Abstract
Strong genetic contributions to drug abuse vulnerability are well documented, but few chromosomal locations for human drug-abuse vulnerability alleles have been confirmed. We now identify chromosomal markers whose alleles distinguish drug abusers from control individuals in each of two samples, on the basis of pooled-sample microarray and association analyses. Reproducibly positive chromosomal regions defined by these markers in conjunction with previous results were especially unlikely to have been identified by chance. Positive markers identify the alcohol dehydrogenase (ADH) locus, flank the brain-derived neurotropic factor (BDNF) locus, and mark seven other regions previously linked to vulnerability to nicotine or alcohol abuse. These data support polygenic contributions of common allelic variants to polysubstance abuse vulnerability.
Introduction
Problems from drug abuse permeate each branch of medicine. Prevention and treatment strategies remain far from optimal. Drug abuse vulnerability is a complex trait with strong genetic influences documented in numerous family and twin studies (Cadoret et al. 1986, 1995; Grove et al. 1990; Pickens et al. 1991; Goldberg 1993; Gynther et al. 1995; Uhl et al. 1995, 1997, 1999; Tsuang et al. 1996, 1998, 1999; Woodward 1996; Kendler and Prescott 1998; Merikangas et al. 1998; Kendler et al. 1999; Stallings et al. 1999; True et al. 1999a, 1999b; Karkowski et al. 2000). Much of the genetic vulnerability to abuse of different legal and illegal addictive substances is shared (Kendler and Prescott 1998; Tsuang et al. 1998; Kendler et al. 1999; True et al. 1999b; Karkowski et al. 2000), and many abusers use multiple addictive substances (Substance Abuse and Mental Health Services Administration 1995). Understanding the genetic bases of drug abuse vulnerability and identifying the particular allelic variants that contribute to this vulnerability can improve our understanding of human addictions and can assist efforts to match vulnerable individuals with the prevention and treatment strategies most likely to work for them.
Linkage-based genome scans for legal addictions have been reported by Long et al. (1998), Reich et al. (1998), Straub et al. (1999), and Faroud et al. (2000). However, few positive findings from any one study were replicated in any other study. With this lack of convergent results, these scans have failed to provide evidence for the existence of common allelic variants, across different human samples, that could predispose to vulnerability to substance abuse.
We approach the problem of identifying chromosomal regions and genes that contain allelic variants that predispose to the abuse of illegal substances by association genome scanning. Association approaches do not require the participation of family members and thus increase the representativeness of our sample, since few abusers allow us to contact their family members. Association studies can gain more power from higher genomic marker densities than can linkage approaches (Risch and Merikangas 1996; Cervino and Hill 2000; Schork et al. 2000; Sham et al. 2000). High marker densities reduce the chance of false-positive results due to unintended ethnic mismatching of the disease and control populations when allele frequencies at many loci in the disease group are indistinguishable from those in control subjects. Association studies can identify smaller genomic regions likely to harbor candidate pathogenic allelic variants than can linkage-based methods, speeding searches for the specific allelic variants that directly contribute to disease vulnerability. Association studies foster pooling strategies that can allow high marker densities to be determined with the maximum possible preservation of confidentiality, since no individuals’ genotypes are identified (Barcellos et al. 1997; Hacia et al. 1999; Germer et al. 2000).
We compare results from unrelated individuals who have substantial histories of illegal substance use and/or dependence with results from matched samples of individuals who have no significant lifetime use of any addictive substance. Relative allelic frequencies of 1,494 single-nucleotide polymorphism (SNP) markers in abuser and control European American samples are assessed. Allelic frequencies for the SNPs that show tentative positive results in the European American sample are tested in a sample of African American abusers and control individuals. We focus analyses on SNPs in chromosomal regions where (a) two nearby SNPs display different allelic frequencies in both European American and African American abusers versus control subjects or (b) one or more of these reproduced positive SNPs are located near markers previously linked with dependence on legal substances (Long et al. 1998; Reich et al. 1998; Straub et al. 1999; Foroud et al. 2000). These data and their convergence with prior results support, for the first time, contributions of common allelic variants to polygenic models of genetic vulnerability to polysubstance abuse. They identify several chromosomal regions that are candidates for harboring common gene variants that alter vulnerability to substance abuse. Finally, they also identify strong candidate genes in several of these regions.
Material and Methods
Research volunteers provided informed consent under assurances of confidentiality, chose the extent of family member contacts that they would authorize, and were reimbursed for time spent. They self-reported their ethnicity and drug histories, using the drug use survey (DUS) (Smith et al. 1992; Persico et al. 1996) and diagnostic interview schedule (DIS). Some results were compared with those obtained separately from family members enrolled in a family pedigree study (G. R. Uhl, J. Hess, D. Walther, and C. Cantoneggi, unpublished data). For extraction of DNA, 10 ml of blood was obtained.
Two hundred thirty-nine “control” unrelated individuals of self-reported European American ancestry had no significant lifetime history of use of any addictive substance, average age 32 years, and 0 or 1+ DUS total drug use scores. Four hundred fifteen unrelated “abusers” averaged 30.6 years old and scored 3+ on the total drug use scale and/or demonstrated dependence, according to DSM-III-R criteria, on at least one illegal abused substance. Individuals with intermediate levels of substance use (2+ DUS scores) who did not reach DSM-III-R dependence criteria for at least one illegal substance were excluded from this study. Average total drug use scores were 2.8 for abusers and 0.57 for control individuals.
Individuals of self-reported African American descent were also studied. Two hundred fifty-two abusers averaged 35.9 years of age and had mean total drug use scores of 2.9. Ninety-nine control individuals averaged 36.5 years of age and displayed average total drug use scores of 0.51.
Leukocyte genomic DNA was prepared as described elsewhere (Smith et al. 1992; Persico et al. 1996). DNA concentrations were quantitated by spectrophotometry and Hoechst dye fluorescence and were diluted to 10 ng/μl. Pools were made by combination of DNAs from 20 individuals in equal amounts. For reproducibility tests, replicate DNA pools were constructed for each determination.
