epilepsy at age 2 years. He started to walk without support at age 2 years 6 mo. He is severely mentally impaired and can speak only a few words (“mama,” “papa”). Patients 7 and 8 also had moderate constipation until age ∼10 years but do not need medication now. Although patients 6, 7, and 8 have constipation, rectal biopsies have never been taken because until now there has been no consideration of the possibility of HSCR.
Patient 9 is a 25-year-old male. He was born at 39 wk gestation weighing 3,260 g (height 48.0 cm), and he had hypertelorism and microcephaly (head circumference 33.0 cm, 2 SD below the mean). He developed epilepsy at age 1 year. He showed delayed motor development, with neck control developing at age 8 mo and walking without support at age 2 years. He is severely mentally retarded, with only a few words of speech.
Patient 10 is a 4-year-old female. She was born at 36 wk gestation weighing 2,690 g (height 46.5 cm), with microcephaly (head circumference 30.5 cm, 2 SD below the mean). She showed delayed psychomotor milestones; neck control was obtained at age 4 mo, she was able to sit down by herself at age 1 year 8 mo, and she cannot walk without support. She is active and shows curiosity but speaks only a few words. Epilepsy has so far not been evident. She shows distinct facial features, such as hypertelorism, prognathism, and a low nasal root.
Patient 11 is a 1.5-year-old male. He was born at 38 wk gestation weighing 2,960 g (height 47.0 cm), with microcephaly (head circumference 32.0 cm, 2 SD below the mean). Neck control was obtained at age 8 mo. He cannot sit down without support and can speak no words at present. He has mild constipation, and his stool is usually hard. His facial appearance is clearly indicative of an abnormality, showing hypertelorism, external strabismus, prognathism, and a low nasal root (fig. 2). There has not been any evidence of epileptic waves on EEG. The findings of microcephaly, delayed motor milestones, and distinct facial features stimulated us to examine ZFHX1B in this baby.
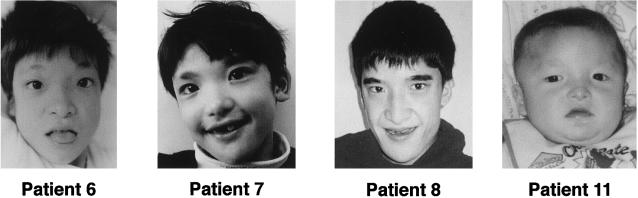
Figure 2.
Facial features of patients. Patients 6, 7, 8, and 11, at age 6, 7, 18, and 1 year(s), respectively. They demonstrate hypertelorism (in all cases), prognathism (in all cases), a low nasal root (in the cases of patients 6, 7, and 11), absence of the internal part of the eyebrows (in all cases), and strabismus (in the case of patient 11). Patients 6 and 7 underwent surgical correction of strabismus at age 3 years.
All patients are children of nonconsanguineous parents. Deafness, pigmentation defects, iris coloboma, ptosis, cleft lip, and cleft palate were not observed. Brain MRI or CT studies were performed on patients 8 and 10 at ages 22 years and 3.5 years, respectively; the results did not show remarkable abnormalities. The clinical features of the patients with ZFHX1B deficiency—except for patient 3, in whom we could not identify any mutation—are summarized in table 1. Photographs of patients 6, 7, 8, and 11 are given in figure 2.
Table 1.
Clinical Features and Mutations Detected in Patients with ZFHX1B Deficiency[Note]
| PatientNumber | Sex | Current Age(years) | Birth Weight(g) | HSCRa | MentalRetardation | Epilepsy | Microcephaly | FacialDysmorphology | Age at Onsetof Walking | HeartDisease | ZFHX1BMutation | ExonMutated | BrainAbnormalityb |
| 1 | F | 6 | 3,000 | + | + | + | + | + | − | PDA | Deletion | All | Atrophy |
| 2 | F | 18 | 3,045 | + | + | + | + | + | 3 years 6 mo | PDA | R549X | 8 | Callosal agenesis |
| 4 | M | 23 | 2,820 | + | + | + | + | + | 8 years | − | R695X | 8 | NE |
| 5 | M | 23 | 3,200 | + | + | + | + | + | 5 years 3 mo | PDA | 392fs394X | 8 | NE |
| 6 | M | 26 | 3,570 | * | + | + | + | + | 6 years | − | R695X | 8 | NE |
| 7 | M | 28 | 3,950 | * | + | + | + | + | 4 years | − | R695X | 8 | NE |
| 8 | M | 23 | 3,410 | * | + | + | + | + | 2 years 6 mo | − | R695X | 8 | Normal |
| 9 | M | 25 | 3,260 | − | + | + | + | + | 2 years | − | 254fs262X | 6 | NE |
| 10 | F | 4 | 2,690 | − | + | − | + | + | − | − | 91fs107X | 3 | Normal |
| 11 | M | 1.5 | 2,960 | * | + | − | + | + | − | − | 727fs754X | 8 | NE |
Note.— + = presence of clinical sign; − = absence of clinical sign.
