Abstract
The identification of a growing number of novel Mendelian disorders and private mutations in the Roma (Gypsies) points to their unique genetic heritage. Linguistic evidence suggests that they are of diverse Indian origins. Their social structure within Europe resembles that of the jatis of India, where the endogamous group, often defined by profession, is the primary unit. Genetic studies have reported dramatic differences in the frequencies of mutations and neutral polymorphisms in different Romani populations. However, these studies have not resolved ambiguities regarding the origins and relatedness of Romani populations. In this study, we examine the genetic structure of 14 well-defined Romani populations. Y-chromosome and mtDNA markers of different mutability were analyzed in a total of 275 individuals. Asian Y-chromosome haplogroup VI-68, defined by a mutation at the M82 locus, was present in all 14 populations and accounted for 44.8% of Romani Y chromosomes. Asian mtDNA-haplogroup M was also identified in all Romani populations and accounted for 26.5% of female lineages in the sample. Limited diversity within these two haplogroups, measured by the variation at eight short-tandem-repeat loci for the Y chromosome, and sequencing of the HVS1 for the mtDNA are consistent with a small group of founders splitting from a single ethnic population in the Indian subcontinent. Principal-components analysis and analysis of molecular variance indicate that genetic structure in extant endogamous Romani populations has been shaped by genetic drift and differential admixture and correlates with the migrational history of the Roma in Europe. By contrast, social organization and professional group divisions appear to be the product of a more recent restitution of the caste system of India.
Introduction
The Roma (Gypsies) became one of the peoples of Europe when they arrived in the Byzantine Empire 900–1,100 years ago (Fraser 1992; Rochow and Matschke 1991). The formation of the present-day Romani populations of European countries is the compound product of the early migrations from the Balkans into western Europe, completed by the 15th century, and three superimposed migration waves: the first during the end of the 19th century, after the abolition of Gypsy slavery in Romania (Hancock 1987; Fraser 1992; Liégeois 1994); the second out of Yugoslavia, during the 1960s and 1970s; and the third during the last decade, following the political and economic changes in eastern Europe (Reyniers 1995). Current estimates of the total Romani population size in Europe range from 4 million to 10 million, with the largest numbers concentrated in central and southeastern Europe (Liégeois 1994; Marushiakova and Popov 2001c).
In recent years, novel single-gene disorders (see Kalaydjieva et al. 1996, 2000; Angelicheva et al. 1999; Tournev et al. 1999; Rogers et al. 2000; Thomas et al., 2001), as well as private mutations causing known Mendelian disorders (see Piccolo et al. 1996; Abicht et al. 1999; Kalaydjieva et al. 1999; Plasilova et al. 1999), have been identified. Large Romani families with psychiatric disorders are being used in an effort to localize susceptibility genes (Kaneva et al. 1998), and epidemiological evidence suggests that there are differences in the prevalence of other complex disorders, such as Parkinson disease and multiple sclerosis, between the Roma and surrounding European populations (Kalman et al. 1991; Milanov et al. 2000). The Roma are thus emerging as an interesting founder population, with a genetic-research potential that is still to be explored.
The complex structure of Romani society, where the Romani Group is the primary unit, has long attracted the attention of cultural anthropologists (Petulengro 1915–16; Fraser 1992; Marushiakova and Popov 1997). Liégeois (1994, p. 61) describes the current social organization of the Roma as a “fluid mosaic of diversified groups.” Group identity and the ensuing social divisions are based on a variety of criteria, such as customs, ethnonyms describing traditional trades, and dialects reflecting the history of migrations. The greatest diversity is found in the Balkans, where numerous Romani populations with well-defined social boundaries exist (Marushiakova and Popov 1997, 2001a). This social organization and its strong impact on rules of endogamy have not been addressed in genetic research. Population-genetic studies of the Roma from different European countries have been performed for nearly 80 years and have mostly sought to compare the Roma to autochthonous Europeans and to identify genetic affinities with proposed parental populations and with other Romani populations. The low resolution of individual classical genetic markers and the random sampling design have often led to contradictory results. Nonetheless, these studies have generally concluded that the Roma are genetically distinct from other European populations, while, at the same time, different Romani populations are separated by larger genetic distances than are their European neighbors (reviewed by Kalaydjieva et al. [2001b]). Recent medical-genetic studies have shown that founder mutations can be shared by socially diverse and geographically dispersed Romani populations, whereas those living in close geographic proximity can display markedly different gene frequencies (reviewed by Kalaydjieva et al. [2001b]). Thus, social practices, as well as genetic data, suggest significant population substructure. The relationship between traditional group divisions and biological affinities, however, is unclear and appears to be complex. Current patterns—genetic as well as social—could be the product of diverse scenarios, with different implications for genetic epidemiology.
In this study, we address the issue of genetic relatedness behind the social and cultural diversity of Romani populations. We have used Y-chromosome and mtDNA markers of different mutability to examine the origins and diversification of paternal and maternal lineages in 14 well-defined Romani populations. The findings point to common Asian origins and suggest that the early history of splits and migrations in Europe has played a major role in shaping current genetic structure.
Subjects and Methods
Study Populations
This study included 275 unrelated males from 14 traditional Romani populations, selected to represent different cultural-anthropological classification criteria (Marushiakova and Popov 1997) and to allow an assessment of their genetic relevance. Group characteristics and numbers sampled are shown in table 1. Most populations are well defined and endogamous relative to each other, except for the Lingurari, Monteni, and Intreni, who are separated by geographic distance rather than by rules of endogamy. The previously described Kalderash, Monteni, and Lom populations (Kalaydjieva et al. 2001a) were typed for additional loci, and the Lom sample size was expanded.
Table 1.
Description of the Romani Populations Included in the Study
| Populationa | Place of Residence | Traditional Trade | Language/Dialect | History of Migrations | Religion | Sample Size |
| Turgovzi (Tu) | Bulgaria, Omurtag | Merchants | Romanes, Balkan dialect; Turkish | Early settlement in Bulgaria | Islam | 36 |
| Feredjelli (Fe) | Bulgaria, Omurtag | Unskilled laborers | Turkish | Early settlement in Bulgaria | Islam | 21 |
| Kalaidjii North (KN) | Bulgaria, Lom | Tinsmiths | Romanes, Balkan dialect; | Early settlement in Bulgaria | Protestant | 20 |
| Koshnichari South Central (KC) | Bulgaria, Plovdiv region | Basket makers | Romanes, Balkan dialect | Early settlement in Bulgaria | Eastern Orthodox | 4 |
| Koshnichari Southwest (KW) | Bulgaria, Gotze Delchev | Basket makers | Romanes, Balkan dialect | Early settlement in Bulgaria | Protestant | 5 |
| Kalaidjii South (KS) | Bulgaria, Gotze Delchev | Tinsmiths | Romanes, Old Vlax dialectb | Wallachia/Moldavia, to Bulgaria in 17th and 18th centuries | Eastern Orthodox | 10 |
| Lom (Lo) | Bulgaria, Lom | Livestock dealers | Romanes, Old Vlax dialectb | Wallachia/Moldavia, to Bulgaria in 17th and 18th centuries | Protestant | 43 |
| Monteni (Mo) | Bulgaria, Balkan Mountain villages | Bowl makers | Archaic Rumanian | Wallachia/Moldavia, to Bulgaria in late 19th century | Eastern Orthodox | 42 |
| Intreni (In) | Bulgaria, Letnitza | Bowl makers | Archaic Rumanian | Wallachia/Moldavia, to Bulgaria in late 19th century | Eastern Orthodox | 17 |
| Lingurari North (LN) | Bulgaria, northern part | Bowl makers | Archaic Rumanian | Wallachia/Moldavia, to Bulgaria in late 19th century | Eastern Orthodox | 18 |
| Lingurari South (LS) | Bulgaria, southern part | Bowl makers | Archaic Rumanian | Wallachia/Moldavia, to Bulgaria in late 19th century | Eastern Orthodox | 9 |
| Kalderash (Ka) | Bulgaria, northern part | Coppersmiths | Romanes, New Vlax dialectb | Wallachia/Moldavia, to Bulgaria in late 19th century | Eastern Orthodox | 23 |
| Spanish Roma (SR) | Madrid | Merchants | Spanish | Early migration to north/Western Europe | Protestant | 27 |
| Lithuanian Roma (LR) | Vilnius, Lithuania | Merchants | Romanes | Early migration to north/Western Europe | Roman Catholic | 20 |
Two-letter abbreviations of population names are used in tables throughout this article.
Vlax dialects are characterized by a strong linguistic influence from Romanian.
The analyses also included samples from 40 males from Asia and the Middle East who were found to carry Y-chromosome haplogroups VI-68 and VI-56, as defined by mutations M82 and M67, respectively (Underhill et al. 2000). These samples were genotyped for the Y-chromosome short-tandem-repeat (Y STR) markers used in this study.
This study is part of an ongoing project, investigating the molecular epidemiology of single-gene disorders and the population structure of the Roma, conducted in collaboration with Romani organizations and local health authorities. Research into genetic epidemiology (to be published separately) involves carrier testing for private founder mutations, with genetic counseling provided to all participating subjects. Informed consent for both aspects of the study has been obtained from all individuals involved. This study complies with the ethical guidelines of the participating institutions.