Multiplex PCR reactions were performed as 24 separate 12-μl reactions in 96-well microtiter plates according to the manufacturer's instructions (Affymetrix). DNA from individuals or pools was amplified in 24 separate reactions that each contained 6 ng of DNA from each individual and 84 multiplex PCR primer pairs. Primary PCR products were subjected to labeling PCR as described by Affymetrix. The 24 labeled PCR samples corresponding to each genomic DNA sample were then pooled, diluted in hybridization buffer, heated to 95°C, cooled to 4°C, and hybridized with Affymetrix HuSNP GeneChip microarrays at 45°C for 18 h. Arrays were washed with two changes of 6× sodium chloride/sodium phosphate EDTA buffer pH 7.4 (SSPE), 0.01% Triton at 25°C and then with six changes of 4× SSPE, 0.01% Triton at 35°C. DNA hybridized to the washed microarrays was stained using 2 μg/ml phycoerythrin-conjugated streptavidin in 6× SSPE, 0.01% Triton, 0.5 mg/ml BSA at 25°C for 30 min. Stained arrays were washed with four changes of 6× SSPE/0.01% Triton at 25°C and then were scanned using a GeneArray Laser Scanner (Hewlett-Packard).
Genotype calls for individual DNAs were made using Affymetrix GeneChip software (3.3). Allelic frequencies in pooled DNA samples were assessed on the basis of data from the “.cell” files and of an algorithm that averaged the hybridization intensity signals from the 8–16 features per chip that provided perfectly complementary hybridization to the alleles termed “A” or “B” for each SNP. First, the “background,” determined as the average of the lowest 5% of intensity values on each microarray, was subtracted from the fluorescence intensity of each cell of that microarray. Second, background-corrected values were normalized by dividing each value by the average of the highest 5% of intensity values on each microarray. Third, normalized hybridization intensities from the microarray elements that corresponded to the perfect match “A” and perfect match “B” alleles for each SNP were each averaged. Fourth, “A/B ratios” were determined by dividing average normalized A values by average normalized B values. Fifth, arctangent transformations were applied to each ratio to improve combination of data from experiments with different absolute intensity values. Lastly, “arctan A/B” ratios for abusers were then divided by “arctan A/B” ratios for control individuals to form an abuser/control ratio, our primary analysis.
Means of three replications for each experiment identified SNPs with abuser/control hybridization ratios in the top and bottom 5% of the distribution for the European American abuser versus control comparisons. Reproducibility of these “candidate positive” SNPs was sought by asking if they were also in the upper or lower 7.5% of the distribution of hybridization ratios in data from two replications in African American abuser versus control samples. Physical locations of these reproducibly positive SNPs and of markers linked to alcohol or nicotine dependence in previous studies were sought in Mapviewer (build 22 [May 2001]) and related NCBI programs by two independent investigators. Additional locations were sought in Mapviewer build 24 [September 2001]).
Allele calls from individual DNA samples were also compared with results from pooled DNAs from the same individuals. Regression analyses compared allele frequencies for SNPs that were called in most of the individuals studied. Test-retest variability was studied in replicated microarray results from DNA pools prepared in duplicate or triplicate from the same DNA stocks from the same individuals. Using Monte Carlo simulations and the C computer language, we compared results with those expected by chance. For the initial results of SNPs that were positive in both European American and African American samples, 100,000 Monte Carlo trials were carried out. In each trial, 149 markers (5%+5% of 1,493) were sampled with replacement, and, subsequently, 222 (7.5%+7.5% of 1,493) were sampled with replacement from all of the 1,493 markers. The number of markers common to both samples were then determined. In only 11 of these trials did we find ⩾42 markers in common (P=.00011). To model the chance probabilities of obtaining positive markers as close to each other as the three closest pairs identified here, we used 1 million simulation trials in which 42 markers were randomly positioned on a 3.2 billion–bp genome and pairwise distances between each marker pair were determined. We assessed the number of simulations in which the smallest distance was <0.7 Mb, the second smallest distance was <1.5 Mb, and the third smallest distance was <2.2 Mb (Mapviewer 22 distances modeled; corresponding build 24 distances are 0.2, 2.8, and 6.6 Mb). To model the chance probabilities of obtaining positive markers as close to markers previously reported to be linked to ethanol or nicotine addictions, we used >100 million simulation trials in which 61 markers were randomly positioned on a 3.2 billion–bp genome, 41 markers were placed randomly on the same genome, and pairwise distances between each marker pair (consisting of one member of the first set and one member of the second set) were determined. We assessed the number of simulations in which at least seven of the distances were <1 Mb (Mapviewer 22 distances modeled; corresponding build 24 distances are 0, 0.1, 0.4, 0.4, 0.4, 2.3, and 2.7 Mb). Linkage disequilibrium was assessed using Arlequin for data from individual genotypes determined for 20 control subjects.
A dinucleotide repeat in the an intron close to the 5′ end of the main coding exon of brain-derived neurotropic factor (BDNF) (Krebs et al. 2000) was amplified using 6-FAM labeled oligonucleotides 5′-GCCACTTTATCTCCTCCAGT (forward) and 5′-AGCACTAGCTGCCTATTCCA (reverse) and AmpliTaq Gold polymerase and buffer in an initial 95°C denaturation for 10 min, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 45 s, and then a final extension at 72°C for 10 min. Markers flanking the BDNF locus, D11S2001 and GATA25B04, were amplified using oligonucleotides 5′-TTGGTTAAGAATGGAAATTCC-3′ (forward) and 5′-TGAAATCACCTAATGGTGGG-3′ (reverse), as well as 5′-TAAGGCACACATCTTCAGCC-3′ (forward) and 5′-GTGTCCTACCAAATAGAAACTTCC-3′ (reverse), respectively, under the same conditions. Alleles were resolved using 10% denaturing polyacrylamide gels, an ABI 373 sequencer, and gene-scan software.
Results
Each research volunteer who was offered participation in the study accepted, although <20% provided permission for family-member contacts. Sibs’ and parents’ drug-use histories for each member of 100 pedigrees were concordant with proband ratings 81% and 73% of the time, respectively. DUS quantity/frequency estimates, when compared with addiction-severity index (ASI) ratings, revealed that no individual with a DUS score of 0 reported alcohol, cocaine, heroin, cannabis, or nicotine “use more than 5 times” by ASI and that >99% of the individuals who scored 3+ on the DUS reported that they “used more than 5 times” by ASI. Good correspondence between DUS 3+ ratings and DSM-III-R criteria for abuse and/or dependence diagnoses is also documented in this population (Smith et al. 1992; Persico et al. 1996).
