Abstract
Forty-seven mtDNAs collected in the Dominican Republic and belonging to the African-specific haplogroup L2 were studied by high-resolution RFLP and control-region sequence analyses. Four sets of diagnostic markers that subdivide L2 into four clades (L2a–L2d) were identified, and a survey of published African data sets appears to indicate that these clades encompass all L2 mtDNAs and harbor very different geographic/ethnic distributions. One mtDNA from each of the four clades was completely sequenced by means of a new sequencing protocol that minimizes time and expense. The phylogeny of the L2 complete sequences showed that the two mtDNAs from L2b and L2d seem disproportionately derived, compared with those from L2a and L2c. This result is not consistent with a simple model of neutral evolution with a uniform molecular clock. The pattern of nonsynonymous versus synonymous substitutions hints at a role for selection in the evolution of human mtDNA. Regardless of whether selection is shaping the evolution of modern human mtDNAs, the population screening of L2 mtDNAs for the mutations identified by our complete sequence study should allow the identification of marker motifs of younger age with more restricted geographic distributions, thus providing new clues about African prehistory and the origin and relationships of African ethnic groups.
Introduction
Even though the term “haplogroup” was not coined until later (Torroni et al. 1993), it had already been known from one of the earliest studies of human mtDNA variation (Johnson et al. 1983) that the cluster of lineages now referred to as “haplogroup L2” (Chen et al. 1995) was a well-defined monophyletic haplotype group (type 2 and derivatives). Early RFLP studies employing five or six rare cutter restriction enzymes showed that haplogroup L2 encompasses about one-third of sub-Saharan African mtDNAs (Johnson et al. 1983; Scozzari et al. 1988, 1994; Soodyall and Jenkins 1992, 1993; Graven et al. 1995). Despite its current high frequency and its high estimated coalescence time, which has been calculated as 59,000–78,000 years on the basis of RFLP data (Chen et al. 1995, 2000) and as ∼56,000 years on the basis of hypervariable segment I (HVS-I) data (Watson et al. 1997), haplogroup L2 was not involved in the process of human expansion out of Africa and remained restricted to that continent. Intriguingly, despite these interesting features, the structure and internal sequence variation of this haplogroup have not been analyzed in detail until now.
In the present study, a group of L2 mtDNAs from the Dominican Republic, a country in which the African population component is predominant and heterogeneous in origin, was first studied by high-resolution RFLP and control-region sequence analyses. Subsequently, one mtDNA from each of the four identified clades within L2 was completely sequenced, reaching the highest possible level of molecular resolution. Unexpectedly, we observed that two of the L2 clades are disproportionately derived compared with the other two.
Subjects and Methods
The population sample consisted of 127 unrelated male subjects from the Dominican Republic who were living in Santo Domingo (n=50) and San Juan de la Maguana (n=77). Appropriate informed consent was obtained from all participants, and genomic DNAs were extracted from blood through use of standard procedures.
To determine high-resolution RFLP haplotypes, the entire mtDNA was amplified using PCR in nine overlapping fragments, by the use of the primer pairs described by Torroni et al. (1997). Each of the nine PCR segments was then digested with 14 restriction endonucleases (AluI, AvaII, BamHI, DdeI, HaeII, HaeIII, HhaI, HincII, HinfI, HpaI, MspI, MboI, RsaI, and TaqI). In addition, all mtDNAs were screened for the presence/absence of the BstOI site at nucleotide position (np) 13704, the AccI sites at nps 14465 and 15254, the BfaI site at np 4914, the NlaIII sites at nps 4216 and 4577, the XbaI site at np 7440, the MseI sites at 14766 and 16297, and the MnlI site at np 10871. The polymorphism at np 12308 was also tested through use of a mismatched primer that generates a HinfI site when the A12308G mutation is present (Torroni et al. 1996). The mtDNA control region was sequenced between nps 16003 and 16474, as described elsewhere (Torroni et al. 1999), and included all of HVS-I (nps 16024–16383).
A new protocol has been developed and optimized to obtain complete mtDNA sequences. The entire mtDNA was amplified in 11 overlapping PCR fragments, using a set of primers with matching annealing temperatures (see Results section). After PCR, the fragments were purified using the QIAquick purification kit (QIAGEN), and Cycle Sequencing was performed by application of BigDye Terminator chemistry associated with the enzyme TaqFS, using a set of 32 nested primers specifically designed for this protocol. An ABI 3700 sequencer with 96 capillaries was employed for separation of the sequencing ladders. The sequencing was performed by the Centro Ricerche Interdipartimentale Biotecnologie Innovative (CRIBI) of the University of Padua (BMR–Servizio Sequenziamento di DNA Web site), where further technical details can be obtained. Complete sequences were aligned, assembled, and compared using the program Sequencher 3.0 (Gene Codes). Since the traces were of excellent quality and were unambiguous, it was only necessary to sequence one strand.