Y-Chromosome Analysis
This part of the study included 252 Romani and 40 non-Romani male subjects. As suggested by de Knijff (2000), we designate Y chromosomes defined by unique-event polymorphisms (UEPs) as “haplogroups,” those defined by Y STRs as “haplotypes,” and those defined by both UEPs and Y STRs as “lineages.” Haplogroup designation follows the nomenclature proposed by Underhill et al. (2000).
Y-Chromosome Haplogroups
Comprehensive analysis of UEPs was performed as described (Underhill et al. 1997, 2000, 2001; Shen et al. 2000) on 94 Romani males, aiming at the identification of the major Y-chromosome haplogroups in the Roma. The remaining 158 samples were typed for the M82 locus, a 2-bp deletion, in derived Y chromosomes, that defines haplogroup VI-68 (Underhill et al. 2000). PCR amplification was done with fluorescently labeled primers 5′-CTGTACTCCTGGGTAGCCTGT-3′ and 5′-AAGAACGATTGAACACACTAACTC-3′. The products were separated by size on a 377 DNA Analyzer (Applied Biosystems).
The 70 samples that carried the ancestral M82 allele were genotyped for specific UEPs on the basis of the identities of their Y STR haplotypes with the common haplotype(s) of the specific haplogroup in the fully characterized Romani samples. These markers included M1, M45, M67, M89, and M170. M1 was analyzed as described elsewhere (Hammer and Horai 1995). The remaining UEPs were analyzed using a modified version of the primer-extension assay (Bray et al. 2001) (protocol available on request) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Mass spectra were collected using a Voyager-DE PRO MALDI-TOF instrument (Applied Biosystems). Genotypes were determined manually by calculation of the mass of the dideoxynucleotide added onto the primer. The above analytical system left five samples for which haplogroup assignment was not possible.
Y STR Haplotypes
A total of 209 Romani and 40 non-Romani individuals were genotyped for eight Y STR loci—namely, DYS19, DYS388, DYS389II, DYS389I, DYS390, DYS391, DYS392, and DYS393. In addition, Y STR data for 43 Roma from three populations described by Kalaydjieva et al. (2001a) were expanded by typing for DYS388. PCR primers were as described elsewhere (Kayser et al. 1997). The products were separated on an ABI 373A DNA Analyzer (Applied Biosystems). Allele sizes were converted to repeat number by ue of allelic ladders, which were analyzed in parallel. We define DYS389CD as equivalent to DYS389I, and we define and DYS389AB as equivalent to DYS389II minus DYS389I (Rolf et al. 1998). Haplotypes were constructed following the ascending numerical order of loci given above.
mtDNA
mtDNA was analyzed in 275 Romani subjects. By analogy to the Y chromosome, mtDNA “haplogroups” are defined by coding-region RFLPs, “haplotypes” are defined by hypervariable segment 1 (HVS1) sequences, and mtDNAs defined by both RFLPs and HVS1 sequences are referred to as “lineages.”
mtDNA Haplogroups
RFLP analysis of coding regions of the mitochondrial genome was performed on 165 samples by use of standard protocols (Passarino et al. 1996; Richards et al. 1998; Macaulay et al. 1999). This analysis provided an indication of the mtDNA haplogroups present in the Roma. In 110 samples, in which RFLP analysis was not performed, haplogroups were inferred from characteristic HVS1 variants (Macaulay et al. 1999; Simoni et al. 2000).
mtDNA Haplotypes
HVS1 sequencing was performed on 194 samples. In addition, 81 HVS1 sequences previously reported in the Roma (Kalaydjieva et al. 2001a) were included in the statistical analyses. PCR amplification of the D-loop segment between positions 15997 and 16400 (Anderson et al. 1981) was performed as described elsewhere (Calafell et al. 1996). The samples were sequenced in both directions and were run on an ABI 373A DNA Analyzer (Applied Biosystems). A 360-bp fragment of HVS1, between positions 16023 and 16384, was analyzed.
Data Analysis
The frequencies of male and female haplotypes, haplogroups, and lineages and the number of shared lineages were determined by direct counting. Diversity indices were determined using ARLEQUIN. Haplotype diversity, h, and its variance, V(h), were calculated according to the method of Nei (1987). Pairwise differences, k, between haplotypes were calculated to provide a measure of the relatedness of haplotypes within haplogroups. Phylogenetic relationships between haplotypes within haplogroups were examined by constructing median-joining networks by use of Network 3.0 (see the Life Sciences and Engineering Technology Solutions web site) (Bandelt et al. 1995).
The age of the founding Y-chromosome haplogroup VI-68 lineage was calculated as described by Kittles et al. (1998), with a Y STR mutation rate of 2.1×10-3 (95% confidence interval [95%CI] 0.6×10-3–4.9×10-3) (Heyer et al. 1997). The age of the mtDNA haplogroup M lineage in the Roma was determined as suggested by Saillard et al. (2000). Given that most of the actual mutated sites appear to have high mutation rates, the average mutation rate used in the calculations was roughly three times that used by Meyer et al. (1999)—that is, one mutation per 6,727 years. The average number of mutations from the ancestral haplotype were computed with Network 3.0 (see the Life Sciences and Engineering Technology Solutions web site) (Bandelt et al. 1995). A generation time of 25 years was used.
Principal-components (PC) analysis was used to examine the differences in the distribution of Y chromosome and mtDNA haplogroups among 11 Romani populations where sample sizes were ⩾10 for both data sets. The analysis was performed using the computer program ANTANA based on Eigenanalysis, where a correlation matrix is generated from standardized frequency data, corrected for sample size.
Analysis of molecular variance (AMOVA; Excoffier et al. 1992) was performed on the Y STR and mtDNA HVS1 data. Different groupings of populations, based on the criteria outlined in table 1, were considered. The apportionment of genetic variance was assessed, between individuals within populations, between populations within groups, and between groups of populations. The analyses were done with ARLEQUIN, using the “sum of squared size difference” setting, for Y STR data, and “pairwise differences,” for mtDNA HVS1 data. Standard Bonferroni corrections were used to account for multiple comparisons.
Results
Y-Chromosome Analysis
The data obtained from the analysis of 252 male Roma are summarized in table 2. A total of nine known haplogroups were identified among the 247 Romani Y chromosomes for which haplogroup assignment was possible. Three haplogroups—namely, VI-68, VI-52, and VI-56—occurred at high frequencies (>10%) and together accounted for ∼80% of all Y chromosomes. Four haplotypes—VI-68A, VI-68B, VI-52A, and VI-56A—together accounted for 57% of all Y chromosomes.
Table 2.
Y-Chromosome Lineages Identified in 14 Romani Populations
|
No. of Y Chromosomes in Population |
||||||||||||||||
| HaplogroupandLineage | Haplotypea | LN | LS | In | Mo | Lo | KS | Ka | KN | KW | KC | Tu | Fe | LR | SR | Total |
| VI-68:b | ||||||||||||||||
| A | 15-12-16-14-22-10-11-12 | 12 | 4 | 9 | 12 | 9 | 3 | 6 | 4 | 3 | 2 | 4 | 4 | 3 | 5 | 80 |
| B | 14-12-16-14-22-10-11-12 | 15 | 5 | 20 | ||||||||||||