Pooled SNP genotyping was performed with acceptable reproducibility and evidence for validity. In three replicate experiments, mean hybridization ratio (arctan A/B) values (± SEM) were 27.2 ± 2.1 in European American control and 27.1 ± 1.9 in abusers. SNPs for which both sense and antisense oligonucleotides provide two independent assessments of hybridization of the same mixed PCR product probe to the same microarrays displayed correlations of .96 between sense and antisense hybridization values. There were correlations of .8–.9 between the hybridization ratios for pools and the ratio between the frequencies of A and B alleles determined individually for each pool member (loci for which we determined these correlations are available at our FTP site). Genotype calls for individuals from the Centre d'Etudes Polymorphisms Humaine show a correlation of >.95 with genotypes determined by sequencing by Affymetrix. Abuser/control hybridization ratios for the 1,494 SNPs examined here fell into nearly Gaussian distributions, with mean values close to 1, providing no evidence for overall ethnic mismatches between abuser and control populations.
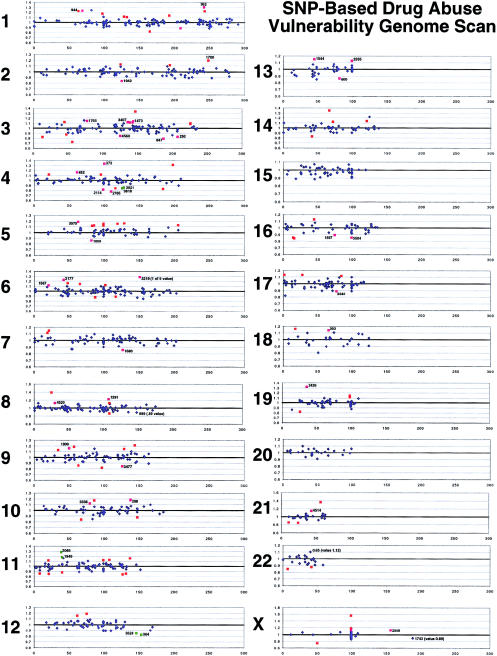
Candidate positive markers, the 5% of SNPs with greatest and smallest abuser/control ratios in European Americans (>1.119 or <0.893; see fig. 1) were tested for their ability to also provide differences between allelic frequencies between abuser and control populations from the African American sample. Of these 150 candidate positive markers, 42 were “reproducibly positive,” with values lying in the upper or lower 7.5% of the African American data (table 1). Chance would have made 22 of the original markers display these features. Monte Carlo simulations suggested that the probability of randomly distributed markers producing ⩾42 markers common to the two populations was .00011.
Figure 1.
Chromosome-by-chromosome distribution (labels 1–22 and X) of abuser/control ratios (y-axis) for SNPs positioned according to their average radiation-hybrid chromosomal distance in centimorgans (x-axis). Abuser/control ratios shown are from the European American sample. Red symbols indicate SNPs with results in the upper or lower 5% of the distribution of values. Labeled symbols indicate SNPs with outlying abuser/control values in both European American and African American samples. Green symbols indicate the positions of reproducibly positive SNPs with adjacent chromosomal positions, on the basis of NCBI Mapviewer coordinates.
Table 1.
Positions of SNP Markers Displaying Reproducibly Positive Association with Polysubstance Abuse in the Current Study and of SSLPs Nominally Positive in Prior Linkage Studies of Alcohol or Nicotine Dependence[Note]
|
Distance |
Abuser/Control Ratio |
|||||
| Chromosomeand WIAFor SSLPNumber | AveragedRadiationHybrid | MapviewerBuild 22 | MapviewerBuild 24 | SubmittedSequenceNumberor Reference | EuropeanAmerican | AfricanAmerican |
| 1: | ||||||
| D1S548 | 5.2 | 7.1 | Reich et al. 1998 | |||
| WIAF-944 | 69 | 30.1 | 32.2 | 3043 | 1.23 | 1.41 |
| D1S1598 | 33.2 | 38.6 | Reich et al. 1998 | |||
| D1S532 | 82.6 | 80.8 | Reich et al. 1998 | |||
| D1S1588 | 109.5 | 95.6 | Reich et al. 1998 | |||
| D1S1592 | 118.3 | 127.8 | Reich et al. 1998 | |||
| WIAF-1018 | 208.6 | 184.3 | 177 | 3117 | .89 | 1.22 |
| WIAF-952 | 242.2 | 194.7 | 211.2 | 3051 | 1.3 | 1.25 |
| 2: | ||||||
| D2S114 | 134.1 | 129.5 | Straub et al. 1999 | |||
| D2S1326 | 142.5 | 135 | Straub et al. 1999 | |||
| D2S426 | 197.3 | 185.2 | Reich et al. 1998 | |||
| WIAF-1942 | 124.7 | 209.6 | 208.1 | 3487 | .84 | 1.38 |
| WIAF-1700 | 248.3 | 234.8 | 229.7 | 2052 | 1.2 | 1.31 |
| 3: | ||||||
| D3S2403 | 16.1 | 15.9 | Long et al. 1998 | |||
| WIAF-1765 | 44.5 | 24.5 | 27.7 | 3312 | 1.12 | 1.4 |
| D3S2432 | 29.5 | 32.4 | Long et al. 1998 | |||
| WIAF-4560 | 125 | 50.9 | 54.3 | 19367 | .84 | .82 |
| WIAF-1473 | 143.3 | 126.6 | 125.4 | 1092 | 1.14 | 1.22 |
| WIAF-3407 | 135.3 | 130.4 | 133.8 | 19294 | 1.12 | 1.31 |
| D3S1746 | 164.1 | 156.5 | Long et al. 1998 | |||
| D3S1763 | 175.8 | 172.9 | Long et al. 1998 | |||
| D3S3053 | 184.4 | 176.6 | Long et al. 1998 | |||
| WIAF-847 | 187.1 | 185.4 | 177 | 2948 | .79 | .78 |
| WIAF-250 | 207.2 | 191.3 | 191.3 | 2605 | .83 | .72 |
| 4: | ||||||
| D4S2382 | 37.3 | 25.1 | Long et al. 1998 | |||
| D4S2632 | 44.3 | 36.4 | Long et al. 1998 | |||
| D4S1647 | 109 | 37.4 | Long et al. 1998 | |||
| D4S2457 | 109 | 37.4 | Reich et al. 1998 | |||
| D4S174 | 49.6 | 41.9 | Long et al. 1998 | |||