* = shows constipation.
NE = not examined.
PCR Amplification and DNA Sequencing
Genomic DNA was isolated from peripheral blood, according to standard protocols. The primer pairs used for the amplification of each exon, exon/intron boundary, and part of the associated introns of ZFHX1B are listed in table 2. Genomic DNA (100–200 ng) from each patient and unaffected control subject was amplified by AmpliTaq Gold (Applied Biosystems, exons 3–10) or a GC-RICH PCR system (Roche Molecular Biochemicals, exons 1 and 2) in a total volume of 40 μl containing 0.4 μg of each primer at the indicated annealing temperature. PCR products were run on 1% agarose gels, were purified with QIAEX II (QIAGEN), and were sequenced directly by methods slightly modified from those for Sequenase Version 2.0 (United States Biochemical Co.), as described elsewhere (Yamada et al. 1992).
Table 2.
Primer Sequences Used for ZFHX1B Analyses
|
Primer Sequence (5′→3′) |
|||||
| Analysis andPrimer Name | Exon | Forward | Reverse | Annealing Temperature(°C) | PCR Fragment Length(bp) |
| PCR direct sequencing: | |||||
| SIP-E1S/1A | 1 | GGAAGGGAGGGAGGTGGAAT | AGGATGGAGGACGAGCACACC | 58 | 535 |
| SIP-E2S/2A | 2 | TGGTTGCTAGATCGAGCCTG | GGTGCCTGACGCTCACTTTG | 58 | 364 |
| SIP-E3S/3A | 3 | GGGTGGCTGATGTTTCTCAAC | GCGATCTGCTAGGTGGAACAG | 58 | 395 |
| SIP-E4S/4A | 4 | GCATGCTTAGTATAGTAAGCCT | TCATTTGAATTCTCAACGACTG | 58 | 521 |
| SIP-E5S/6A | 5, 6 | AGAAGGCCCGGAAACAACAT | ATATCGTTTCTCTAAGGGGTTA | 58 | 1,209 |
| SIP-E7S/7A | 7 | TTTTCTTTGTTCCTCCTGCAC | TTTTTCATGGGTAGGTCTCA | 58 | 523 |
| SIP-E8.1S/8.1A | 8 | AATGTAGAGGTACCCCATTGTG | CTCAGGTTGAGAGCATGGAT | 58 | 700 |
| SIP-E8.2S/8.2A | 8 | AAAAATGGACTGCAAGGCTG | GTGGCTATAATACTTTTGGG | 58 | 1,077 |
| SIP-E8.3S/8.3A | 8 | CTTCTGAGGAGCTCCAGGCT | CATAATCAAAATAATTGCCACCTC | 58 | 526 |
| SIP-E9S/9A | 9 | GCACAAAGAGAAGCCACTGTA | GCCATCTGACTGTCCCTTTG | 58 | 494 |
| SIP-E10S/10A | 10 | TAGTGGAAAGAGACTTCATGCA | TGAACAGCTTAACACAGCAGTG | 58 | 723 |
| Restriction enzyme analysis: | |||||
| SIP-E8R1S/1A | 8 | CTAATATTCCGCCTGTCGGT | TTTTCATGAGGCTGCAGGAC | 60 | 340 |
| SIP-E8R2S/2A | 8 | ATGAAGAGATCAAGGCGGTC | CGCTTTGAGGTGGAAGAGCT | 58 | 888 |
| SIP-E8R3S/3A | 8 | ACCCATACAAGGACCACATG | TTTGAGTACTGGTAGACTaTgCa | 56 | 157 |
Mismatch primer (T→a, T→g).