| C | 15-12-16-14-23-10-11-12 | 1 | 1 | 2 | ||||||||||||
| D | 15-12-16-14-22-10-11-13 | 2 | 2 | |||||||||||||
| E | 14-12-16-14-22-9-11-12 | 2 | 2 | |||||||||||||
| F | 15-12-17-14-22-10-11-12 | 1 | 1 | |||||||||||||
| G | 15-12-16-13-22-10-11-12 | 1 | 1 | |||||||||||||
| H | 15-12-16-14-21-10-11-12 | 1 | 1 | |||||||||||||
| I | 15-12-16-15-22-10-11-12 | 1 | 1 | |||||||||||||
| J | 15-12-15-14-22-10-11-12 | 1 | 1 | |||||||||||||
| K | 15-10-16-14-22-10-11-12 | 1 | 1 | |||||||||||||
| L | 14-12-17-14-22-10-11-12 | 1 | 1 | |||||||||||||
| VI-52:c | ||||||||||||||||
| A | 14-14-16-12-22-10-11-13 | 1 | 1 | 3 | 14 | 5 | 24 | |||||||||
| B | 17-13-17-13-24-10-11-13 | 2 | 8 | 1 | 11 | |||||||||||
| C | 14-14-16-13-22-10-11-13 | 1 | 4 | 1 | 6 | |||||||||||
| D | 15-13-18-13-25-11-11-13 | 3 | 2 | |||||||||||||
| E | 14-14-16-12-21-10-11-13 | 3 | 3 | |||||||||||||
| F | 17-13-16-14-23-10-11-13 | 1 | 1 | |||||||||||||
| G | 16-13-18-13-24-11-11-13 | 1 | 1 | |||||||||||||
| H | 16-13-17-13-24-11-11-13 | 1 | 1 | |||||||||||||
| I | 15-13-18-14-23-9-12-14 | 1 | 1 | |||||||||||||
| J | 14-13-17-13-23-10-11-13 | 1 | 1 | |||||||||||||
| K | 13-13-18-14-23-10-12-12 | 1 | 1 | |||||||||||||
| L | 17-13-17-13-24-10-13-13 | 1 | 1 | |||||||||||||
| M | ?-14-16-12-22-10-11-13 | 1 | 1 | |||||||||||||
| N | 15-13-18-13-24-11-11-13 | 1 | 1 | |||||||||||||
| O | 15-14-16-13-22-10-11-13 | 1 | 1 | |||||||||||||
| VI-56:d | ||||||||||||||||
| A | 14-15-17-14-23-10-11-12 | 5 | 4 | 1 | 3 | 6 | 19 | |||||||||
| B | 14-15-17-14-22-10-11-12 | 4 | 4 | |||||||||||||
| C | 14-15-16-14-23-10-11-12 | 1 | 1 | 1 | 3 | |||||||||||
| D | 14-16-17-14-23-11-11-12 | 1 | 1 | |||||||||||||
| E | 14-15-17-15-23-10-11-12 | 1 | 1 | |||||||||||||
| F | 14-15-17-14-23-11-11-12 | 1 | 1 | |||||||||||||
| G | 15-15-17-14-23-10-11-12 | 1 | 1 | |||||||||||||
| H | 14-15-16-14-22-10-11-12 | 1 | 1 | |||||||||||||
| I | 13-15-17-14-23-10-11-12 | 1 | 1 | |||||||||||||
| IX-104:e | ||||||||||||||||
| A | 14-12-16-13-25-10-13-13 | 1 | 1 | 1 | 3 | |||||||||||
| B | 14-12-16-13-24-11-13-13 | 1 | 2 | 3 | ||||||||||||
| C | 14-12-16-14-24-11-13-13 | 1 | 1 | 2 | ||||||||||||
| D | 14-12-16-13-24-11-13-12 | 2 | 2 | |||||||||||||
| E | 14-12-17-13-24-11-13-12 | 1 | 1 | |||||||||||||
| F | 14-12-16-14-24-11-11-13 | 1 | 1 | |||||||||||||
| G | 14-12-16-13-23-11-13-13 | 1 | 1 | |||||||||||||
| H | 14-12-15-13-24-10-14-13 | 1 | 1 | |||||||||||||
| I | 15-14-16-13-24-11-11-13 | 1 | 1 | |||||||||||||
| J | 15-14-16-13-24-11-13-13 | 1 | 1 | |||||||||||||
| K | ?-12-17-13-26-10-11-13 | 1 | 1 | |||||||||||||
| III-36:f | ||||||||||||||||
| A | 13-12-17-13-24-10-11-13 | 3 | 1 | 1 | 1 | 6 | ||||||||||
| B | 13-12-19-14-24-10-11-14 | 1 | 1 | |||||||||||||
| C | ?-12-17-13-24-9-11-13 | 1 | 1 | |||||||||||||
| D | 13-12-19-13-24-10-11-13 | 1 | 1 | |||||||||||||
| VI-71:g | ||||||||||||||||
| A | 14-15-17-14-25-10-11-13 | 3 | 3 | |||||||||||||
| B | 14-15-16-14-25-10-11-13 | 2 | 2 | |||||||||||||
| C | 15-16-16-13-23-10-11-12 | 1 | 1 | |||||||||||||
| D | 14-14-16-12-23-10-11-13 | 1 | 1 | |||||||||||||
| E | 14-15-15-14-25-10-11-13 | 1 | 1 | |||||||||||||
| F | 14-12-16-14-22-10-12-14 | 1 | 1 | |||||||||||||
| VI-57h | ||||||||||||||||
| A | 16-15-19-13-22-10-11-12 | 1 | 1 | 1 | 3 | |||||||||||
| B | 16-15-18-13-22-10-11-12 | 1 | 1 | |||||||||||||
| C | 16-15-20-14-22-10-11-12 | 1 | 1 | |||||||||||||
| V-52i,j | ||||||||||||||||
| A | 15-13-16-13-24-10-11-13 | 2 | 2 | |||||||||||||
| B | 15-13-16-13-25-10-11-13 | 1 | 1 | |||||||||||||
| C | 15-13-16-13-24-11-11-12 | 1 | 1 | |||||||||||||
| IX-108k | ||||||||||||||||
| A | 14-12-17-13-24-11-11-13 | 1 | 1 | |||||||||||||
| Unknownl | ||||||||||||||||
| A | 17-12-16-13-24-10-11-13 | 1 | 1 | |||||||||||||
| B | 16-15-19-13-22-11-11-12 | 1 | 1 | |||||||||||||
| C | 16-13-16-13-23-10-11-13 | 1 | 1 | |||||||||||||
| D | 15-12-18-13-25-11-11-13 | 1 | 1 | |||||||||||||
| E | 13-12-16-14-23-?-13-14 | 1 | 1 | |||||||||||||
| Total | 16 | 6 | 17 | 17 | 39 | 10 | 11 | 20 | 8 | 4 | 36 | 21 | 20 | 27 | 252 | |
Constructed using the marker order DYS19-DYS388-DYS389AB-DYS389CD-DYS390-DYS391-DYS392-DYS393.
Defined by UEP delAT at locus M82 and accounts for 44.8% of the total population in this study.
Defined by UEP A→C at locus M170 and accounts for 22.6% of the total population in this study.
Defined by UEP A→T at locus M67 and accounts for 12.7% of the total population in this study.
Defined by UEP A→C at locus M173 and accounts for 6.7% of the total population in this study.
Defined by UEP T→G at locus M35 and accounts for 3.6% of the total population in this study.
Defined by UEP C→T at locus M89 and accounts for 3.6% of the total population in this study.
Defined by UEP T→C at locus M92 and accounts for 2.0% of the total population in this study.
Defined by UEP A→C at locus M217 and accounts for 1.6% of the total population in this study.
The M217 locus was first reported, by Underhill et al. (2001), as defining haplogroup V-52 .
Defined by UEP delG at locus M17 and accounts for 0.4% of the total population in this study.
Unknown haplogroups account for 2.0% of the total population in this study.
Major Paternal Founding Lineage
VI-68 was by far the most common haplogroup. It was observed in all 14 Romani populations and comprised 113 chromosomes, or 44.8% of the overall study population. Haplogroup VI-68 has been found previously at low frequencies in the Indian subcontinent and central Asia but, so far, has not been observed in other European populations (Underhill et al. 2000), with the exception of one individual in the Ukraine (Semino et al. 2000).
Y STR analysis of haplogroup VI-68 chromosomes identified 12 haplotypes (VI-68A–VI-68L). In a median-joining network (fig. 1A), these haplotypes clustered tightly together, with a single inferred node. The two high-frequency haplotypes, VI-68A and VI-68B, are centrally located in the network, with the remaining haplotypes radiating from them. The high frequency of these two haplotypes is reflected in the low diversity within this haplogroup (h=0.47; k=0.56).
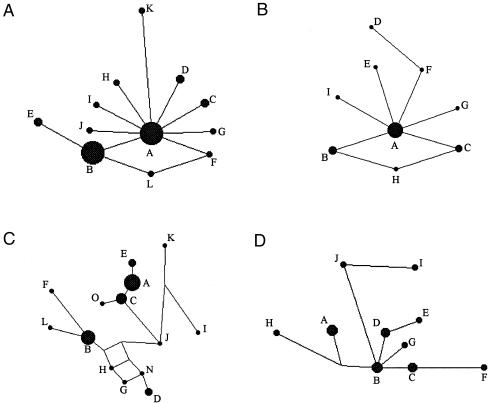
Figure 1.
Median-joining networks of Y STR haplotypes within four haplogroups. A, Haplogroup VI-68 (N=113; h=0.47; k=0.56). B, Haplogroup VI-56 (N=32; h=0.87; k=0.64). C, Haplogroup VI-52 (N=57; h=0.76; k=3.15). D, Haplogroup IX-104 (N=17; h=0.94; k=2.50). The sizes of the nodes are proportional to the relative frequency of that haplotype within the haplogroup. Branch lengths within each network are proportional to the number of mutations separating haplotypes.
The distribution of VI-68 haplotypes in the Roma was compared with that of non-Romani haplogroup VI-68 chromosomes from different Asian populations. The 22 non-Romani chromosomes presented with 22 different Y STR haplotypes (table 3), including a haplotype that was one mutational step away from the most common Romani VI-68A lineage. A median-joining network, constructed from all 34 haplogroup VI-68 haplotypes (12 Romani and 22 Asian non-Romani) displayed a complex topology, in which the Romani Y chromosomes represented a limited subset of closely related haplotypes within the overall diversity of haplogroup VI-68 (data not shown). The non-Romani haplotypes were widely dispersed across the network, with many inferred nodes.
Table 3.