| D4S2456 | 81.1 | 43.5 | Long et al. 1998 | |||
| D4S1627 | 51.4 | 45.8 | Long et al. 1998 | |||
| WIAF-452 | 60.9 | 51.6 | 45.9 | 2739 | 1.16 | 1.57 |
| D4S1645 | 66.4 | 60.7 | Long et al. 1998 | |||
| D4S244 | 71.5 | 64.2 | Reich et al. 1998 | |||
| D4S2393 | 81.8 | 74.3 | Reich et al. 1998 | |||
| WIAF-2114 | 98.5 | 93.8 | 83.4 | 1227 | .81 | .71 |
| WIAF-373 | 100 | 102.8 | 90 | 2688 | 1.34 | 1.44 |
| ADH3 | 107.1 | 97 | Reich et al. 1998 | |||
| D4S3256 | 118.6 | 104.5 | Long et al. 1998 | |||
| D4S3240 | 116.8 | 106.6 | Long et al. 1998 | |||
| WIAF-2765 | 109.7 | 110.7 | 110.9 | 19501 | .78 | .73 |
| WIAF-3818 | 125.8 | 139 | 122.6 | 1879 | .84 | 1.29 |
| WIAF-3821 | 127.8 | 139 | 122.6 | 1882 | .85 | 1.63 |
| 5: | ||||||
| WIAF-3575 | 62.8 | 54 | 49.8 | 1636 | 1.19 | 1.32 |
| WIAF-1869 | 81.5 | 68.3 | 69.5 | 3414 | .86 | 1.32 |
| D5S2501 | 105.6 | 105.9 | Long et al. 1998 | |||
| 6: | ||||||
| WIAF-1567 | 20.5 | 16.9 | 12.1 | 1124 | 1.12 | 1.56 |
| WIAF-2177 | 41.9 | 37.7 | 36.4 | 1290 | 1.22 | 1.28 |
| D6S1018 | 51.4 | 51.2 | Reich et al. 1998 | |||
| 7: | ||||||
| D7S531 | 2.1 | 2.9 | Long et al. 1998 | |||
| D7S1793 | 49.2 | 50.3 | Reich et al. 1998 | |||
| WIAF-1680 | 126.5 | 104.6 | 104.6 | 1168 | .85 | 1.25 |
| D7S1809 | 127.4 | 121.1 | Reich et al. 1998 | |||
| 8: | ||||||
| D8S549 | 14.9 | 15.7 | Reich et al. 1998 | |||
| D8S280 | 20.8 | 20.6 | Reich et al. 1998 | |||
| WIAF-1291 | 107 | 100.1 | 93.6 | 934 | 1.23 | 1.25 |
| WIAF-989 | 147.7 | 131.3 | 130.5 | 3088 | .89 | .75 |
| 9: | ||||||
| WIAF-1900 | 49.8 | 30.3 | 28.5 | 3445 | 1.16 | 1.27 |
| D9S319 | 28.3/31.4 | 30.8 | Long et al. 1998 | |||
| WIAF-3477 | 125.8 | 108.6 | 109.9 | 1538 | .85 | 1.35 |
| 10: | ||||||
| D10S1435 | 2.2 | 2.1 | Long et al. 1998 | |||
| WIAF-3336 | 85.6 | 52.5 | 60.3 | 19256 | 1.17 | .77 |
| D10S677 | 102.4 | 98 | Straub et al. 1999 | |||
| D10S1239 | 108.7 | 105.6 | Straub et al. 1999 | |||
| WIAF-268 | 137.3 | 114.4 | 109.5 | 19487 | 1.18 | 1.21 |
| D10S2469 | 114.4 | 109.5 | Straub et al. 1998 | |||
| 11: | ||||||
| D11S1984 | 1.4 | .3 | Long et al. 1998 | |||
| D11S2368 | 15.3 | 17.8 | Long et al. 1998 | |||
| AFM333TH1 | 17.7 | 17.1/18.1 | Reich et al. 1998 | |||
| WIAF-2046 | 39.1 | 25.9 | 25.5 | 3585 | 1.28 | 1.36 |
| WIAF-1949 | 40 | 28.1 | 28.3 | 3494 | 1.18 | 1.47 |
| D11S1392 | 35.4 | 34.4 | Long et al. 1998 | |||
| D11S976 | 128.1 | 122.4 | Long et al. 1998 | |||
| D11S2359 | 144.8 | 140.5 | Reich et al. 1998 | |||
| 12: | ||||||
| D12S393 | 104.1 | 100.7 | Reich et al. 1998 | |||
| WIAF-364 | 153.2 | 133.9 | 129.3 | 2681 | .83 | .71 |
| WIAF-3624 | 146.3 | 134.6 | 129.1 | 1685 | .85 | 1.22 |
| D12S1045 | 140 | 136 | Reich et al. 1998 | |||
| 13: | ||||||
| D13S321 | 58.2 | 56.1 | Reich et al. 1998 | |||
| D13S762 | 96.5 | 90.8 | Reich et al. 1998 | |||
| WIAF-4520 | 29.9 | 94.8 | 93.5 | 19336 | 1.13 | 1.42 |
| WIAF-2555 | 96.6 | 91.3 | 114.1/115.5 | 2068 | 1.13 | .78 |
| D13S285 | 92.2 | 115.1 | Long et al. 1998 | |||
| WIAF-600 | 80.8 | 102.5 | 101.5 | 2197 | .86 | 1.23 |
| D13S895 | 111.5 | 110.6 | Long et al. 1998 | |||
| 15: | ||||||
| D15S153 | 65.2 | 62.5 | Long et al. 1998 | |||
| D15S642 | 99 | 99 | Reich et al. 1998 | |||
| 16: | ||||||
| D16S675 | 9.13 | 7 | Reich et al. 1998 | |||
| GATA5H07 | NA | NA | Long et al. 1998 | |||
| D16S422 | 84.4 | 86.1 | Straub et al. 1999 | |||
| 17: | ||||||
| D17S1308 | 1.7 | .2 | Long et al. 1998 | |||
| WIAF-3343 | 77 | 61.9 | 67.4 | 1416 | .88 | .8 |
| D17S2059 | 70.7 | 71.3 | Straub et al. 1999 | |||
| D17S1535 | 74.8 | 75.6 | Long et al. 1998 | |||
| 18: | ||||||
| D18S869 | 21.4 | 19 | Straub et al. 1999 | |||
| WIAF-303 | 65.9 | 51.1 | 50.7 | 2127 | 1.14 | 1.52 |
| D18S844 | 76.2 | 77.4 | Long et al. 1998 | |||
| 19: | ||||||
| WIAF-2426 | 35 | NA | NA | 3698 | 1.32 | 1.46 |
| D19S49 | 40.7 | 44.1 | Reich et al. 1998 | |||
| 21: | ||||||
| WIAF-4514 | 43 | 31.7 | NA | 19393 | 1.14 | 1.21 |
| 22: | ||||||
| WIAF-3065 | 43.2 | 42.8 | 43.3 | 19273 | 1.12 | 1.55 |
| X: | ||||||
| WIAF-2589 | 156.1 | 117.4 | 123.1 | 3824 | 1.13 | 1.28 |
| DXS8061 | 138.4 | 147.5 | Straub et al. 1998 | |||
| WIAF-1743 | 187.7 | 138.8 | 147.8 | 3290 | .89 | .75 |
| Not placed: | ||||||
| D4S3242 | NA | Long et al. 1998 | ||||