Phylogeny construction was performed by hand and was confirmed using Network 2.0e (Bandelt et al. 1995), for the reduced median network, and PAUP* (Swofford 2000), for the most parsimonious tree. The likelihood-ratio test of the molecular clock was performed using TREE-PUZZLE 5.0 (Strimmer and von Haeseler 1996).
Results
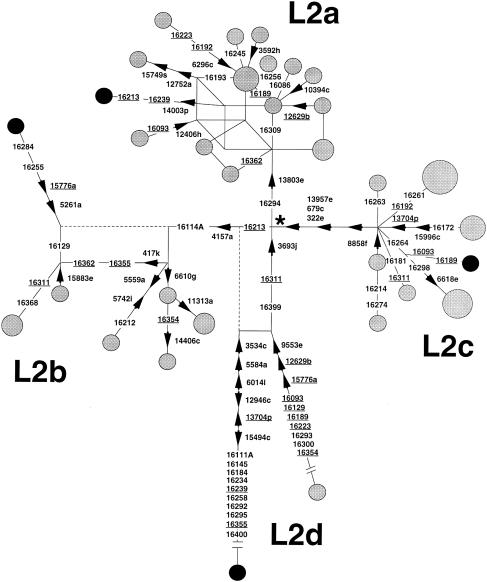
High-resolution RFLP analysis and control-region sequencing revealed that 47 of the 127 Dominican subjects (37%) harbored L2 haplotypes (table 1) and that the remainder belonged to other known African (L1, L3b, L3d, L3e, L3*, and U6), American Indian (A, B, C, and D), and western Eurasian (J and U2) haplogroups (data not shown). As reported elsewhere, L2 mtDNAs are characterized by the RFLP motif +3592 HpaI, +10394 DdeI, −10871 MnlI, +16390 HinfI/−16390 AvaII, and by the HVS-I motif 16223-16278-16390 (Chen et al. 1995, 2000; Watson et al. 1997; Quintana-Murci et al. 1999; Alves-Silva et al. 2000; Pereira et al., in press). However, our survey shows that additional RFLP markers subdivide L2 into four clades that have been termed “L2a,” “L2b,” “L2c,” and “L2d” (table 1). Clades L2a (+13803 HaeIII), L2b (+4157 AluI), and L2c (−322 HaeIII, −679 DdeI, and −13957 HaeIII) were previously identified by Chen et al. (2000), and L2d (−3693 MboI and a transition at np 16399) is described here for the first time. Diagnostic mutations in HVS-I further distinguish the four clades from each other in some cases (table 1 and fig. 1). The clade L2d, although represented by only two subjects in our sample, is by far the most divergent clade within L2 (fig. 1).
Table 1.
RFLP and HVS-I Variation of L2 mtDNAs
|
Haplotypea |
|||
| L2Clade | RFLPb | HVS-Ic | No. of Subjects |
| L2a | +13803e | 223-278-294-390-189-362 | 1 |
| L2a | +13803e | 223-278-294-390-362 | 1 |
| L2a | +13803e | 223-278-294-390-309 | 1 |
| L2a | +13803e | 223-278-294-390-086-309 | 1 |
| L2a | +13803e | 223-278-294-390-189-245-309 | 1 |
| L2a | +13803e | 223-278-294-390-189-309 | 3 |
| L2a | +13803e, [−10394c] | 223-278-294-390-309 | 1 |
| L2a | +13803e, +16517e | 223-278-294-390-256-309 | 1 |
| L2a | +13803e, +16517e | 223-278-294-390-189-309, | 1 |
| L2a | +13803e, +12752a, +15749s, +16517e | 223-278-294-390-189-193-309 | 1 |
| L2a | +13803e, −12629b/+12629j | 223-278-294-390 | 2 |
| L2a | +13803e, −12629b/+12629j, +16517e | 223-278-294-390-309 | 1 |
| L2a | +13803e, +14003p,+16239s, +16517e | 223-278-294-390-193-213-239-309 | 1 |
| L2a | +13803e, −6296c; +16517e | 278-294-390-189-192-309 | 1 |
| L2a | +13803e, −12406h | 223-278-294-390-093-189-193 | 1 |
| L2a | +13803e, [−3592h], +16517e | 223-278-294-390-189-309 | 1 |
| L2b | +4157a, +6610g, +14406c | 114A-129-213-223-278-390-354 | 1 |
| L2b | +4157a, +6610g, +11313a | 114A-129-213-223-278-390 | 2 |
| L2b | +4157a, +6610g | 114A-129-213-223-278-390 | 1 |
| L2b | +4157a, +417kd, −16310k | 114A-129-213-223-278-390-311-355-362-368 | 2 |
| L2b | +4157a, +417kd, −15883e | 114A-129-213-223-278-390-355-362-465 | 1 |
| L2b | +4157a, +417kd, −5261e, −15776a | 114A-213-223-278-390-255-284-355-362 | 1 |
| L2b | +4157a, +5559a, −5742i | 114A-129-213-223-278-390-212 | 1 |
| L2c | −322e, −679c, −13957e | 223-278-390-192-261 | 5 |
| L2c | −322e, −679c, −13957e | 223-278-390-263 | 1 |
| L2c | −322e, −679c, −13957e | 223-278-390-093-189-264 | 1 |
| L2c | −322e, −679c, −13957e, −8858f | 223-278-390-214-274 | 1 |
| L2c | −322e, −679c, −13957e, −8858f, +16517e | 223-278-390 | 1 |
| L2c | −322e, −679c, −13957e, +6618e, −16297s | 223-278-390-264-298 | 4 |
| L2c | −322e, −679c, −13957e, −16310k, +16517e | 223-278-390-181-311 | 1 |
| L2c | −322e, −679c, −13957e, −13704p, −15996c/−16000g | 223-278-390-172 | 3 |
| L2d | −3693j, −3534c/−3537a, −5584a, −6014l, +12946c/+12949n/+12950f, −13704p, +15494c, +16143s, +16239s, −16310k, +16517e, COII-tRNALys 6-bp insertion | 223-278-390-399-111A-145-184-213-234-239-258-292-295-311-355-400 | 1 |
| L2d | −3693j, −9553e, −12629b/+12629j; −15776a, +16296c/−16297s, −16310k, +16398e, +16517e | 278-390-399-093-129-189-293-300-311-354 | 1 |
States diagnostic of each of the L2 clades are underlined.