RFLP Analysis
To identify mutations, PCR-dependent restriction enzyme diagnostic tests were established.
Identification of 1645A→T (R549X)
Genomic DNA from family members of patient 2, as well as from an unaffected control, was amplified using specific primers for exon 8 (SIP-8R1S/1A). The PCR products were digested with HphI and were separated in 1.5% low-melting-point agarose gels (Gibco-BRL).
Identification of 2083C→T (R695X)
Since PCR-dependent diagnosis is very difficult with one-step PCR (SIP-8R3S/3A), we chose two steps. First, genomic DNA containing exon 8 of ZFHX1B was amplified with specific primers (SIP-8R2S/2A) for 20 cycles. Then, the PCR products were diluted 500 times with TE (10 mM Tris-HCl, pH 7.5 and 1mM EDTA) and were reamplified with an internal mismatch primer pair (SIP-8R3S/3A) for 30 cycles, to detect creation of a NsiI site (ATGCAT) in the mutant DNA (table 2). The PCR products were digested with NsiI, were electrophoresed in 1.5% low-melting-point agarose gels, and were stained with ethidium bromide.
Expression of ZFHX1B in tissue
Multiple Tissue Northern (MTN) Blots (Clontech, #7780-1) and Multiple Tissue Expression Arrays (Clontech, #7775-1) were hybridized with 32P-labeled 4.1-kb and 685-bp (nt 1422–2106, from ATG) ZFHX1B cDNA probes, according to the respective user's manuals. Filters were washed with solution 1 (2× saline sodium citrate [SSC], 1% SDS) at 65°C and with solution 2 (0.1× SSC, 0.5% SDS) at 55°C, for exposure of X-ray films (RX-U; FUJIFILM).
Results
Identification of Mutations in ZFHX1B
Ten exons, splice junctions, and parts of introns from patients 6–11 were amplified in 11 portions by PCR, using primers specific for ZFHX1B. By direct-sequencing analysis of PCR-amplified ZFHX1B fragments, R695X nonsense mutations in exon 8 were identified in three patients (6–8), a 2-bp insertion (760insCA resulting in 254fs262X) was identified in exon 6 of patient 9, a single-base deletion (270delG resulting in 91fs107X) was identified in exon 3 of patient 10, and a 2-bp deletion (2178delTT resulting in 727fs754X) was identified in exon 8 of patient 11. These are summarized, with the previously reported ZFHX1B mutations, in table 1 and figure 1.
Detection of Disease Mutations by RFLP
To determine whether the mutations we identified also appeared in patients’ family members, we established diagnostic tests specific to each mutation; these tests were based on performance of restriction-enzyme cleavage of PCR products, either directly or by primers flanking each mutation site (table 2). The assays demonstrated that patient 2, as well as four others (patients 4 and 6–8), were heterozygous for their identified mutations—R549X and R695X, respectively. However, all of their parents showed normal alleles (data not shown). Direct-sequencing analysis for the parents of patient 5 (1173delAACA), patient 9 (760insCA), patient 10 (270delG), and patient 11 (2178delTT) also showed them to have normal alleles (data not shown). Thus, the ZFHX1B mutations identified in the patients were all de novo events.
ZFHX1B Tissue Expression and Transcripts
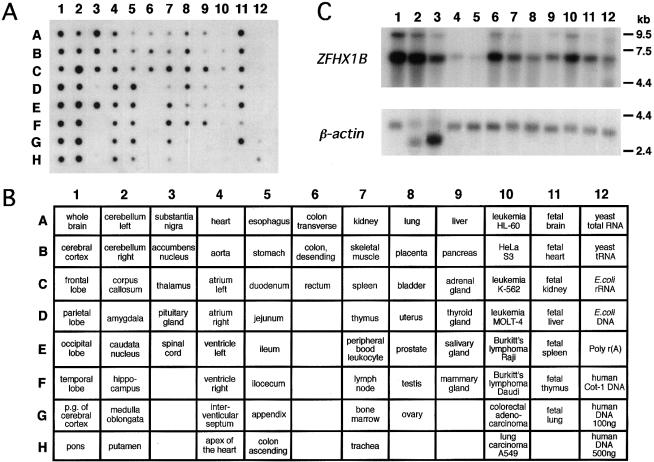
Hybridization of a ZFHX1B-specific probe to mRNA samples derived from many human tissues revealed ubiquitous expression, predominantly in brain (except in the pituitary gland), with small amounts in esophagus, kidney, thymus, pancreas, thyroid gland, salivary gland, and in tumor cell lines (fig. 3A). Northern blot analysis revealed one dominant ZFHX1B transcript (∼6 kb; fig. 3B).