Note.— Reproducibly positive SNPs from the current study and simple sequence–length repeat markers displaying nominally significant linkage in prior affected sib-pair linkage studies of alcohol or nicotine dependence. Reproducibly positive SNPs with abuser/control ratios in the upper or lower 5% of values in the European American sample and in the upper or lower 7.5% in the African American sample are listed here. SNP markers are identified by WIAF numbers (column 1). Markers are listed along with their chromosomal assignments (column 1), averaged radiation hybrid distances (column 2) in cM, ratio of A/B alleles in substance abusers divided by those for control individuals in the European American (column 7) and African American (column 8) sample pools (see Methods). Mapviewer chromosomal positions (expressed in Mb) of SNPs use build 22 and build 24. Markers whose data are shown in boldface italics lie near other positive markers in build 24. Two pairs of markers, D4S2457/D4S1647 and WIAF-3818/WIAF-3821, sample the same two underlying polymorphisms. Underlining indicates differently named assays that sample the same underlying sequence variations.
When the positions of these 42 reproducibly positive markers were determined, one pair of independent markers were adjacent to each other, one other pair of apparent SNPs actually queried the same underlying SNP, and a pair on chromosome 11 flanked a strong candidate-gene locus. Monte Carlo simulations indicated a .043 probability that the members of three closest marker pairs would lie at least as close to each other as they do here by chance (Mapviewer, build 22; P=.036 for build 24 positions).
The positive chromosome 4 markers WIAF-3818 and WIAF-3821 both display positive abuser/control hybridization ratios in European American and African American samples. These WAIF markers use different oligonucleotides to amplify overlapping sequences that identify the same sequence tagged site, stsG67686; this congruent result thus helps to validate the technique used here.
Nearby reproducibly positive markers on chromosome 11, WIAF-2046 and WIAF-1949, are located at Mapviewer build 22 coordinates 25.9 and 28.08 Mb, respectively. The most strikingly positive SNP, WIAF-2046, borders the location (25.9 Mb) of the strong candidate gene encoding BDNF (Jones and Reichardt 1990; Maisonpierre et al. 1991; Shintani et al. 1992; Timmusk et al. 1993; Horger et al. 1999; Lyons et al. 1999; Krebs et al. 2000). This gene contains a dinucleotide-repeat polymorphism located close to the 5′ end of the gene’s main coding exon (Krebs et al. 2000). In initial studies with sample pools, this BDNF gene marker also displayed association with drug-abuse vulnerability. The long forms of this repeat were found with 21.3%±6% frequency in the 12 pooled samples of the 240 European American individuals free from substance abuse and in 16.3%±2% of the 20 pooled samples of 400 drug abusers (mean ± SEM of three repeated comparisons, differences between abuser and control P=.001, t-test). Simple sequence–repeat markers that flank WIAF-2046 and WIAF (and thus flank this locus), D11S2001 and GATA25B04, display statistically significant D′ values, ranging from 0.3 to 1, for their different alleles.
The reproducibly positive markers on chromosome 12, WIAF-364 and WIAF-3624, lie within ∼0.2 Mb of each other and flank genes, including that for the T6J protein, as well as several EST clusters (Mapviewer builds 22 and 24). These display strong linkage disequilibrium, with D′ values ∼1 in control subjects.
The positions of these reproducibly positive SNPs from the current study can also be compared with localizable positions of 61 of the microsatellite markers that were nominally positive in at least one linkage study of alcohol or nicotine dependence (Long et al. 1998; Reich et al. 1998; Straub et al. 1999; Foroud et al. 2000). Nine of these markers lie in six regions within 2 Mb of the location of one of the confirmed positive SNPs in the current data set (Mapviewer build 22). Seven of these nine lie within 1 Mb of one of these SNPs (table 1; Mapviewer build 22; in build 24, five of them are within that distance). Monte Carlo simulations indicate that nine randomly distributed microsatellite markers would lie this near to one of the confirmed positive SNP markers in <1 in 108 simulation trials (Mapviewer build 22; P<.006 for build 24 positions).
D3S3053, linked to alcoholism by Long et al. (1998), is located 0.4–1 Mb from WIAF-452, a marker reproducibly associated in the current study with polysubstance abuse and is positioned near the phospholipase D1 gene. D4S1627 is linked to alcoholism by Long et al. (1998) and located at 51.4 Mb on chromosome 4 within 0.1–0.2 Mb of the reproducibly positive marker WIAF-452. These markers lie near genes encoding ras and GABAA receptor–family members.