All L2 mtDNAs harbor the RFLP motif +3592h, +10394c, −10871z, +16389g/−16390b, except for those in which square brackets indicate reverted RFLP sites. Sites are numbered from the first nucleotide of the recognition sequence. A “+” indicates the presence of a restriction site, a “−” the absence. The explicit indication of the presence/absence of a site implies the absence/presence in haplotypes not so designated. The restriction enzymes used in the analysis are designated by the following single-letter codes: a, AluI; b, AvaII; c, DdeI; e, HaeIII; f, HhaI; g, HinfI; h, HpaI; i, MspI; j, MboI; k, RsaI; l, TaqI; m, BamHI; n, HaeII; o, HincII; p, BstOI; q, NlaIII; r, BfaI; s, MseI; z, MnlI. A slash (/) separating states indicates the simultaneous presence or absence of restriction sites that can be correlated with a single-nucleotide substitution.
Only those nucleotide positions (minus 16000) between 16003 and 16474 that differ from the Cambridge Reference Sequence (CRS) (Andrews et al. 1999) are shown. Mutations are transitions, unless the base change is specified explicitly.
Incorrectly mapped as +762k by Chen et al. (1995).
Figure 1.
Unweighted reduced median network (Bandelt et al. 1995) of the 47 L2 samples from the Dominican Republic, showing the four clades L2a–L2d. The circles represent combined high-resolution RFLP and HVS-I haplotypes, with their areas proportional to the frequency. The smallest circles are singletons, whereas the largest have frequency 5. The black circles denote the four mtDNAs (one for each clade) that have been completely sequenced. RFLP mutations are indicated next to the branches, with the arrow pointing in the direction of a site gain. The label is the nucleotide at the beginning of the recognition sequence (in the numbering of the reference sequence of Anderson et al. [1981]); the letter suffix indicates the enzyme (see table 1). HVS-I mutations (between 16003 and 16474) are shown on the branches; they are transitions unless the base change is explicitly indicated. Underlining indicates resolved recurrent mutations, and unresolved events are shown by reticulation. Implausible links are shown with a dotted line. The node marked with an asterisk (*) has the RFLP motif +3592h, +10394c, −10871z, +16390g/−16390b and the HVS-I motif 223-278-390. The lengths of branches in L2a and L2d are shown distorted, for convenience of display. The hypervariable RFLP 16517e was not considered, nor were indel events.
To better define the relationships between the four L2 clades, one mtDNA (denoted by a black circle in fig. 1) from each of the four clades was completely sequenced. For the present analysis, we developed an efficient sequencing strategy that minimizes time and expense. First, the mtDNA was PCR amplified into 11 fragments by means of primer pairs with almost identical melting temperatures (table 2), so that the 11 PCR reactions could be performed simultaneously at the same annealing temperature (55°C) in the same thermocycler. Only 32 nested primers were then employed for the cycle sequencing procedure (table 3).
Table 2.