Y STR Haplotypes Observed in Non-Romani Y-Chromosome Haplogroups VI-68 and VI-56
| Haplogroup | Frequency | DYS19 | DYS388 | DYS389AB | DYS389CD | DYS390 | DYS391 | DYS392 | DYS393 |
| VI-68 (N=22) | 1 | 14 | 12 | 14 | 11 | 23 | 10 | 11 | 11 |
| 1 | 14 | 12 | 15 | 13 | 23 | 10 | 11 | 11 | |
| 1 | 14 | 12 | 16 | 13 | 22 | 10 | 11 | 11 | |
| 1 | 15 | 12 | 14 | 13 | 22 | 10 | 11 | 11 | |
| 1 | 15 | 12 | 14 | 13 | 21 | 10 | 11 | 11 | |
| 1 | 15 | 12 | 15 | 13 | 23 | 10 | 11 | 12 | |
| 4 | 15 | 12 | 15 | 13 | 23 | 10 | 11 | 11 | |
| 1 | 15 | 12 | 15 | 13 | 21 | 10 | 11 | 12 | |
| 1 | 15 | 12 | 16 | 14 | 22 | 11 | 11 | 11 | |
| 1 | 15 | 12 | 16 | 14 | 22 | 10 | 11 | 11 | |
| 1 | 15 | 12 | 17 | 12 | 24 | 10 | 11 | 11 | |
| 1 | 15 | 12 | 17 | 13 | 21 | 10 | 11 | 12 | |
| 1 | 15 | 12 | 17 | 14 | 23 | 10 | 10 | 11 | |
| 1 | 15 | 13 | 15 | 13 | 22 | 10 | 11 | 11 | |
| 1 | 15 | 13 | 16 | 13 | 21 | 10 | 11 | 11 | |
| 1 | 15 | 13 | 17 | 13 | 22 | 10 | 11 | 11 | |
| 1 | 16 | 12 | 14 | 14 | 22 | 10 | 11 | 11 | |
| 1 | 16 | 13 | 16 | 14 | 22 | 10 | 11 | 11 | |
| 1 | 17 | 12 | 14 | 13 | 22 | 10 | 11 | 12 | |
| VI-56 (N=18) | 3 | 14 | 14 | 15 | 13 | 22 | 10 | 11 | 11 |
| 1 | 14 | 14 | 15 | 14 | 22 | 10 | 11 | 11 | |
| 2 | 14 | 15 | 15 | 13 | 22 | 9 | 11 | 11 | |
| 5 | 14 | 15 | 15 | 13 | 22 | 10 | 11 | 11 | |
| 1 | 14 | 15 | 15 | 13 | 22 | 9 | 11 | 11 | |
| 1 | 14 | 15 | 16 | 13 | 22 | 10 | 11 | 13 | |
| 1 | 14 | 15 | 17 | 14 | 23 | 10 | 11 | 11 | |
| 1 | 14 | 15 | 17 | 13 | 22 | 10 | 11 | 11 | |
| 1 | 15 | 15 | 17 | 14 | 21 | 10 | 11 | 11 | |
| 1 | 15 | 15 | 17 | 13 | 24 | 10 | 11 | 12 | |
| 1 | 15 | 16 | 16 | 13 | 23 | 11 | 11 | 11 |
A single male lineage, VI-68A, defined by the 2-bp deletion at M82 and by Y STR haplotype 15-12-16-14-22-10-11-12, was shared by 80 individuals from all Romani populations. This common lineage accounted for 71% of haplogroup VI-68 chromosomes and for 32% of all Romani Y chromosomes examined. It was separated by one mutational step (at marker DYS19) from the second most common VI-68 lineage (VI-68B). VI-68B was not as widespread as VI-68A and occurred mostly in the Lom and the Lithuanian Roma (table 2). The remaining haplogroup VI-68 lineages were rare and confined to individual Romani populations. When we considered the most frequent haplotype within haplogroup VI-68 to be the founding lineage, a coalescent date of 992 years ago (95%CI 425–3,472 years) was estimated.
Additional Y-Chromosome Lineages
Haplogroup VI-56 accounted for 12.7% (32 chromosomes) of all Romani males (table 2). It was identified in 6 of the 14 Romani populations and occurred at high frequency in the Lithuanian (25%) and Spanish (33%) Roma. This haplogroup has been found in Pakistan, central Asia, and the Middle East (Underhill et al. 2000). Within Europe, haplogroup VI-56 has been identified in a single male individual from Sardinia (Underhill et al. 2000). In the Roma, the 32 haplogroup VI-56 chromosomes fell into nine Y STR haplotypes, VI-56A–VI-56I (table 2). The pattern of the median-joining network for these haplotypes (fig. 1B) was similar to that described for haplogroup VI-68, with tight clustering of haplotypes and no inferred nodes. Haplogroup-diversity indices were h=0.87 and k=0.64. By comparison, 18 non-Romani haplogroup VI-56 chromosomes displayed 11 Y STR haplotypes (table 3), of which one was a single mutational step away from the Romani VI-56A lineage.
Haplogroups VI-52 and IX-104, referred to as “Eu7” and “Eu18” by Semino et al. (2000), accounted for 22.6% and 6.7%, respectively, of all Romani Y chromosomes. These two haplogroups are common in Europe (Underhill et al. 2000), where reverse clinal distributions have been reported (Semino et al. 2000), with higher frequencies of VI-52 in eastern Europe and of IX-104 in the western part of the continent.
Haplogroup VI-52 was identified in 57 males from 11 of the 14 Romani populations (table 2). The majority (52 of 57) were Roma resident in Bulgaria. Y STR analysis identified 15 haplotypes within this haplogroup. Two common haplotypes (VI-52A and VI-52B), contributed primarily by Romani groups that were early settlers in Bulgaria, accounted for 61% of the chromosomes of this haplogroup and for nearly 14% of all Romani Y chromosomes. Haplogroup VI-52 diversity indices were h=0.76 and k=3.15. The median-joining network (fig. 1C) contained inferred nodes, with many haplotypes differing from each other by multiple mutational steps.
Haplogroup IX-104 was found in 8 of the 14 Romani populations, with 8 of 17 chromosomes coming from the Lithuanian and Spanish Roma (table 2). Y STR analysis revealed 11 different haplotypes that connect to each other in a median-joining network with a number of inferred nodes (fig. 1D). The diversity indices in haplogroup IX-104 were h=0.94 and k=2.50.
The remaining five characterized haplogroups (table 2) were rare, each accounting for <4% of the total sample. Haplogroups VI-57, V-52, and IX-108 have been found in different parts of Asia, and III-36 has been identified in Ethiopia and South Africa (Underhill et al. 2000, 2001). Haplogroup VI-71 has no specific geographic association and is widely distributed throughout the world (Underhill et al. 2000).
mtDNA Diversity
The results of the mtDNA analysis of 275 Roma are shown in table 4. A total of 12 mtDNA haplogroups were identified, of which 2—haplogroups M and H—accounted for 62% of the overall study population. Analysis of HVS1 revealed 72 unique sequences. Four common lineages—two of haplogroup H and one each of haplogroups M and U3—accounted for 36% of all Romani individuals.
Table 4.
mtDNA Lineages Identified in Roma
|
No. of mtDNA Lineages in Population |
|||||||||||||||
| Haplogroup and HVS1 Variant(s)a | LN | LS | In | Mo | Lo | KS | Ka | KN | KW | KC | Fe | Tu | LR | SR | Total |
| M:b | |||||||||||||||
| 129, 223, 291, 298 | 1 | 2 | 2 | 3 | 4 | 2 | 3 | 1 | 3 | 4 | 4 | 29 | |||
| 129, 223, 291 | 1 | 2 | 3 | 2 | 2 | 2 | 12 | ||||||||
| 129, 223, 230, 233, 304 | 1 | 1 | |||||||||||||
| 129, 223, 230, 233, 304, 344 | 1 | 3 | 3 | 2 | 1 | 10 | |||||||||
| 129, 223, 230, 233, 304, 344, 355 | 3 | 1 | 4 | ||||||||||||
| 129, 148, 223, 291, 298 | 1 | 1 | 2 | 4 | |||||||||||
| 129, 223, 291, 298, 311 | 1 | 1 | 1 | 3 | |||||||||||
| 129, 223, 256, 291 | 1 | 1 | 1 | 3 | |||||||||||
| 223, 291, 298 | 2 | 2 | |||||||||||||
| 129, 223, 234, 291, 298 | 1 | 1 | |||||||||||||
| 129, 223, 291, 298, 362 | 1 | 1 | |||||||||||||
| 129, 223, 266, 291 | 1 | 1 | |||||||||||||
| 223, 290, 318T | 1 | 1 | |||||||||||||
| 223, 304 | 1 | 1 | |||||||||||||
| H:c | |||||||||||||||
| 261, 304 | 3 | 2 | 8 | 4 | 1 | 3 | 2 | 23 | |||||||
| 186, 304 | 6 | 5 | 3 | 8 | 22 | ||||||||||
| 218, 278 | 3 | 3 | 1 | 1 | 8 | ||||||||||
| 354 | 6 | 2 | 8 | ||||||||||||
| Cambridge reference sequence | 2 | 3 | 1 | 6 | |||||||||||
| 192A, 320 | 2 | 3 | 5 | ||||||||||||
| 189 | 1 | 2 | 3 | ||||||||||||
| 168 | 3 | 3 | |||||||||||||
| 223 | 1 | 2 | 3 | ||||||||||||
| 93 | 1 | 1 | 2 | ||||||||||||
| 67 | 2 | 2 | |||||||||||||
| 51, 145, 304 | 1 | 1 | |||||||||||||
| 304 | 1 | 1 | |||||||||||||
| 278, 293, 311 | 1 | 1 | |||||||||||||
| 187, 189 | 1 | 1 | |||||||||||||
| 189, 311 | 1 | 1 | |||||||||||||
| 93, 291 | 1 | 1 | 2 | ||||||||||||
| 174 | 1 | 1 | |||||||||||||
| 261 | 1 | 1 | |||||||||||||
| 242 | 1 | 1 | |||||||||||||
| 260 | 1 | 1 | |||||||||||||
| 362 | 1 | 1 | |||||||||||||
| 93, 223 | 1 | 1 | |||||||||||||
| U3:d | |||||||||||||||
| 343 | 1 | 3 | 1 | 10 | 11 | 26 | |||||||||