ADH3 and D4S2457—as well as D4S2457's equivalent, D4S1647—have been linked to alcoholism by two different groups (Long et al. 1998; Reich et al. 1998). These markers lie near WIAF-2765, a reproducibly positive SNP from the current study (Mapviewer build 22), and near the alcohol dehydrogenase gene cluster. Altered risk of alcoholism and/or effects on alcohol metabolism from allelic variants at ADH3 or ADH2 have been reported in several studies (Osier et al. 1999; Borras et al. 2000).
D9S319 is linked to alcoholism (Long et al. 1998) and is positioned at 28.4 and 31.4 Mb (Mapviewer build 22). These positions flank the 30.3–30.8 Mb (Mapviewer builds 22 and 24) location for WIAF-1900 that was reproducibly positive in the current study. This marker is near an interesting gene encoding a phospholipase A2–activating protein.
D10S2469, a chromosome 10 marker located at 114.4–109.5 Mb and linked to nicotine dependence (Mapviewer builds 22 and 24) (Straub et al. 1999) has a position difficult to distinguish from that of the reproducibly positive SNP WIAF-268. D13S285, a 91.3- to 115.1-Mb marker linked to alcoholism (Long et al. 1998) lies near the reproducibly positive SNP WIAF-2555, located at 92.2–114.1/115.5 Mb (Mapviewer build 22 and two locations in build 24). Genes encoding the endothelin B receptor and several meiotic proteins are nearby. DXS8061 is linked to nicotine dependence with a modestly positive LOD score and is located on chromosome X at 138.4–147.5 Mb (Mapviewer builds 22 and 24) (Straub et al. 1999). WIAF-1743 is also located at 138.8–147.8 Mb, near genes that encode zinc finger proteins.
Discussion
This initial genome-scanning study of polysubstance-abuse vulnerability provides 41 candidate chromosomal regions nominated in a European American sample and reproduced in an African American sample. A candidate region on chromosome 12 contains two closely adjacent positive SNP markers. Two markers on chromosome 11 identify the BDNF locus where initial studies, with additional locus-specific markers, also identify association. Other regions—on chromosomes 3, 4, 10, 13, and X—are sites for both positive SNPs from the current data and positive markers from previous linkage studies of alcohol or nicotine dependence. These results are unlikely to all be due to chance. They identify regions containing candidate genes that have also been implicated in other populations, in studies of the genetics of vulnerability to addiction.
Clinical and laboratory results provide evidence for the reliability and validity of this approach. The extent of study participation and the good fit between clinical histories supplied by different family members support the validity and representativeness of the clinical sample studied here. Features of the SNP-analysis method used here also indicate sufficient reliability and validity to test our working hypotheses. Test-retest correlations, the correlations between sense and antisense hybridization patterns, and the correlations between pooled and individually obtained genotypes each support the pooling strategy used here. Finding reproducible positive results from two sets of oligonucleotides that interrogate the same sequence-tagged site further validates the current technique. The extent to which markers are positive in both our European American sample and our African American one, the extent to which some of these doubly positive markers lie next to each other, and the extent to which they are located next to markers defined in previous studies of legal addictions are each greater than is likely to be due to chance. These observations each add to confidence in the methods used.
The chromosome 11 region between WIAF-2046 and WIAF-1949 contains the strong candidate-gene locus that encodes BDNF. Long alleles of the BDNF dinucleotide repeat found in the intron just 5′ to the main coding exon are also associated in our sample with vulnerability to polysubstance abuse. These same alleles distinguish French substance-abusing schizophrenic patients from schizophrenic individuals who are free of substance disorders (Krebs et al. 2000). These data from several populations add to other genomic and neurobiological information supporting the likelihood that BDNF variants could contribute to vulnerabilities to polysubstance abuse. This locus displays a wealth of variation (Timmusk et al. 1993). BDNF influences dopamine and serotonin neurotransmitters that are heavily linked to addiction (Dluzen et al. 1999; Horger et al. 1999; Lyons et al. 1999; Kernie et al. 2000). Cocaine preference and alcohol consumption can be altered with BDNF administration or heterozygous gene deletion (Dluzen et al. 1999; Horger et al. 1999; Lyons et al. 1999; Kernie et al. 2000). Although the exact distances from WIAF-2046 to BDNF and from BDNF to WIAF-1949 vary in recent Mapviewer versions, there is substantial linkage disequilibrium between the GATA25B04/D11S2364 and D11S2001 markers that flank the BDNF/WIAF-2046/WIAF-1949 interval (significant D′ values 0.3−1; χ2=17.9; n=54; P<.001). As we study this region further, we will be able to determine whether variation in BDNF and/or adjacent genes contributes to our initial observations of association.
Other reproducibly positive SNPs lie near markers previously linked to legal addictions much more often than is likely by chance. Each of the regions defined by these convergent data sets contains interesting candidate genes worthy of further study. Striking findings at the chromosome 4 region of ∼107–110.7 Mb (Mapviewer build 22) containing WIAF-2765, D4S2457/ D4D1647, and ADH3 each fit with the large body of evidence that alcohol dehydrogenase gene variants alter predisposition to alcohol dependence. Other studies have also identified altered risk of alcoholism and/or effects on alcohol metabolism from ADH allelic variants (Xu et al. 1988; Osier et al. 1999; Borras et al. 2000). Our current data, however, provide the first association between these loci and vulnerability to illegal substance abuse. Primary and/or secondary effects of ADH alleles on polysubstance abuse are possible. For example, alcohol could serve as a gateway drug, and/or smoking could interact with ADH activities (Marselos et al. 1991; Bhagwat et al. 1998).
These initial genome-scanning studies of polysubstance abuse–vulnerability genes provide an approach that can be extended. Modeling studies demonstrate increasing power of association approaches with increasing marker densities (D. Naiman and G. R. Uhl, unpublished data). Studying additional markers in these and other samples will help us to identify more of the alleles that distinguish individuals with substance vulnerabilities from those who remain free from substance abuse. Although few studies may individually provide sufficient power to unequivocally identify disease alleles (Lander and Kruglyak 1995), careful nomination and confirmation in multiple clinical samples provides stronger and stronger cumulative assurance against false-positive or poorly generalizable results. We report the current results and their congruence with others’ data in this spirit.