Oligonucleotides Used to Amplify the Entire Human mtDNA in 11 PCR Fragments[Note]
|
Oligonucleotidea |
||||||
| PCR IDNumber | Fragment Length(bp) | Name | 5′ np | 3′ np | Sequence (5′→3′) | MeltingTemperature(°C) |
| 1 | 1,845 | 14897for | 14897 | 14918 | ctagccatgcactactcaccag | 59.96 |
| 155rev | 155 | 134 | aataggatgaggcaggaatcaa | 59.93 | ||
| 2 | 1,759 | 16488for | 16488 | 16509 | ctgtatccgacatctggttcct | 59.93 |
| 1677rev | 1677 | 1656 | gtttagctcagagcggtcaagt | 60.08 | ||
| 3 | 1,832 | 1404for | 1404 | 1425 | acttaagggtcgaaggtggatt | 60.23 |
| 3235rev | 3235 | 3214 | cttaacaaaccctgttcttggg | 59.90 | ||
| 4 | 1,784 | 2900for | 2900 | 2921 | caataacttgaccaacggaaca | 59.90 |
| 4683rev | 4683 | 4662 | ttagaaggattatggatgcggt | 59.83 | ||
| 5 | 1,771 | 4381for | 4381 | 4402 | acctatcacaccccatcctaaa | 59.59 |
| 6151rev | 6151 | 6130 | actagtcagttgccaaagcctc | 59.95 | ||
| 6 | 1,747 | 5871for | 5871 | 5892 | gcttcactcagccattttacct | 59.79 |
| 7617rev | 7617 | 7596 | tcttgtagacctacttgcgctg | 59.72 | ||
| 7 | 1,980 | 7239for | 7239 | 7260 | gcatacaccacatgaaacatcc | 60.13 |
| 9218rev | 9218 | 9197 | ttggtgggtcattatgtgttgt | 60.02 | ||
| 8 | 1,740 | 8910for | 8910 | 8931 | cttaccacaaggcacacctaca | 60.09 |
| 10649rev | 10649 | 10628 | aggcacaatattggctaagagg | 59.65 | ||
| 9 | 1,769 | 10457for | 10457 | 10478 | tcatatttaccaaatgcccctc | 60.04 |
| 12225rev | 12225 | 12204 | agttcttgtgagctttctcggt | 59.57 | ||
| 10 | 1,816 | 12014for | 12014 | 12035 | ctcacccaccacattaacaaca | 60.70 |
| 13829rev | 13829 | 13808 | agtcctaggaaagtgacagcga | 60.44 | ||
| 11 | 1,873 | 13477for | 13477 | 13498 | gcaggaatacctttcctcacag | 60.13 |
| 15349rev | 15349 | 15328 | gtgcaagaataggaggtggagt | 59.64 | ||
Note.— The annealing temperature for all PCR reactions is 55°C;
nps correspond to the CRS (Anderson et al. 1981). The length of each oligonucleotide was 22 nucleotides.
Table 3.
Oligonucleotides Used for Sequencing the Entire Human mtDNA
|
Sequencing Oligonucleotidea |
||||||
| TemplatePCR IDNumber | Name | Length(nucleotides) | 5′ np | 3′ np | Sequence (5′→3′) | MeltingTemperature(°C) |
| 1 | 14948for | 20 | 14948 | 14967 | cacatcactcgagacgtaaa | 54.92 |
| 1 | 15564for | 20 | 15564 | 15583 | atttcctattcgcctacaca | 54.93 |
| 1 | 131rev | 20 | 131 | 112 | acagatactgcgacataggg | 55.28 |
| 2 | 16522for | 20 | 16522 | 16541 | taaagcctaaatagcccaca | 55.27 |
| 2 | 584for | 20 | 584 | 603 | tagcttacctcctcaaagca | 55.46 |
| 2 | 1060for | 20 | 1060 | 1079 | aagacccaaactgggattag | 55.74 |
| 3 | 1445for | 20 | 1445 | 1464 | gagtgcttagttgaacaggg | 55.02 |
| 3 | 2047for | 20 | 2047 | 2066 | tttaaatttgcccacagaac | 55.39 |
| 3 | 2509for | 20 | 2509 | 2528 | atcacctctagcatcaccag | 55.23 |
| 4 | 3085for | 20 | 3085 | 3104 | atccaggtcggtttctatct | 54.24 |
| 4 | 3598for | 20 | 3598 | 3617 | ctcaacctaggcctcctatt | 55.17 |
| 4 | 4010for | 20 | 4010 | 4029 | acaccctcaccactacaatc | 54.77 |
| 5 | 4410for | 20 | 4410 | 4429 | cagctaaataagctatcggg | 54.58 |
| 5 | 5014for | 20 | 5014 | 5033 | cctcaattacccacatagga | 55.02 |
| 5 | 5544for | 20 | 5544 | 5563 | tcaaagccctcagtaagttg | 55.63 |
| 6 | 6041for | 20 | 6041 | 6060 | ccttctaggtaacgaccaca | 55.33 |
| 6 | 6600for | 20 | 6600 | 6619 | cacctattctgatttttcgg | 54.91 |
| 7 | 7336for | 20 | 7336 | 7355 | cgaagcgaaaagtcctaata | 55.00 |
| 7 | 7937for | 21 | 7937 | 7957 | ttcaactcctacatacttccc | 53.49 |
| 7 | 8459for | 20 | 8459 | 8478 | aactaccacctacctccctc | 54.74 |
| 8 | 8975for | 18 | 8975 | 8992 | tcattcaaccaatagccc | 54.27 |
| 8 | 9589for | 20 | 9589 | 9608 | aagtcccactcctaaacaca | 54.68 |
| 8 | 10147for | 20 | 10147 | 10166 | acatagaaaaatccacccct | 55.09 |
| 9 | 10498for | 22 | 10498 | 10519 | tagcatttaccatctcacttct | 53.48 |
| 9 | 11081for | 20 | 11081 | 11100 | ataacattcacagccacaga | 54.03 |
| 9 | 11644for | 20 | 11644 | 11663 | cctcgtagtaacagccattc | 54.99 |
| 10 | 12114for | 19 | 12114 | 12132 | acatcattaccgggttttc | 54.81 |
| 10 | 12600for | 20 | 12600 | 12619 | attcatccctgtagcattgt | 54.56 |
| 10 | 13134for | 20 | 13134 | 13153 | agcagaaaatagcccactaa | 54.42 |
| 11 | 13568for | 20 | 13568 | 13587 | ttactctcatcgctacctcc | 55.02 |
| 11 | 14103for | 20 | 14103 | 14122 | ctctttcttcttcccactca | 54.61 |
| 11 | 14603for | 20 | 14603 | 14622 | gaaggcttagaagaaaaccc | 54.87 |
nps correspond to the CRS (Anderson et al. 1981).