| 343, 260 | 2 | 2 | |||||||||||||
| J:e | |||||||||||||||
| 69, 126 | 2 | 3 | 1 | 4 | 1 | 11 | |||||||||
| 69, 126, 145, 222, 261, 311 | 2 | 2 | 1 | 5 | |||||||||||
| 69, 126, 145, 222, 235, 261, 271 | 1 | 1 | |||||||||||||
| 69, 126, 145, 222, 235, 261 | 1 | 1 | |||||||||||||
| 69, 126, 261 | 1 | 1 | |||||||||||||
| 69, 93, 126, | 1 | 1 | |||||||||||||
| 39C, 69, 126 | 1 | 1 | |||||||||||||
| 69, 126, 193 | 1 | 1 | |||||||||||||
| 69, 126, 278, 366 | 1 | 1 | |||||||||||||
| 69, 126, 300 | 1 | 1 | |||||||||||||
| 69, 126, 311 | 1 | 1 | |||||||||||||
| X:f | |||||||||||||||
| 126, 189A, 223, 278 | 1 | 3 | 2 | 2 | 1 | 1 | 2 | 12 | |||||||
| 93, 189, 223, 241, 278 | 2 | 1 | 2 | 5 | |||||||||||
| 92, 126, 189A, 223, 278 | 2 | 2 | |||||||||||||
| 93, 96T, 189, 223, 241, 278 | 1 | 1 | |||||||||||||
| 92, 189A, 223, 278 | 1 | 1 | |||||||||||||
| I:g | |||||||||||||||
| 129, 172, 223, 311 | 3 | 1 | 1 | 5 | |||||||||||
| N:1bh | |||||||||||||||
| 86, 129, 145, 176G, 223 | 1 | 3 | 1 | 5 | |||||||||||
| T:i | |||||||||||||||
| 126, 294, 296 | 1 | 1 | 1 | 3 | |||||||||||
| 126, 294, 324 | 1 | 1 | 2 | ||||||||||||
| 126, 294, 352 | 1 | 1 | |||||||||||||
| U5:j | |||||||||||||||
| 28G, 192, 224, 261, 270 | 1 | 1 | |||||||||||||
| 192, 224, 261, 270 | 1 | 1 | |||||||||||||
| 189, 270, 311, 336 | 1 | 1 | |||||||||||||
| 189, 270 | 1 | 1 | |||||||||||||
| 167, 192, 270, 311, 356 | 1 | 1 | |||||||||||||
| 256, 270 | 1 | 1 | |||||||||||||
| U(K):k | |||||||||||||||
| 224, 261, 311 | 1 | 1 | |||||||||||||
| 222, 224, 261, 311 | 1 | 1 | |||||||||||||
| 224, 311 | 1 | 1 | |||||||||||||
| 224, 311, 344 | 1 | 1 | |||||||||||||
| U1:l | |||||||||||||||
| 183C, 189, 249 | 1 | 1 | |||||||||||||
| W:m | |||||||||||||||
| 172, 223, 231, 292 | 2 | 1 | 3 | ||||||||||||
| Total | 18 | 9 | 16 | 42 | 43 | 10 | 23 | 20 | 5 | 3 | 18 | 25 | 18 | 25 | 275 |
Numbers are those given by Anderson et al. (1981), plus 16,000. All variants are transitions from the reference sequence, unless indicated with a letter.
Accounts for 26.5% of all mtDNA lineages in this study.
Accounts for 35.6% of all mtDNA lineages in this study.
Accounts for 10.2% of all mtDNA lineages in this study.
Accounts for 9.1% of all mtDNA lineages in this study.
Accounts for 7.6% of all mtDNA lineages in this study.
Accounts for 1.8% of all mtDNA lineages in this study.
Accounts for 1.8% of all mtDNA lineages in this study.
Accounts for 2.2% of all mtDNA lineages in this study.
Accounts for 2.2% of all mtDNA lineages in this study.
Accounts for 1.4% of all mtDNA lineages in this study.
Accounts for 0.4% of all mtDNA lineages in this study.
Accounts for 1.1% of all mtDNA lineages in this study.
Diversity of Maternal Lineages
Haplogroup M was identified in all 14 Romani populations and accounted for 73 individuals, or 26.5% of the total sample (table 4). Haplogroup M is rare in Europe (Richards et al. 1998; Simoni et al. 2000) but is common in Asia and eastern Africa (Quintana-Murci et al. 1999). HVS1 sequence analysis did not identify the motif characterizing the African subhaplogroup M1, defined by variants at positions 16129, 16189, 16223, 16249, and 16311 (Quintana-Murci et al. 1999), thereby pointing to the Asian origin of these Romani lineages.
HVS1 analysis of haplogroup M samples revealed 14 sequences. The two most common haplogroup M lineages differed by a single mutation step, at position 16298 (table 4). These two lineages were present in 13 of the 14 Romani populations and accounted for 14.9% of all samples.
A transition at position 16129, which defines subhaplogroup M5 (Bamshad et al. 2001), was present in 11 of the 14 HVS1 sequences of Romani haplogroup M. One of the three lineages that do not bear the 16129 variant—namely, the lineage defined by variants at positions 16223, 16291, and 16298—are closely related to haplogroup M5 lineages and may represent a back mutation at position 16129, a known mutational hotspot (Stoneking 2000). Subhaplogroup M5 was thus found to account for 97.3% of haplogroup M. A modified median-joining network (fig. 2) was used to compare haplogroup M lineages in the Roma to those observed in India (Kivisild et al. 1999; Quintana-Murci et al. 1999). All but two Romani lineages clustered together as a small subset of the overall diversity present within the Indian haplogroup M. The coalescence of haplogroup M lineages in the Roma was estimated to be 4,625 years ago (95%CI 2,000–7,250 years). This date was obtained by considering that an average of 0.6896 mutations have accumulated from the putative ancestral haplotype—that is, the haplotype with variants at positions 16129, 16223, 16291, and 16298.
Figure 2.
Modified median-joining network of mtDNA haplogroup M, constructed from data presented in studies by Quintana-Murci et al. (1999) and Kivisild et al. (1999) and in the present study. All numbers are those given by Anderson et al. (1981), plus 16,000. Sequences identified in the Roma are shown in red; sequences reported for Indian samples are shown in blue. Subhaplogroup designations are as proposed by Bamshad et al. (2001), plus additional subclades defined by frequent variants at positions 16189, 16318, and 16093. Branches are proportional to the number of mutations separating sequence types, except those that connect subhaplogroups.
Haplogroup H was the most frequent mtDNA haplogroup among the Roma (table 4). It was detected in 13 of 14 Romani populations and represented 35.6% (98 individuals) of the total sample. Haplogroup H is most common in Europe (Simoni et al. 2000) and the Near East (Richards et al. 2000) but is also found in India (Kivisild et al. 1999). HVS1 analysis of haplogroup H identified 23 sequences, 2 of which (defined by variants at positions 16261 and 16304 and at positions 16218 and 16278, respectively) each accounted for ∼22% of haplogroup H and together comprised 20% of the overall sample. These two lineages have not been found in a large survey of Near Eastern and European individuals (Richards et al. 2000).
Haplogroup U3 was identified in 28 subjects (10.2% of the entire sample), most of whom (23 of 28) were Spanish and Lithuanian Roma. Only two lineages were identified by HVS1 sequencing, with one of them accounting for 93% of all U3 samples (table 4). Haplogroup U3 is distributed throughout the Middle East and Europe (Richards et al. 2000).
Haplogroup X occurred in 7.6% of Romani samples and could be subdivided into five lineages by HVS1 sequencing. Three of these lineages, bearing a transversion at position 16189, have not been seen in Europe and the Middle East, where haplogroup X is widely distributed (Kivisild et al. 1999; Richards et al. 2000).
The remaining haplogroups—J, I, N1b, T, U5, U(K), U1, and W—accounted for 20% of Romani samples. Varying numbers of Romani lineages were identified by HVS1 sequencing in each haplogroup. These haplogroups have been observed in Europe, the Middle East, and India (Kivisild et al. 1999; Richards et al. 2000; Simoni et al. 2000).
Genetic Structure
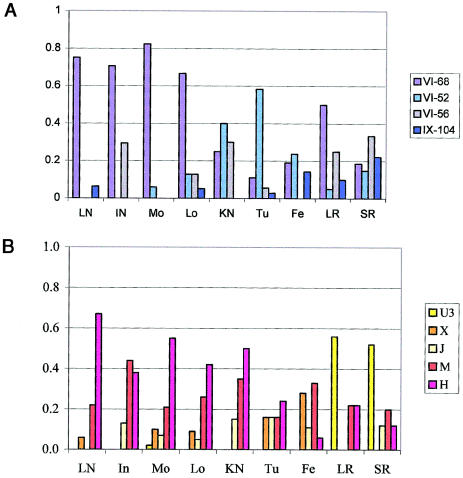
As shown in tables 2 and 4, a total of 13 paternal and 25 maternal lineages were found to occur in more than one Romani group. The male VI-68A lineage was shared by Roma from all populations, and two pairs of closely related mtDNA lineages, of haplogroups M and H, were common to 13 and 8 Romani populations respectively. At the same time, the frequency distribution of both major and rare male and female lineages differed dramatically between Romani populations (fig. 3).