Figure 3.
Expression patterns of ZFHX1B. A, Autoradiograph showing hybridization with a 685-bp ZFHX1B cDNA against a Multiple Tissue Expression Array mRNA dot blot. The order and arrangement of applied samples are given in panel B. C, Autoradiographs of northern blots hybridized with a 4.1-kb ZFHX1B full-length cDNA (containing all exons) and a 2.0-kb human β-actin cDNA as a control: lane 1, brain; lane 2, heart; lane 3, skeletal muscle; lane 4, colon; lane 5, thymus; lane 6, spleen; lane 7, kidney; lane 8, liver; lane 9, small intestine; lane 10, placenta; lane 11, lung; and lane 12, peripheral blood leukocytes.
Discussion
In the present article, we describe a mutation analysis of ZFHX1B, encoding human SIP1, in six patients with profound mental retardation, delayed motor development, epilepsy, and mild facial dysmorphology, as well as in four previously reported patients with ZFHX1B deficiency (Wakamatsu et al. 2001). Comparison of the clinical presentations of the 10 patients indicates that ZFHX1B deficiency is a complex developmental disorder with a wide spectrum of clinical features suggestive of neurocristopathies at the cephalic (distinct facial features and microcephaly), cardiac (PDA), and vagal levels (HSCR) (table 1). RFLP studies or direct-sequencing analysis of identified mutations of family members demonstrated the ZFHX1B deficiency to be caused by de novo mutations that cause a complete deletion (in patient 1) or null functions (in patients 2 and 4–11). Thus, ZFHX1B deficiency is an autosomal dominant complex developmental disorder caused by a decrease to half of the normal amount of active SIP1 during embryogenesis.
In the present study, we found a nonsense mutation, R695X, in 4 of 10 independent patients, generating a C→T transition at the CpG doublet as a de novo event. Data from studies on the molecular evolution of various vertebrate DNA sequences suggest that the CpG doublet is a favored target for mutations. Many mutant sequences feature CpG-to-TpG or CpG-to-CpA substitutions (Cooper and Youssoufian 1988), and this site is likely to be a hot spot for ZFHX1B mutations. Furthermore, RFLP analysis to identify R695X in the present study might be considered a useful tool for newborn screening for ZFHX1B deficiency.
Although all of our patients have mental retardation, delayed motor development, microcephaly, and distinct facial features in common, eight have epilepsy (not evident in patients 10 and 11), only three have PDA, and four have HSCR. The constipation observed in four patients (6, 7, 8, and 11) is suggestive of a partial defect in ganglion cells of the gastrointestinal tract. However, intensive studies, including rectal biopsies, have not yet been performed (table 1). This finding that the penetrance of possible neurocristopathies at the cardiac and vagal levels is variable suggests that the associated phenotypes may be less sensitive to a decrease in the normal amount of SIP1. The fact that four patients (4 and 6–8) with the R695X mutation showed different clinical presentations, with and without HSCR, further indicates that unknown factors, possibly environmental in nature and occurring during embryonic development, may exert important influences (table 1). Other disease-causing genes may also be present, because we did not find any ZFHX1B mutations in patient 3. The identification of 10 patients in our center who have ZFHX1B deficiency and show a wide spectrum of clinical variation, however, suggests that this is one common candidate gene for syndromic mental retardation associated with delayed motor development, epilepsy, and neurocristopathies. Patients having missense mutations—for example, in functional domains (SBD or ZF) of ZFHX1B—have yet to be identified, but it would be of interest to clarify what clinical features they would have.
Very recently, Comijn et al. (2001) clearly demonstrated that E-cadherin is one target gene of SIP1 in epithelial tumor cell lines. SIP1 has an important role in the invasion of these tumors, by downregulating the transcription of E-cadherin. In the present study, we provide evidence that SIP1 is essential for normal development of brain and, most likely, for development of the neural crest.