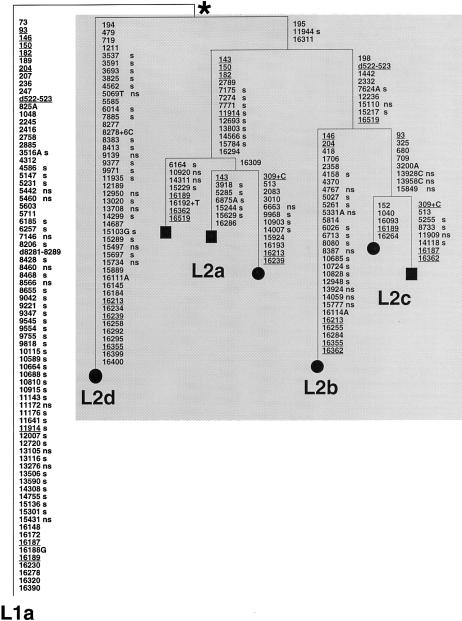
A phylogeny of the four L2 complete sequences is shown in figure 2. Consistent with L2d being the most divergent clade, the tree (rooted using a complete sequence from L1a as an outgroup) shows that L2d branched earliest within haplogroup L2. This first branching was followed by that giving rise to L2a, and L2b and L2c are the most closely related.
Figure 2.
Most-parsimonious reconstruction of the character evolution on a most-parsimonious tree of complete L2 sequences, rooted by use of a complete haplogroup L1a sequence. This tree includes the four L2 mtDNAs sequenced in the course of the present study (blackened circles) and the three L2 complete sequences (blackened squares) previously reported by Ingman et al. (2000). The L1a sequence used as an outgroup, as suggested by the phylogenies of Watson et al. (1997) and Ingman et al. (2000), was also obtained in the course of the present study and is from a Dominican subject. Mutations are shown on the branches; they are transitions, unless the base change is explicitly indicated. Deletions are indicated by a “d” preceding the deleted nucleotides. Insertions are indicated by a plus sign (+) preceding the number and type of inserted nucleotides. Underlining indicates recurrent mutations. “s” indicates synonymous mutations, whereas “ns” indicates nonsynonymous mutations. The asterisk (*) indicates the most recent common ancestor of the L2 mtDNAs in our sample. This differs from the revised CRS (Andrews et al. 1999) by mutations (transitions unless otherwise indicated) at the following positions: 73, 146, 150, 152, 182, 263, 315+C, 750, 769, 1018, 1438, 2416, 2706, 3594, 4104, 4769, 7028, 7256, 7521, 8206, 8701, 8860, 9221, 9540, 10115, 10398, 10873, 11719, 12705, 13590, 13650, 14766, 15301, 15326, 16223, 16278, 16311, 16390, and 16519.