Figure 3.
Frequency distributions of the common (overall frequency >5%) male (A) and female (B) haplogroups in Romani populations. Populations in which sample size was <15 for either Y-chromosome or mtDNA haplogroup data were excluded from the analysis.
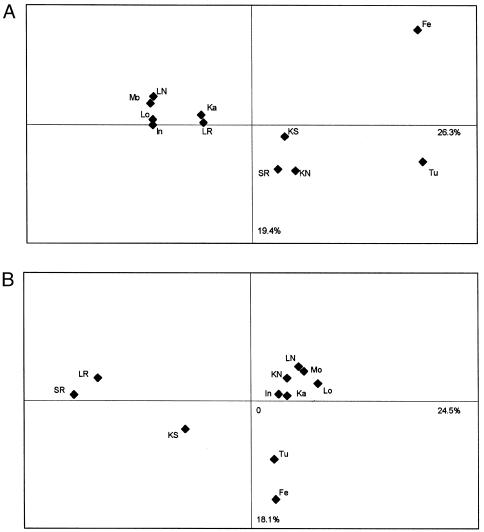
PC analysis was based on Y-chromosome and mtDNA haplogroup frequencies in Romani populations. The resultant PC plots provided better resolution of the genetic structure than was provided by a neighbor-joining tree (Nei 1987) using Y STR haplotypes (not shown). The PC plots are presented in figure 4.
Figure 4.
Two-dimensional PC plots based on Y STR haplotype frequencies (A) and mtDNA haplogroup frequencies (B). The population affinities shown are based on 51% and 42.6%, respectively, of the variation that, on the basis of Y-chromosome and mtDNA data, is present within the entire sample.
Two clusters, consistently present in both Y-chromosome and mtDNA analysis, were formed by the Monteni, Intreni, Lingurari, Kalderash, and Lom on one hand and by the Feredjelli and Turgovtzi on the other. The Spanish and Lithuanian Roma clustered together in the mtDNA analysis, and the Kalaidjii North and South clustered together in the Y-chromosome comparisons.
To examine the relevance of different cultural, historical, and geographic classification criteria to the genetic structure of the Roma, we used AMOVA based on Y STR data and mtDNA HVS1 sequences (table 5). The country-of-residence, in which all Roma from Bulgaria were compared versus those from Lithuania versus those from Spain, showed no significant intergroup differences. The same result was obtained with comparisons based on place of residence, in which three pairs of Romani populations living in close proximity in three small towns in Bulgaria were examined. In the analysis based on ethnonym reflecting traditional trade, the comparison of bowl makers, tinsmiths, traders, and livestock dealers showed no significant intergroup differences.
Table 5.
AMOVA Using Y STR and mtDNA Data for Romani Populations
|
Variation (Pa) |
||||||
| Among Groups |
Among Populations within Groups |
Within Populations |
||||
| Grouping Criterion | Y STR | mtDNA | Y STR | mtDNA | Y STR | mtDNA |
| Total sample | 13.0% (<.00001) | 6.2% (<.00001) | 87.0% (<.00001) | 93.8% (<.00001) | ||
| Country of residenceb | 5.1% (.79277) | 4.0% (.01760) | 15.2% (<.00001) | 4.8% (<.00001) | 89.9% (<.00001) | 91.2% (<.00001) |
| Town of residencec | 6.7% (.21408) | .5% (.32551) | 7.5% (.00391) | .8% (.26686) | 85.8% (<.00001) | 98.7% (.16618) |
| Trade/group (ethnonym)d | 7.9% (.08113) | 4.7% (.01622) | 8.5% (<.00001) | 2.1% (.05083) | 83.6% (<.00001) | 93.2% (<.00001) |
| Religione | 6.2% (.03617) | 4.3% (.00196) | 8.0% (<.00001) | 2.9% (<.00001) | 85.8% (<.00001) | 92.8% (<.00001) |
| Languagef | 6.5% (.07234) | 6.3% (<.00001) | 7.2% (<.00001) | 0.7% (<.00001) | 86.3% (<.00001) | 92.9% (<.00001) |
| Historical migrationg | 10.5% (<.00001) | 5.0% (<.00001) | 5.3% (<.00001) | 3.0% (<.00001)A | 84.2% (<.00001) | 92.0% (<.00001) |
With Bonferroni correction, P<.0083.
For Group 1 populations Tu, Fe, KN, KC, KW, Mo, In, Lo, Ka, LN, LS, and KS; Group 2 population SR; and Group 3 population LR.
For Group 1 populations Lo and KN; Group 2 populations Tu and Fe; and Group 3 populations KS and KW.
For Group 1 populations Mo, In, LN, and LS; Group 2 populations Tu and SR; Group 3 populations KN and KS; and Group 4 population Lo.
For Group 1 populations Tu, Fe, KS, and KC; Group 2 populations Mo, In, Ka, LN, and LS; Group 3 populations Lo, SR, KN, and KW; and Group 4 population LR.
For Group 1 populations Tu, KN, KC, and KW; Group 2 population Fe; Group 3 populations KS, Lo, and Ka; Group 4 populations Mo, In, LN, and LS; Group 5 population LR; and Group 6 population SR.
For Group 1 populations Tu, Fe, KN, KW, and KC; Group 2 populations Lo, Ka, KS, LN, LS, Mo, and In; and Group 3 populations SR and LR.
Intergroup differences accounted for a significant proportion of the variance only when language and the history of migrations were used for classification of Romani populations. In the language-based classification, the comparisons included speakers of (a) Balkan dialects of Romanes, (b) Vlax dialects (Old as well as New Vlax), (c) Romanian, and (d) the languages of the surrounding majority populations. The major difference between these two groupings was related to the Lingurari, Monteni, and Intreni; they formed the group of Romanian speakers in the language classification, whereas, in the classification based on migrational history, they were placed together with the speakers of Vlax Romanes dialects. The language division resulted in significant intergroup differences for the female lineages only. Highly significant intergroup differences for both paternal and maternal lineages were observed only when classification was based on the history of migrations, comparing the old settlers in the Balkans to the migrants to Wallachia and Moldavia and to those moving to northern and western Europe. This comparison showed that ∼10% of the variance for Y chromosome and 5% for mtDNA (P<.00001 for both) was due to differences between the migrational groups.
Discussion
The Roma do not have their own written history; therefore, theories about their origins and migrations are based on legends or on linguistics and cultural anthropology. Early European historical records refer to the Roma as Egyptians, and the term “Gypsy” is thought to reflect that assumption (Fraser 1992). Another popular legend is derived from an 11th-century chronicle by a Persian historian, describing a group of 10,000–12,000 musicians and entertainers given as a gift to the ruler of Persia, Shah Bahram Gur, by an Indian Maharaja, during the 5th century (Fraser 1992). The theory of the Indian origins of the Roma (reviewed in Fraser 1992) is based on the similarities between Romanes and languages of the Indian subcontinent. However, the lack of close relationship with any specific living language or dialect in India has given rise to the concept of Romanes resulting from the “mixing of linguistic subsystems in the context of increased interaction among speakers of these varieties” (Hancock 2000, p. 2). This linguistic theory has been linked to the historical period of the Islamic invasions of India and proposes that the Roma derive from the ethnically diverse martial society of the Rajputs, as well as from camp followers drawn from the lowest Varna and the out-caste or untouchable groups (Hancock 2000). The argument of diverse origins rooted in India is supported by the social organization of the Roma, whose multiple endogamous populations with professional ethnonyms bear close resemblance to the jatis of India (Fraser 1992; Marushiakova and Popov 1997). The endogamous professional-group organization could thus have been an inherent social characteristic of the proto-Roma at the time of the exodus from India. It is also conceivable that the fragmentation into small populations has occurred, within Europe, as a means of higher mobility—and, thus, survival in the face of repressive legislation and persecution (Hancock 1987; Fraser 1992; Liégeois 1994)—and has been consolidated further by geographic dispersal and cultural and linguistic diversification. These scenarios could have a different impact on present genetic structure, with implications for genetic research, especially into complex disorders.
This study has demonstrated the sharing of identical Asian-specific paternal and maternal lineages between all Romani populations. Nearly 45% of Y chromosomes belong to haplogroup VI-68, and a single lineage within that haplogroup, found across Romani populations, accounts for almost one-third of Romani males. A similar preservation of a highly resolved male lineage has been reported elsewhere only for Jewish priests (Thomas et al. 1998). Similarly, Asian-specific mtDNA haplogroup M is found in 13 of 14 Romani populations and accounts for 26.5% of maternal lineages in the Roma. The data provide strong evidence of Asian origins, in contrast with claims that the Roma are a socially defined population of European descent (Okely 1983; Wexler 1997).