The current data and their convergence with others’ results now provide support for the likelihood that common allelic variants play significant roles in vulnerabilities to substance abuse. Such support was not evident in prior linkage studies that did not identify similar linked chromosomal regions. Even common allelic variants, however, could each provide distinct influences on different clinical features of the disorder. Vulnerability alleles could also provide pleiotropic manifestations in other behavioral disorders. As the alleles that contribute to vulnerability to substance abuse are elucidated in more repeated studies, identification of the ways in which they change specific features of vulnerability to addiction will help us better understand addictions themselves.
Acknowledgments
We acknowledge exceptional clinical data collection by Fely Carillo-London, Brenda Campbell, Linda Kahler, and Fred Snyder, assistance with informed-consent forms for some research volunteers by Carlo Contoreggi, Larry Rodriguez, and David Gorelick, assistance with initial microarray experiments from Zicheng Lin, spirited help with data analyses from Andrew Shapiro, Leslie Cope, and Cheryl Mayo, help from the Johns Hopkins-Bayview support staff, and financial support from the National Institute on Drug Abuse, National Institutes of Health.
Electronic-Database Information
The URL for data in this article is as follows:
- Authors' FTP site, ftp://137.187.144.38/loci1.xls (for data that support )
References
- Barcellos LF, Klitz W, Field LL, Tobias R, Bowcock AM, Wilson R, Nelson MP, Nagatomi J, Thomson G (1997) Association mapping of disease loci, by use of a pooled DNA genomic screen. Am J Hum Genet 61:734–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat SV, Vijayasarathy C, Raza H, Mullick J, Avadhani NG (1998) Preferential effects of nicotine and 4-(N-methyl-N-nitrosamine)-1-(3-pyridyl)-1-butanone on mitochondrial glutathione S-transferase A4-4 induction and increased oxidative stress in the rat brain. Biochem Pharmacol 56:831–839 [DOI] [PubMed] [Google Scholar]
- Borras E, Coutelle C, Rosell A, Fernandez-Muixi F, Broch M, Crosas B, Hjelmqvist L, Lorenzo A, Gutierrez C, Santos M, Szczepanek M, Heilig M, Quattrocchi P, Farres J, Vidal F, Richart C, Mach T, Bogdal J, Jornvall H, Seitz HK, Couzigou P, Pares X (2000) Genetic polymorphism of alcohol dehydrogenase in Europeans: the ADH2*2 allele decreases the risk for alcoholism and is associated with ADH3*1. Hepatology 31:984–989 [DOI] [PubMed] [Google Scholar]
- Cadoret RJ, Troughton E, O'Gorman TW, Heywood E (1986) An adoption study of genetic and environmental factors in drug abuse. Arch Gen Psychiatry 43:1131–1136 [DOI] [PubMed] [Google Scholar]
- Cadoret RJ, Yates WR, Troughton E, Woodworth G, Stewart MA (1995) Adoption study demonstrating two genetic pathways to drug abuse. Arch Gen Psychiatry 52:42–52 [DOI] [PubMed] [Google Scholar]
- Cervino AC, Hill AV (2000) Comparison of tests for association and linkage in incomplete families. Am J Hum Genet 67:120–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dluzen DE, Story GM, Xu K, Kucera J, Walro JM (1999) Alterations in nigrostriatal dopaminergic function within BDNF mutant mice. Exp Neurol 160:500–507 [DOI] [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, Li TK, Hesselbrock V, Crowe R, Schuckit M, Porjesz B, Begleiter H, Reich T (2000) Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res 24:933–945 [PubMed] [Google Scholar]
- Germer S, Holland MJ, Higuchi R (2000) High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res 10:258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J, Lyons MJ, Eisen SA, True W, Tsuang M (1993) Genetic influence on drug use: preliminary analysis of 2674 Vietnam era veteran twins. Behav Genet 1993:552 [Google Scholar]
- Grove WM, Eckert ED, Heston L, Bouchard TJ Jr, Segal N, Lykken DT (1990) Heritability of substance abuse and antisocial behavior: a study of monozygotic twins reared apart. Biol Psychiatry 27:1293–1304 [DOI] [PubMed] [Google Scholar]
- Gynther LM, Carey G, Gottesman, II, Vogler GP (1995) A twin study of non-alcohol substance abuse. Psychiatry Res 56:213–220 [DOI] [PubMed] [Google Scholar]
- Hacia JG, Fan JB, Ryder O, Jin L, Edgemon K, Ghandour G, Mayer RA, Sun B, Hsie L, Robbins CM, Brody LC, Wang D, Lander ES, Lipshutz R, Fodor SP, Collins FS (1999) Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat Genet 22:164–167 [DOI] [PubMed] [Google Scholar]
- Horger BA, Iyasere CA, Berhow MT, Messer CJ, Nestler EJ, Taylor JR (1999) Enhancement of locomotor activity and conditioned reward to cocaine by brain-derived neurotropic factor. J Neurosci 19:4110–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KR, Reichardt LF (1990) Molecular cloning of a human gene that is a member of the nerve growth factor family. Proc Natl Acad Sci USA 87:8060–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkowski LM, Prescott CA, Kendler KS (2000) Multivariate assessment of factors influencing illicit substance use in twins from female-female pairs. Am J Med Genet 96:665–670 [PubMed] [Google Scholar]
- Kendler KS, Karkowski LM, Corey LA, Prescott CA, Neale MC (1999) Genetic and environmental risk factors in the aetiology of illicit drug initiation and subsequent misuse in women. Br J Psychiatry 175:351–356 [DOI] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA (1998) Cocaine use, abuse and dependence in a population-based sample of female twins. Br J Psychiatry 173:345–350 [DOI] [PubMed] [Google Scholar]
- Kernie SG, Liebl DJ, Parada LF (2000) BDNF regulates eating behavior and locomotor activity in mice. Embo J 19:1290–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs MO, Guillin O, Bourdell MC, Schwartz JC, Olie JP, Poirier MF, Sokoloff P (2000) Brain derived neurotropic factor (BDNF) gene variants association with age at onset and therapeutic response in schizophrenia. Mol Psychiatry 5:558–562 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D (1998) Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet 81:216–221 [DOI] [PubMed] [Google Scholar]