Discussion
The first studies with high-resolution restriction mapping divided global mtDNA variation into a number of major ancient clades, called haplogroups (Wallace 1995; Torroni et al. 1996; Macaulay et al. 1999). In recent years, the dissection of these “old haplogroups” into smaller and younger monophyletic units, characterized by a more restricted geographic/ethnic distribution, has begun. For instance, haplogroups U and M are now subdivided into numerous clades (Kivisild et al. 1999; Macaulay et al. 1999; Richards et al. 2000), and even rather recent haplogroups, such as the European pre-V, have been dissected to identify spatial frequency patterns (Torroni et al. 2001). However, the intrahaplogroup clades identified so far in Eurasian haplogroups do not generally encompass all of the haplogroup members—that is, there is often a “leftover bag” of unclassified mtDNAs within each haplogroup. Our data in table 1 suggest that this situation may not apply to the African haplogroup L2, since all L2 members from a country—the Dominican Republic—that has been populated by Africans of very different ethnic ancestry are classifiable into four well-defined clades. Indeed, a survey of our data and those published elsewhere (Chen et al. 1995, 2000; Mateu et al. 1997; Watson et al. 1997; Rando et al. 1998; Krings et al. 1999; Alves-Silva et al. 2000; Pereira et al., in press; A. Brehm, L. Pereira, H.-J. Bandelt, M. J. Prata, and A. Amorim, unpublished data) suggests that only 2 of 503 L2 mtDNAs do not fit into any of the four clades. These are 2 Biaka L2 mtDNAs, detected in a sample of 17 subjects, which harbored the RFLP motif +1899 HaeIII, −5261 HaeIII (Chen et al. 1995). Unfortunately, these two mtDNAs have apparently not been included among the 17 Biaka (4 belonging to L1a and 13 belonging to L1c) whose control-region sequences have been reported by Vigilant et al. (1991), even though both studies used the Biaka cell lines from L. Cavalli-Sforza’s laboratory as the DNA source. Thus, at the moment, it is not possible to determine whether the two L2 Biaka mtDNAs are members of L2a or L2b that have reverted at the diagnostic RFLP marker, or whether they form an additional very rare L2 clade.
The survey of available L2 HVS-I and RFLP data also suggests that the four L2 clades display different geographic/ethnic distributions. L2a, the most common clade (62% of the total L2), is the only one widespread all over Africa and appears to be subdivided into two major widespread subsets by the 16309 mutation. The derived form at 16309 appears to be more concentrated in western Africa, but distribution studies are hampered by likely reversions at this position. In contrast, L2b appears to be absent in eastern Africans (Watson et al. 1997; Krings et al. 1999) and in Biaka and Mbuti Pygmies (Vigilant et al. 1991; Chen et al. 1995), rare in southern Africans (2.9%) (Vigilant et al. 1991; Chen et al. 2000; Pereira et al., in press), but is common in some Senegalese populations (9.5%) (Chen et al. 1995; Rando et al. 1998). A similar distribution is shown by L2c, which is very common in Senegal (13.5%) (Chen et al. 1995; Rando et al. 1998) and Cabo Verde (16.7%) (A. Brehm, L. Pereira, H.-J. Bandelt, M. J. Prata, and A. Amorim, unpublished data) but is virtually absent in eastern and southern Africans (Watson et al. 1997; Krings et al. 1999; Pereira et al., in press), the Pygmies, and the !Kung (Vigilant et al. 1991; Chen et al. 1995, 2000). The fourth, newly-defined clade, L2d, is rather rare. Including the mtDNAs of two subjects from the Dominican Republic, only 19 L2d mtDNAs can be identified in a total of 503 L2 subjects (3.8%): 7 in Equatorial Guinea, 2 in West Saharans, 3 in the Wolof, 1 in the Mandenka, 1 in Nigeria, 1 in the Lake Chad Kanuri, 1 in southern Sudan, and 1 in Brazil (Chen et al. 1995, 2000; Mateu et al. 1997; Watson et al. 1997; Rando et al. 1998; Krings et al. 1999; Alves-Silva et al. 2000; Pereira et al., in press; A. Brehm, L. Pereira, H.-J. Bandelt, M. J. Prata, and A. Amorim, unpublished data). Seven of these belong to the subset defined by the HVS-I motif 16111A-16145-16184-16239-16292-16355, and the other 12 harbor the distinguishing HVS-I motif 16129-16189-16300-16354. Overall, L2d appears to be mainly restricted to western Africa, like L2b and L2c.
It is worth mentioning that the less common clades L2b and L2d were not sampled in the study by Ingman et al. (2000). This is because their mtDNAs were not preselected on the basis of haplogroup affiliation, and a random sampling obviously tends to miss less-common haplogroups. To provide the widest and most-detailed coverage of the human mtDNA phylogeny, an alternative strategy—namely, selection of mtDNAs on the basis of some haplotype information, ideally both control-region and RFLP data—was pursued here, for one major haplogroup.
The phylogeny in figure 2 is striking in at least one regard: the two subjects from L2b and L2d seem disproportionately derived compared with those from L2a and L2c. This highlights a risk in using a small number of complete sequences to access the divergence time of haplogroups. A small sample of sequences might capture only some of the variation; in this case, perhaps just that of the most common clades, L2a and L2c (see Ingman et al. 2000). In this case, a point estimate of the divergence of L2 would be an underestimate for two reasons: first, the sample would not coalesce on the likely most recent common ancestor of L2 (since it lacks L2d), and, second, the sample would lack the longer branches (in L2b and L2d). Indeed, the average number of mutations (outside of the control region) from the inferred most recent common ancestor of the L2a and L2c sequences in our sample is 14.8, whereas the same statistic evaluated for all seven L2 sequences is 19.4.