Analysis of diversity within haplogroups VI-68 and M provides an insight into the genetic composition of the ancestral population. The Y-chromosome haplogroup VI-68 Y STR haplotypes are closely related, suggesting recent diversification by mutational processes, and cluster as a subset of the overall diversity of Asian haplogroup VI-68. Detailed comparisons between the diversity in the Romani VI-68 lineage and that in the Asian haplogroup VI-68 will become possible when more information about male lineages in the Indian subcontinent becomes available. Most mtDNA haplogroup M lineages belong to subhaplogroup M5 (Bamshad et al. 2001) and form a small subset of the diversity within Indian haplogroup M. Again, close genealogical relationship suggests that diversity has arisen by mutation rather than by diverse origins or admixture. The relatively recent ages determined for haplogroup VI-68 and M in this study suggest that the ethnogenesis of the Roma can be understood as a profound bottleneck event. Although identification of the parental population of the proto-Roma has to await better understanding of genetic diversity in the Indian subcontinent, our results suggest a limited number of related founders, compatible with a small group of migrants splitting from a distinct caste or tribal group.
The present findings and the published data on global diversity do not allow a distinction between additional founding lineages and early admixture for Y-chromosome haplogroup VI-56 and the less common haplogroups, shown to occur in Asia and the Middle East (Underhill et al. 2000, 2001), and for mtDNA haplogroups H and X, widely distributed from Europe to India (Kivisild et al. 1999; Simoni et al. 2000; Richards et al. 2000). Both the close relationship between haplotypes within haplogroup VI-56 and its frequency distribution among the Roma point to introduction by a small number of related males. The fact that the common Romani mtDNA haplogroup H and X lineages have not been found among a large number of Middle Eastern and European individuals (Richards et al. 2000) suggests that they might be founding lineages of Indian origin. Regardless of the history of these lineages, the observed pattern points to greater female diversity in the early Romani population, compared with the male component.
Although the sharing of genetic lineages supports the common origins of the Roma, differentiation between Romani populations is evidenced by the distribution of male and female lineages (fig. 3). The results of the AMOVA and PC analysis provide an insight into the contribution that different factors make to the shaping of the genetic structure of Romani populations. The irrelevance of geographic criteria for studying the Roma has been emphasized repeatedly by cultural anthropologists (Petulengro 1915–16; Fraser 1992; Liégeois 1994; Marushiakova and Popov 1997), yet country of residence has been used consistently as the descriptor in genetic studies of the Roma (reviewed by Kalaydjieva et al. [2001b]). Our present results indicate that geography has no relevance to genetic structure, even when Romani populations living in close proximity in the same small town are considered. This is in contrast to the findings for other European populations, in which geographic distance (rather than culture and language) has been found to play the major role (Rosser et al. 2000). The lack of genetic correlation with recently acquired religions (Muslim or Christian) is hardly surprising. Interestingly, traditional trade reflected in the ethnonym, an important factor in defining self-identity of Romani populations, was found to be a poor grouping criterion. By far the most significant differences between groups of populations were observed when language and especially history of migrations were used as the classification criteria in the AMOVA comparisons. These two indicators are closely related, since the classification of Romanes dialects is based mainly on external linguistic influences and borrowings. The significant difference between language groups, for female (but not male) lineages, possibly reflects the strict endogamy rules practiced by the Romanian-speaking Roma toward females from other populations. Strong support for the migrational grouping of populations was provided also by the results of the PC analysis.
The European migrations of the Roma have followed three major streams. Whereas the majority settled within the Balkan provinces of the Ottoman Empire, some headed to the autonomous principalities of Wallachia and Moldavia, north of the Danube (in present-day Romania), and others continued the journey north and west. Ottoman tax registries suggest that the number of Roma initially settling in the Empire would have been small (Marushiakova and Popov 1997), and early historical records from Western Europe invariably describe Gypsies arriving as a group of 50–300 individuals led by an elder (Colocci 1889). The early-settled Romani population south of the Danube and the superimposed migrations, from Wallachia and Moldavia, of small groups of runaway slaves during the 17th and 18th centuries and of larger numbers after the abolition of Gypsy slavery during the 19th century (Marushiakova and Popov 2001b) have spawned >50 socially diverse Romani populations in Bulgaria alone (Marushiakova and Popov 1997). Our data indicate that current genetic structure results mainly from the early splits and divergent routes within Europe. Two processes, genetic drift and different levels and sources of admixture, appear to have played a role in the subsequent differentiation of populations. The effects of differential admixture are illustrated by the distribution of Y-chromosome haplogroups VI-52 and IX-104, whose occurrence among the Roma reflects the reported clinal distribution in Europe (Semino et al. 2000). Intrahaplogroup diversities in the Roma are consistent with multiple independent admixture events. Similar examples are provided by mtDNA haplogroups H (excluding the two common lineages), X, T, and U5. The effects of drift are likely to account for the different frequencies of the major common lineages in the diverse Romani populations (fig. 3), such as the uneven representation of Y-chromosome haplogroup VI-56 and mtDNA haplogroup U3, both of which occur in multiple Romani populations.
Application of the knowledge of the origins and diversification of the Roma should prove useful in the design of future medical genetic studies. Our results are in need of further confirmation through the study of larger sample sizes, with wider representation of western-European Roma and of populations speaking the two major varieties of Balkan dialects of Romanes. One should also note that current genetic data may not mirror accurately the original composition of the migrant proto-Romani population; the profound effect of genetic drift due to small population size would have been complemented by the history of violent persecution of the Roma in Europe, culminating in the death camps of the Second World War (Fings et al. 1997). Nonetheless, the findings point to an interesting difference in the biological and cultural history of the Roma. Whereas genetic differentiation appears to carry the imprint of the early European history of the Roma, social diversification seems to be the product of a recent restitution of the traditions of the ancient country of origin.
Acknowledgments
Funding for this project was provided by the Australian Research Council and The Wellcome Trust. We thank L. L. Cavalli-Sforza for support of this project, S. Qasim Mehdi for the DNA samples from Asian males, and P. de Knijff for providing Y STR allelic ladders.
Electronic-Database Information
URLs for data in this article are as follows:
- ARLEQUIN, http://lgb.unige.ch/arlequin/
- Forensic Laboratory for DNA Research, http://www.medfac.leidenuniv.nl/fldo/ (for Y STR genotyping information)
- Life Sciences and Engineering Technology Solutions, http://www.fluxus-engineering.com/ (for Network 3.0 software)
References
- Abicht A, Stucka R, Karcagi V, Herczegfalvi A, Horvath R, Mortier W, Schara U, Ramaekers V, Jost W, Brunner J, Janssen G, Seidel U, Schlotter B, Muller-Felber W, Pongratz D, Rudel R, Lochmuller H (1999) A common mutation (epsilon1267delG) in congenital myasthenic patients of Gypsy ethnic origin. Neurology 53:1564–1569 [DOI] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Angelicheva D, Turnev I, Dye D, Chandler D, Thomas PK, Kalaydjieva L (1999) Congenital cataracts facial dysmorphism neuropathy (CCFDN) syndrome: a novel developmental disorder in Gypsies maps to 18qter. Eur J Hum Genet 7:560–566 [DOI] [PubMed] [Google Scholar]
- Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BV, Reddy PG, Rasanayagam A, Papiha SS, Villems R, Redd AJ, Hammer MF, Nguyen SV, Carroll ML, Batzer MA, Jorde LB (2001) Genetic evidence on the origins of indian caste populations. Genome Res 11:994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray MS, Boerwinkle E, Doris PA (2001) High-throughput multiplex SNP genotyping with MALDI-TOF mass spectrometry: practice, problems and promise. Hum Mutat 17:296–304 [DOI] [PubMed] [Google Scholar]
- Calafell F, Underhill P, Tolun A, Angelicheva D, Kalaydjieva L (1996) From Asia to Europe: mitochondrial DNA sequence variability in Bulgarians and Turks. Ann Hum Genet 60:35–49 [DOI] [PubMed] [Google Scholar]
- Colocci A (1889) Gli zingara: storia di un popolo errante. Torino [Google Scholar]
- de Knijff P (2000) Messages through bottlenecks: on the combined use of slow and fast evolving polymorphic markers on the human Y chromosome. Am J Hum Genet 67:1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fings K, Heuss H, Sparing F (1997) From “race science” to the camps: the Gypsies during the Second Wold War. University of Hertfordshire Press, Hatfield, England [Google Scholar]
- Fraser A (1992) The Gypsies. Blackwell Publishers, Oxford [Google Scholar]