- Lyons WE, Mamounas LA, Ricaurte GA, Coppola V, Reid SW, Bora SH, Wihler C, Koliatsos VE, Tessarollo L (1999) Brain-derived neurotropic factor-deficient mice develop aggressiveness and hyperphagia in conjunction with brain serotonergic abnormalities. Proc Natl Acad Sci USA 96:15239–15244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonpierre PC, Le Beau MM, Espinosa R, 3rd, Ip NY, Belluscio L, de la Monte SM, Squinto S, Furth ME, Yancopoulos GD (1991) Human and rat brain-derived neurotropic factor and neurotrophin-3: gene structures, distributions, and chromosomal localizations. Genomics 10:558–568 [DOI] [PubMed] [Google Scholar]
- Marselos M, Vasiliou V, Malamas M, Alikaridis F, Kefalas T (1991) Effects of cannabis and tobacco on the enzymes of alcohol metabolism in the rat. Rev Environ Health 9:31–37 [DOI] [PubMed] [Google Scholar]
- Merikangas KR, Stolar M, Stevens DE, Goulet J, Preisig MA, Fenton B, Zhang H, O'Malley SS, Rounsaville BJ (1998) Familial transmission of substance use disorders. Arch Gen Psychiatry 55:973–979 [DOI] [PubMed] [Google Scholar]
- Osier M, Pakstis AJ, Kidd JR, Lee JF, Yin SJ, Ko HC, Edenberg HJ, Lu RB, Kidd KK (1999) Linkage disequilibrium at the ADH2 and ADH3 loci and risk of alcoholism. Am J Hum Genet 64:1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persico AM, Bird G, Gabbay FH, Uhl GR (1996) D2 dopamine receptor gene TaqI A1 and B1 restriction fragment length polymorphisms: enhanced frequencies in psychostimulant-preferring polysubstance abusers. Biol Psychiatry 40:776–784 [DOI] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS, McGue M, Lykken DT, Heston LL, Clayton PJ (1991) Heterogeneity in the inheritance of alcoholism: a study of male and female twins. Arch Gen Psychiatry 48:19–28 [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI Jr, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H (1998) Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet 81:207–215 [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Schork NJ, Nath SK, Fallin D, Chakravarti A (2000) Linkage disequilibrium analysis of biallelic DNA markers, human quantitative trait loci, and threshold-defined case and control subjects. Am J Hum Genet 67:1208–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Cherny SS, Purcell S, Hewitt JK (2000) Power of linkage versus association analysis of quantitative traits, by use of variance-components models, for sibship data. Am J Hum Genet 66:1616–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani A, Ono Y, Kaisho Y, Igarashi K (1992) Characterization of the 5′-flanking region of the human brain-derived neurotropic factor gene. Biochem Biophys Res Commun 182:325–332 [DOI] [PubMed] [Google Scholar]
- Smith SS, O'Hara BF, Persico AM, Gorelick DA, Newlin DB, Vlahov D, Solomon L, Pickens R, Uhl GR (1992) Genetic vulnerability to drug abuse: the D2 dopamine receptor TaqI B1 restriction fragment length polymorphism appears more frequently in polysubstance abusers. Arch Gen Psychiatry 49:723–727 [DOI] [PubMed] [Google Scholar]
- Stallings MC, Hewitt JK, Beresford T, Heath AC, Eaves LJ (1999) A twin study of drinking and smoking onset and latencies from first use to regular use. Behav Genet 29:409–421 [DOI] [PubMed] [Google Scholar]
- Straub RE, Sullivan PF, Ma Y, Myakishev MV, Harris-Kerr C, Wormley B, Kadambi B, Sadek H, Silverman MA, Webb BT, Neale MC, Bulik CM, Joyce PR, Kendler KS (1999) Susceptibility genes for nicotine dependence: a genome scan and followup in an independent sample suggest that regions on chromosomes 2, 4, 10, 16, 17 and 18 merit further study. Mol Psychiatry 4:129–144 [DOI] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Services Administration (1995) National household survey on drug abuse: population estimates. U.S. Department of Health and Human Services, Public Health Service, Washington, DC [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H (1993) Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron 10:475–489 [DOI] [PubMed] [Google Scholar]
- True WR, Heath AC, Scherrer JF, Xian H, Lin N, Eisen SA, Lyons MJ, Goldberg J, Tsuang MT (1999a) Interrelationship of genetic and environmental influences on conduct disorder and alcohol and marijuana dependence symptoms. Am J Med Genet 88:391–397 [DOI] [PubMed] [Google Scholar]
- True WR, Xian H, Scherrer JF, Madden PA, Bucholz KK, Heath AC, Eisen SA, Lyons MJ, Goldberg J, Tsuang M (1999b) Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry 56:655–661 [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Eisen SA, Goldberg J, True W, Lin N, Meyer JM, Toomey R, Faraone SV, Eaves L (1996) Genetic influences on DSM-III-R drug abuse and dependence: a study of 3,372 twin pairs. Am J Med Genet 67:473–477 [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Harley RM, Xian H, Eisen S, Goldberg J, True WR, Faraone SV (1999) Genetic and environmental influences on transitions in drug use. Behav Genet 29:473–479 [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L (1998) Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry 55:967–972 [DOI] [PubMed] [Google Scholar]
- Uhl GR (1999) Molecular genetics of substance abuse vulnerability: a current approach. Neuropsychopharmacology 20:3–9 [DOI] [PubMed] [Google Scholar]
- Uhl GR, Elmer GI, Labuda MC, Pickens RW (1995) Genetic influences on drug abuse. In: Bloom F, Kupfer DJ (ed) Psychopharmacology: the fourth generation of progress. Raven Press, New York, pp 1793–1806 [Google Scholar]
- Uhl GR, Gold LH, Risch N (1997) Genetic analyses of complex behavioral disorders. Proc Natl Acad Sci USA 94:2785–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward CE MH, Silberg JL, Meyer JM, Eaves LJ (1996) Tobacco, alcohol and drug use in 8–16 year old twins. NIDA Res Monograph 1996:309 [Google Scholar]
- Xu YL, Carr LG, Bosron WF, Li TK, Edenberg HJ (1988) Genotyping of human alcohol dehydrogenases at the ADH2 and ADH3 loci following DNA sequence amplification. Genomics 2:209–214 [DOI] [PubMed] [Google Scholar]