This pattern raises the question as to whether the variation at sites outside the control region (neglecting indels) is consistent with a neutral model with a uniform molecular clock. To test this, we evaluated the likelihood of the reconstructed character evolution shown on the tree in figure 2 under two models: one in which a uniform rate was enforced and another where each branch could evolve at its own rate. This calculation was made by coding the mutations inferred in the maximum-parsimony tree as binary characters and by use of a two-state model. Using the likelihood-ratio test, we could reject the uniform clock model at the 5% level (log likelihood L0=-11835.4, for uniform clock; L1=-11842.5, for variable rate model; test statistic 2[L0-L1]=14.4, a value that is exceeded in only 2.6% of cases under the null hypothesis, assuming that the test statistic is distributed as χ2 with 6 df).
Our observation suggests that the mutation process has not been adequately modeled, and this could be for several reasons. First, we may have reconstructed the phylogeny imperfectly—that is, an unfortunate set of recurrent mutations could have distorted the tree topology and the reconstruction of character evolution. This seems unlikely: the L2 sequences are not highly divergent, and we have had to infer only a single recurrent mutation within the coding sequence. In addition, the tree is broadly consistent with the picture that emerges from the variation in the control region, as discussed above. Second, we may not have accounted fully for the stochastic variation in our very small sample of sequences. For instance, another example of L2d may emerge which falls on a shorter branch, more consistent with the variation in L2a and L2c; however, this might in itself be additional evidence of rate variation, since the branches within L2d would then be very different. Only more data can really settle this issue. Third, a succession of founder events and bottlenecks could perhaps generate rather extreme patterns, such as those observed in L2; however, only simulations could test this possibility. Fourth, there may be different selective pressures acting on different lineages. This latter effect might be apparent in the pattern of synonymous and nonsynonymous changes (“s” and “ns” in fig. 2) within protein-coding genes. There do appear to be differences in the proportions of these changes in different parts of L2. L2a appears impoverished in nonsynonymous changes, in comparison with the other parts of L2 and with L2bc in particular (one-tailed Fisher’s exact test for L2a versus the rest of L2: P=.031; this result should be treated with caution, since there is a potential issue concerning multiple comparisons).
This hint of a role for selection in the evolution of human mtDNA follows previous work on its role in the divergence of the mtDNA of humans and chimpanzees (Nachman et al. 1996). It remains to be seen whether stronger evidence can be found in other parts of the human mtDNA phylogeny, in other geographical regions. If so, the challenge of disentangling the effects of the various evolutionary forces that have shaped human mtDNA will be renewed. In any case, it is likely that the screening of members of the L2 clades for the mutations identified by our complete sequence study will identify markers of younger age with more-restricted geographic and ethnic distributions. A detailed analysis of these subclades should provide new clues about African prehistory and the origin and relationships of African populations.
Acknowledgments
This research received support from Telethon-Italy grants E.0890 (to A.T.) and B.57 (to G.V.); Italian Consiglio Nazionale delle Ricerche grant 99.02620.CT04 (to A.T.); Fondo d’Ateneo per la Ricerca 2001 dell'Università di Pavia (to A.T.); Progetto Finalizzato C.N.R. “Beni Culturali” (Cultural Heritage, Italy) (to R.S. and A. C.); Grandi Progetti Ateneo Università di Roma “La Sapienza” (to R.S.); the Italian Ministry of the University, Progetti Ricerca Interesse Nazionale 1999 and 2001 (to A.T., R.S., and A. C.); the “Istituto Pasteur Fondazione Cenci Bolognetti,” Università di Roma “La Sapienza” (to R.S.), and a Research Career Development Fellowship from the Wellcome Trust (to V.M.).