- Hancock I (1987) The Pariah syndrome. Karoma Publishers, Ann Arbor [Google Scholar]
- ——— (2000) The emergence of Romani as a koine outside of India. In: Acton T (ed) Scholarship and Gypsy struggle: commitment in Romani studies: essays in honour of Donald Kenrick on the occasion of his seventieth birthday. University of Hertfordshire Press, Hatfield, England, pp 1–13 [Google Scholar]
- Hammer MF, Horai S (1995) Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 56:951–962 [PMC free article] [PubMed] [Google Scholar]
- Heyer E, Puymirat J, Dieltjes P, Bakker E, de Knijff P (1997) Estimating Y chromosome specific microsatellite mutation frequencies using deep rooting pedigrees. Hum Mol Genet 6:799–803 [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Calafell F, Jobling MA, Angelicheva D, de Knijff P, Rosser ZH, Hurles ME, Underhill P, Tournev I, Marushiakova E, Popov V (2001a) Patterns of inter- and intra-group genetic diversity in the Vlax Roma as revealed by Y chromosome and mitochondrial DNA lineages. Eur J Hum Genet 9:97–104 [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Gresham D, Calafell F (2001b) Genetic studies of the Roma (Gypsies): a review. BMC Med Genet 2:5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaydjieva L, Gresham D, Gooding R, Heather L, Baas F, de Jonge R, Blechschmidt K, Angelicheva D, Chandler D, Worsley P, Rosenthal A, King RH, Thomas PK (2000) N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy-Lom. Am J Hum Genet 67:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaydjieva L, Hallmayer J, Chandler D, Savov A, Nikolova A, Angelicheva D, King RH, Ishpekova B, Honeyman K, Calafell F, Shmarov A, Petrova J, Turnev I, Hristova A, Moskov M, Stancheva S, Petkova I, Bittles AH, Georgieva V, Middleton L, Thomas PK (1996) Gene mapping in Gypsies identifies a novel demyelinating neuropathy on chromosome 8q24. Nat Genet 14:214–217 [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Perez-Lezaun A, Angelicheva D, Onengut S, Dye D, Bosshard NU, Jordanova A, Savov A, Yanakiev P, Kremensky I, Radeva B, Hallmayer J, Markov A, Nedkova V, Tournev I, Aneva L, Gitzelmann R (1999) A founder mutation in the GK1 gene is responsible for galactokinase deficiency in Roma (Gypsies). Am J Hum Genet 65:1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman B, Takacs K, Gyodi E, Kramer J, Fust G, Tauszik T, Guseo A, Kuntar L, Komoly S, Nagy C, Palffy G, Petranyi GG (1991) Sclerosis multiplex in gypsies. Acta Neurol Scand 84:181–185 [DOI] [PubMed] [Google Scholar]
- Kaneva R, Milanova V, Onchev G, Stoyanova V, Chakarova CH, Nikolova A, Hallmayer J, Belemezova M, Milenska T, Kirov G, Kremensky I, Kalaydjieva L, Jablensky A (1998) A linkage study of affective disorders in two Bulgarian Gypsy families: results for candidate regions on chromosomes 18 and 21. Psychiatr Genet 8:245–249 [DOI] [PubMed] [Google Scholar]
- Kayser M, Caglia A, Corach D, Fretwell N, Gehrig C, Graziosi G, Heidorn F, Herrmann S, Herzog B, Hidding M, Honda K, Jobling M, Krawczak M, Leim K, Meuser S, Meyer E, Oesterreich W, Pandya A, Parson W, Penacino G, Perez-Lezaun A, Piccinini A, Prinz M, Schmitt C, Roewer L (1997) Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med 110:125–133 [DOI] [PubMed] [Google Scholar]
- Kittles RA, Perola M, Peltonen L, Bergen AW, Aragon RA, Virkkunen M, Linnoila M, Goldman D, Long JC (1998) Dual origins of Finns revealed by Y chromosome haplotype variation. Am J Hum Genet 62:1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Bamshad MJ, Kaldma K, Metspalu M, Metspalu E, Reidla M, Laos S, Parik J, Watkins WS, Dixon ME, Papiha SS, Mastana SS, Mir MR, Ferak V, Villems R (1999) Deep common ancestry of indian and western-Eurasian mitochondrial DNA lineages. Curr Biol 9:1331–1334 [DOI] [PubMed] [Google Scholar]
- Liégeois J-P (1994) Roma, Gypsies, Travellers. Council of Europe, Strasbourg [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonne-Tamir B, Sykes B, Torroni A (1999) The emerging tree of west Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 64:232–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marushiakova E, Popov V (1997) Gypsies (Roma) in Bulgaria. Peter Lang, Frankfurt am Main [Google Scholar]
- ——— (2001a) Bulgaria: ethnic diversity—a common struggle for equality. In: Guy W (ed) Between past and future: the Roma of Central and Eastern Europe. University of Hertfordshire Press, Hatfield, England, pp 370–388 [Google Scholar]
- ——— (2001b) Gypsies in the Ottoman Empire. University of Hertfordshire Press, Hatfield, England [Google Scholar]
- ——— (2001c) Historical and ethnological background. In: Guy W (ed) Between past and future: the Roma of Central and Eastern Europe. University of Hertfordshire Press, Hatfield, England, pp 33–53 [Google Scholar]
- Meyer S, Weiss G, von Haeseler A (1999) Pattern of nucleotide substitution and rate heterogeneity in the hypervariable regions I and II of human mtDNA. Genetics 152:1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanov I, Kmetski TS, Lyons KE, Koller WC (2000) Prevalence of Parkinson's disease in Bulgarian Gypsies. Neuroepidemiology 19:206–209 [DOI] [PubMed] [Google Scholar]
- Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York [Google Scholar]
- Okely J (1983) The traveller-gypsies. Cambridge University Press, Cambridge [Google Scholar]
- Passarino G, Semino O, Bernini LF, Santachiara-Benerecetti AS (1996) Pre-Caucasoid and Caucasoid genetic features of the Indian population revealed by mtDNA polymorphisms. Am J Hum Genet 59:927–934 [PMC free article] [PubMed] [Google Scholar]
- “Petulengro” (1915–16) Report on the Gypsy tribes of north-east Bulgaria. J Gypsy Lore Soc 9:1–109 [Google Scholar]
- Piccolo F, Jeanpierre M, Leturcq F, Dode C, Azibi K, Toutain A, Merlini L, Jarre L, Navarro C, Krishnamoorthy R, Tome FM, Urtizberea JA, Beckmann JS, Campbell KP, Kaplan JC (1996) A founder mutation in the gamma-sarcoglycan gene of gypsies possibly predating their migration out of India. Hum Mol Genet 5:2019–2022 [DOI] [PubMed] [Google Scholar]
- Plasilova M, Stoilov I, Sarfarazi M, Kadasi L, Ferakova E, Ferak V (1999) Identification of a single ancestral CYP1B1 mutation in Slovak Gypsies (Roms) affected with primary congenital glaucoma. J Med Genet 36:290–294 [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS (1999) Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23:437–441 [DOI] [PubMed] [Google Scholar]
- Reyniers A (1995) Gypsy populations and their movements within central and eastern Europe and towards some OECD countries. In: International migration and labour market policies: occasional papers, No 1. Organisation for Economic Co-operation and Development, Paris, p 8 [Google Scholar]
- Richards MB, Macaulay VA, Bandelt HJ, Sykes BC (1998) Phylogeography of mitochondrial DNA in western Europe. Ann Hum Genet 62:241–260 [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, et al (2000) Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- Rochow I, Matschke K (1998) Neues zu den zigeunern im Byzantinischen reich um die wende von 13. Zum 14. Jahrhundert. Jahrbuch der Österreichischen Byzantinistik 41:241–254 [Google Scholar]
- Rogers T, Chandler D, Angelicheva D, Thomas PK, Youl B, Tournev I, Gergelcheva V, Kalaydjieva L (2000) A novel locus for autosomal recessive peripheral neuropathy in the EGR2 region on 10q23. Am J Hum Genet 67:664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolf B, Meyer E, Brinkmann B, de Knijff P (1998) Polymorphism at the tetranucleotide repeat locus DYS389 in 10 populations reveals strong geographic clustering. Eur J Hum Genet 6:583–588 [DOI] [PubMed] [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, et al (2000) Y-chromosomal diversity in Europe is clinal and influenced primarily by geography rather than by language. Am J Hum Genet 67:1526–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt HJ, Norby S (2000) mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 67:718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290:1155–1159 [DOI] [PubMed] [Google Scholar]
- Shen P, Wang F, Underhill PA, Franco C, Yang WH, Roxas A, Sung R, Lin AA, Hyman RW, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ (2000) Population genetic implications from sequence variation in four Y chromosome genes. Proc Natl Acad Sci USA 97:7354–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni L, Calafell F, Pettener D, Bertranpetit J, Barbujani G (2000) Geographic patterns of mtDNA diversity in Europe. Am J Hum Genet 66:262–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M (2000) Hypervariable sites in the mtDNA control region are mutational hotspots. Am J Hum Genet 67:1029–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Skorecki K, Ben-Ami H, Parfitt T, Bradman N, Goldstein DB (1998) Origins of Old Testament priests. Nature 394:138–140 [DOI] [PubMed] [Google Scholar]
- Thomas PK, Kalaydjieva L, Youl B, Rogers T, Angelicheva D, King RHM, Guergueltcheva V, Colomer J, Lupu C, Corches A, Popa G, Merlini L, Shmarov A, Nourallah M, Muddle JR, Tournev I. Hereditary motor and sensory neuropathy Russe (HMSN-R): new autosomal recessive neuropathy in Balkan gypsies. Ann Neurol 50:452–457 [DOI] [PubMed] [Google Scholar]
- Tournev I, King RH, Workman J, Nourallah M, Muddle JR, Kalaydjieva L, Romanski K, Thomas PK (1999) Peripheral nerve abnormalities in the congenital cataracts facial dysmorphism neuropathy (CCFDN) syndrome. Acta Neuropathol 98:165–170 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ (1997) Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 7:996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oefner PJ, Cavalli-Sforza LL (2001) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 [DOI] [PubMed] [Google Scholar]
- Wexler P (1997) Could there be a Rotwelsch origin for the Romani lexicon? Paper presented at Third International Conference on Romani Linguistics, Prague, December 1996 [Google Scholar]