Although ZFHX1B was here found to be expressed ubiquitously, albeit with very different levels in different tissues (fig. 3), strictly regulated precise expression would be essential for normal embryonic neural and neural crest development at the correct stages of embryogenesis. Identification and analysis of target genes of SIP1, including E-cadherin, especially in neural and neural crest cells, during embryonic development should provide insights into the molecular mechanisms underlying mental retardation, epilepsy, and a variety of neurocristopathies.
Note.—During the review of this article, Cacheux et al. (2001) reported the ZFHX1B mutations in patients with a syndromic HSCR.
Acknowledgments
We are grateful to the patients and their families for agreeing to participate in this study. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (C)–Advanced Brain Science Project, from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (to N.W. and K.K.) and by a grant from the Tsubaki Memorial Neuroscience Research Foundation (to N.W.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for ZFHX1B mRNA [accession number AB 056507] and Zfx1b [Mus musculus] [accession number AF 033116])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HSCR [MIM 142623], HSCR with microcephaly, mental retardation, and distinct facial features [MIM 235730], and ZFHX1B, encoding SIP1 [MIM 605802])
References
- Angrist M, Bolk S, Halushka M, Lapchak PA, Chakravarti A (1996) Germline mutations in glial cell line-derived neurotrophic factor (GDNF) and RET in a Hirschsprung disease patient. Nat Genet 14:341–344 [DOI] [PubMed] [Google Scholar]
- Brooks AS, Breuning MH, Osinga J, vd Smagt JJ, Catsman CE, Buys CHCM, Meijers C, Hofstra RMW (1999) A consanguineous family with Hirschsprung disease, microcephaly, and mental retardation (Goldberg-Shprintzen syndrome). J Med Genet 36:485–489 [PMC free article] [PubMed] [Google Scholar]
- Cacheux V, Dastot-Le Moal F, Kääriäinen H, Bondurand N, Rintala R, Boissier B, Wilson M, Mowat D, Goossens M (2001) Loss-of-function mutations in SIP1 Smad interacting protein 1 result in a syndromic Hirschsprung disease. Hum Mol Genet 10:1503–1510 [DOI] [PubMed] [Google Scholar]
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F (2001) The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell 7:1267–1278 [DOI] [PubMed] [Google Scholar]
- Cooper DN, Youssoufian H (1988) The CpG dinucleotide and human genetic disease. Hum Genet 78:151–155 [DOI] [PubMed] [Google Scholar]
- Edery P, Attié T, Amiel J, Pelet A, Eng C, Hofstra RMW, Martelli H, Bidaud C, Munnich A, Lyonnet S (1996) Mutation of the endothelin-3 gene in the Waardenburg-Hirschsprung disease (Shah-Waardenburg syndrome). Nat Genet 12:442–444 [DOI] [PubMed] [Google Scholar]
- Edery P, Lyonnet S, Mulligan LM, Pelet A, Dow E, Abel L, Holder S, Nihoul-Fékété C, Ponder BAJ, Munnich A (1994) Mutations of the RET proto-oncogene in Hirschsprung’s disease. Nature 367:378–380 [DOI] [PubMed] [Google Scholar]
- Fryer AE (1998) Goldberg-Shprintzen syndrome: report of a new family and review of the literature. Clin Dysmorphol 7:97–101 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Shprintzen RJ (1981) Hirschsprung megacolon and cleft palate in two sibs. J Craniofac Genet Dev Biol 1:185–189 [PubMed] [Google Scholar]
- Halal F, Morel J (1990) The syndrome of Hirschsprung disease, microcephaly, unusual face, and mental retardation. Am J Med Genet 37:106–108 [DOI] [PubMed] [Google Scholar]
- Hofstra RMW, Osinga J, Tan-Sindhunata G, Wu Y, Kamsteeg EJ, Stulp RP, van Ravenswaaij-Arts C, Majoor-Krakauer D, Angrist M, Chakravarti A, Meijers C, Buys CHCM (1996) A homozygous mutation in the endothelin-3 gene associated with a combined Waardenburg type 2 and Hirschsprung phenotype (Shah-Waardenburg syndrome). Nat Genet 12:445–447 [DOI] [PubMed] [Google Scholar]