Electronic-Database Information
The URL for data in this article is as follows:
- BMR–Servizio Sequenziamento di DNA, http://bmr.cribi.unipd.it/ (for technical details regarding mtDNA sequencing)
References
- Alves-Silva J, Santos MDS, Guimarães PEM, Ferreira ACS, Bandelt H-J, Pena SDJ, Prado VF (2000) The ancestry of Brazilian mtDNA lineages. Am J Hum Genet 67:444–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organisation of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-S, Olckers A, Schurr TG, Kogelnik AM, Huoponen K, Wallace DC (2000) mtDNA variation in the South African Kung and Khwe—and their genetic relationships to other African populations. Am J Hum Genet 66:1362–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-S, Torroni A, Excoffier L, Santachiara-Benerecetti AS, Wallace DC (1995) Analysis of mtDNA variation in African populations reveals the most ancient of all human continent-specific haplogroups. Am J Hum Genet 57:133–149 [PMC free article] [PubMed] [Google Scholar]
- Graven L, Passarino G, Semino O, Boursot P, Santachiara-Benerecetti S, Langaney A, Excoffier L (1995) Evolutionary correlation between control region sequence and restriction polymorphisms in the mitochondrial genome of a large Senegalese Mandenka sample. Mol Biol Evol 12:334–345 [DOI] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408:708–713 [DOI] [PubMed] [Google Scholar]
- Johnson MJ, Wallace DC, Ferris SD, Rattazzi MC, Cavalli-Sforza LL (1983) Radiation of human mitochondrial DNA types analyzed by restriction endonuclease cleavage patterns. J Mol Evol 19:255–271 [DOI] [PubMed] [Google Scholar]
- Kivisild T, Bamshad M J, Kaldma K, Metspalu M, Metspalu E, Reidla M, Laos S, Parik J, Watkins WS, Dixon ME, Papiha SS, Mastana SS, Mir M R, Ferak V, Villems R (1999) Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages. Curr Biol 9:1331–1334 [DOI] [PubMed] [Google Scholar]
- Krings M, Salem AE, Bauer K, Geisert H, Malek AK, Chaix L, Simon C, Welsby D, Di Rienzo A, Utermann G, Sajantila A, Pääbo S, Stoneking M (1999) mtDNA analysis of Nile River Valley populations: a genetic corridor or a barrier to migration? Am J Hum Genet 64:1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonné-Tamir B, Sykes B, Torroni A (1999) The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 64:232–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateu E, Comas D, Calafell F, Pérez-Lezaun A, Abade A, Bertranpetit J (1997) A tale of two islands: population history and mitochondrial DNA sequence variation of Bioko and São Tomé, Gulf of Guinea. Ann Hum Genet 61:507–518 [DOI] [PubMed] [Google Scholar]
- Nachman MW, Brown WM, Stoneking M, Aquadro CF (1996) Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics 142:953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Macaulay V, Torroni A, Scozzari R, Prata MJ, Amorim A. Prehistoric and historic traces in the mtDNA of Mozambique: insights into the Bantu expansions and the slave trade. Ann Hum Genet (in press) [DOI] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt H-J, Passarino G, McElreavey K, Santachiara-Benerecetti AS (1999) Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23:437–441 [DOI] [PubMed] [Google Scholar]
- Rando JC, Pinto F, González AM, Hernández M, Larruga JM, Cabrera VM, Bandelt H-J (1998) Mitochondrial DNA analysis of northwest African populations reveals genetic exchanges with European, Near-Eastern, and sub-Saharan populations. Ann Hum Genet 62:531–550 [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, et al (2000) Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- Scozzari R, Torroni A, Semino O, Cruciani F, Spedini G, Santachiara Benerecetti AS (1994) Genetic studies in Cameroon: Mitochondrial DNA polymorphisms in Bamileke. Hum Biol 66:1–12 [PubMed] [Google Scholar]
- Scozzari R, Torroni A, Semino O, Sirugo G, Brega A, Santachiara-Benerecetti AS (1988) Genetic studies on the Senegal population. I. Mitochondrial DNA polymorphisms. Am J Hum Genet 43:534–544 [PMC free article] [PubMed] [Google Scholar]
- Soodyall H, Jenkins T (1992) Mitochondrial DNA polymorphisms in Khoisan populations from southern Africa. Ann Hum Genet 56:315–324 [DOI] [PubMed] [Google Scholar]
- ——— (1993) Mitochondrial DNA polymorphisms in Negroid populations from Namibia: new light on the origins of the Dama, Herero and Ambo. Ann Hum Biol 20:477–485 [DOI] [PubMed] [Google Scholar]
- Strimmer K, von Haeseler A (1996) Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol Biol Evol 13:964–969 [Google Scholar]
- Swofford DL (2000) PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, Massachusetts [Google Scholar]
- Torroni A, Bandelt H-J, Macaulay V, Richards M, Cruciani F, Rengo C, Martinez-Cabrera V, et al (2001) A signal, from human mtDNA, of postglacial recolonization in Europe. Am J Hum Genet 69:844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Cruciani F, Rengo C, Sellitto D, López-Bigas N, Rabionet R, Govea N, López de Munain A, Sarduy M, Romero L, Villamar M, del Castillo I, Moreno F, Estivill X, Scozzari R (1999) The A1555G mutation in the 12S rRNA gene of human mtDNA: recurrent origins and founder events in families affected by sensorineural deafness. Am J Hum Genet 65:1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus ML, Wallace DC (1996) Classification of European mtDNAs from an analysis of three European populations. Genetics 144:1835–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Petrozzi M, D'Urbano L, Sellitto D, Zeviani M, Carrara F, Carducci C, Leuzzi V, Carelli V, Barboni P, De Negri A, Scozzari R (1997) Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet 60:1107–1121 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, Vullo CM, Wallace DC (1993) Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet 53:563–590 [PMC free article] [PubMed] [Google Scholar]
- Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC (1991) African populations and the evolution of human mitochondrial DNA. Science 253:1503–1507 [DOI] [PubMed] [Google Scholar]
- Wallace DC (1995) Mitochondrial DNA variation in human evolution, degenerative disease, and aging. Am J Hum Genet 57:201–223 [PMC free article] [PubMed] [Google Scholar]
- Watson E, Forster P, Richards M, Bandelt H-J (1997) Mitochondrial footprints of human expansions in Africa. Am J Hum Genet 61:691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]