- Hofstra RMW, Valdenaire O, Arch E, Osinga J, Kroes H, Löffler BM, Hamosh A, Meijers C, Buys CHCM (1999) A loss-of-function mutation in the endothelin-converting enzyme 1 (ECE-1) associated with Hirschsprung disease, cardiac defects, and autonomic dysfunction. Am J Hum Genet 64:304–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst JA, Markiewicz M, Kumar D, Brett EM (1988) Unknown syndrome: Hirschsprung's disease, microcephaly, and iris coloboma: a new syndrome of defective neuronal migration. J Med Genet 25:494–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumasaka K, Clarren SK (1988) Familial patterns of central nervous system dysfunction, growth deficiency, facial clefts and congenital megacolon: a specific disorder? Am J Med Genet 31:465–466 [DOI] [PubMed] [Google Scholar]
- Lurie IW, Supovitz KR, Rosenblum-Vos LS, Wulfsberg EA (1994) Phenotypic variability of del(2) (q22-q23): report of a case with a review of the literature. Genet Couns 5:11–14 [PubMed] [Google Scholar]
- Mowat DR, Croaker GDH, Cass DT, Kerr BA, Chaitow J, Adès LC, Chia NL, Wilson MJ (1998) Hirschsprung disease, microcephaly, mental retardation, and characteristic facial features: delineation of a new syndrome and identification of a locus at chromosome 2q22-q23. J Med Genet 35:617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingault V, Bondurand N, Kuhlbrodt K, Goerich DE, Préhu MO, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, Amiel J, Lyonnet S, Ceccherini I, Romeo G, Smith JC, Read AP, Wegner M, Goossens M (1998) SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet 18:171–173 [DOI] [PubMed] [Google Scholar]
- Postigo AA, Dean DC (2000) Differential expression and function of members of the zfh-1 family of zinc finger/homeodomain repressors. Proc Natl Acad Sci USA 97:6391–6396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puffenberger EG, Hosoda K, Washington SS, Nakao K, deWit D, Yanagisawa M, Chakravarti A (1994) A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung's disease. Cell 79:1257–1266 [DOI] [PubMed] [Google Scholar]
- Remacle JE, Kraft H, Lerchner W, Wuytens G, Collart C, Verschueren K, Smith JC, Huylebroeck D (1999) New mode of DNA binding of multi-zinc finger transcription factors: δEF1 family members bind with two hands to two target sites. EMBO J 18:5073–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo G, Ronchetto P, Luo Y, Barone V, Seri M, Ceccherini I, Pasini B, Bocciardi R, Lerone M, Kääriäinen H, Martucciello G (1994) Point mutations affecting the tyrosine kinase domain of the RET proto-oncogene in Hirschsprung’s disease. Nature 367:377–378 [DOI] [PubMed] [Google Scholar]
- Salomon R, Attié T, Pelet A, Bidaud C, Eng C, Amiel J, Sarnacki S, Goulet O, Ricour C, Nihoul-Fékété C, Munnich A, Lyonnet S (1996) Germline mutations of the RET ligand GDNF are not sufficient to cause Hirschsprung disease. Nat Genet 14:345–347 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Ito J, Cho K, Mikawa M (1993) Hirschsprung disease, unusual face, mental retardation, epilepsy, and congenital heart disease: Goldberg-Shprintzen syndrome. Pediatr Neurol 9:479–481 [DOI] [PubMed] [Google Scholar]
- Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su MT, Bodmer R, Smith JC, Huylebroeck D (1999) SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J Biol Chem 274:20489–20498 [DOI] [PubMed] [Google Scholar]
- Wakamatsu N, Yamada Y, Yamada K, Ono T, Nomura N, Taniguchi H, Kitoh H, Mutoh N, Yamanaka T, Mushiake K, Kato K, Sonta S, Nagaya M (2001) Mutations in SIP1, encoding Smad interacting protein-1, cause a form of Hirschsprung disease. Nat Genet 27:369–370 [DOI] [PubMed] [Google Scholar]
- Yamada Y, Goto H, Suzumori K, Adachi R, Ogasawara N (1992) Molecular analysis of five independent Japanese mutant genes responsible for hypoxanthine quanine phosphoribosyltransferase (HPRT) deficiency. Hum Genet 90:379–384 [DOI] [PubMed] [Google Scholar]
- Yomo A, Taira T, Kondo I (1991) Goldberg-Shprintzen syndrome: Hirschsprung disease, hypotonia, and ptosis in sibs. Am J Med Genet 41:188–191 [DOI] [PubMed] [Google Scholar]