Abstract
Catecholamine-induced polymorphic ventricular tachycardia (PVT) is characterized by episodes of syncope, seizures, or sudden death, in response to physical activity or emotional stress. Two modes of inheritance have been described: autosomal dominant and autosomal recessive. Mutations in the ryanodine receptor 2 gene (RYR2), which encodes a cardiac sarcoplasmic reticulum (SR) Ca2+-release channel, were recently shown to cause the autosomal dominant form of the disease. In the present report, we describe a missense mutation in a highly conserved region of the calsequestrin 2 gene (CASQ2) as the potential cause of the autosomal recessive form. The CASQ2 protein serves as the major Ca2+ reservoir within the SR of cardiac myocytes and is part of a protein complex that contains the ryanodine receptor. The mutation, which is in full segregation in seven Bedouin families affected by the disorder, converts a negatively charged aspartic acid into a positively charged histidine, in a highly negatively charged domain, and is likely to exert its deleterious effect by disrupting Ca2+ binding.
Catecholamine-induced polymorphic ventricular tachycardia (PVT [MIM 604772]) was first described as a distinct clinical entity by Leenhardt et al. (1995). The disease is characterized by a reproducible form of ventricular tachycardia, inducible by physical activity or catecholamine infusion (fig. 1), which can rapidly deteriorate into ventricular fibrillation. Patients present with recurrent syncope and/or seizures or sudden death following physical activity or emotional stress (Viskin and Belhassen 1998). The disease, which appears in structurally normal hearts, has a strong genetic basis, and both autosomal dominant and recessive modes of inheritance have been described (Swan et al. 1999; Lahat et al. 2001). In its autosomal dominant form, mutations in the ryanodine receptor 2 gene (RYR2), located on chromosome 1q42-q43, were shown to cause the disease (Laitinen et al. 2001; Priori et al. 2001). RYR2 encodes a cardiac sarcoplasmic reticulum (SR) Ca2+-release channel that couples the excitation of myocardial cells to their actin/myosin contractile apparatus, by a mechanism involving Ca2+ release (Stokes and Wagenknecht 2000). Recently, the spectrum of catecholamine-induced PVT has been expanded, when Tiso et al. (2001) described RYR2 mutations in patients with arrhythmogenic right ventricular cardiomyopathy type 2 (ARVD2).
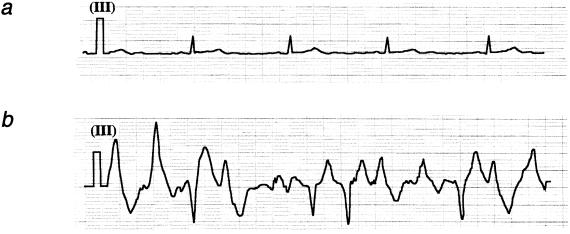
Figure 1.
a, Baseline EKG strip (lead 3) in a patient showing normal sinus rhythm. b, EKG strip (lead 3) in the same patient during an exercise test, showing sustained PVT.
We have previously described an autosomal recessive form of catecholamine-induced PVT in seven families from a Bedouin tribe in the north of Israel (Lahat et al. 2001). Affected family members are characterized by an early age at onset (mean±SD = 7±4 years), a penetrance of 100% by age 10 years, and a high mortality rate when left untreated. The parents and carrier siblings do not show any disease manifestations. Patients with autosomal recessive PVT exhibit a resting bradycardia and a mild prolongation of the QTc segment compared with their unaffected siblings; however, they can be clearly distinguished from patients with long QT syndrome on the basis of the clinical presentation, the mode of inheritance, and the length of the QTc segment, which is still within normal limits (Viskin and Belhassen 1998). Patients with the autosomal recessive disease apparently have a more malignant course, compared with those with the autosomal dominant form, and they do not show any pathological or clinical features of ARVD. (Swan et al. 1999; Lahat et al. 2001). We have previously mapped the disease-causing gene to chromosome 1p13-p21, between the markers D1S187 and D1S534, and have found a common haplotype in all of the carrier chromosomes, suggesting that a single mutation is segregating in these families (Lahat et al. 2001).
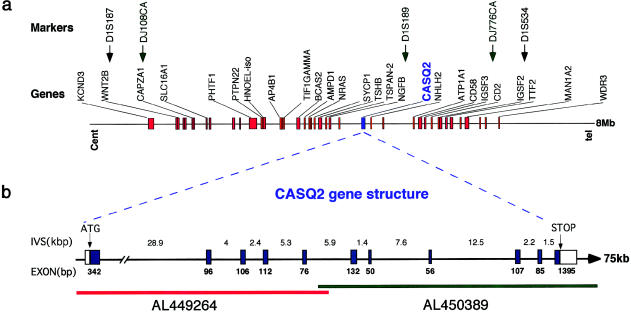
Through electronic screening of DNA clones from the linkage interval, we have identified two microsatellite CA repeats, DJ108CA (clone RP11-114K10) and DJ776CA (clone AL139345), amplified with the primer pairs 5′-GTTCAGGGACACACTGCTCCTC-3′ and 5′-CAAAGTCCTTGAATTCTTACAC-3′, and 5′-CAATAACTTGTATTGTATAATG-3′ and 5′-GATATTTTGATATCCATATAC-3′, respectively. Recombination and haplotype analysis with these markers refined the linkage interval to 8 Mb (fig. 2a). Currently, 46 known transcripts map to this interval (Human Genome Working Draft Web site; see the Weizman Institute of Science Crown Human Genome Center Web site for a list of genes).
Figure 2.
a, Physical map of the PVT locus. Landmark genes are shown as red boxes with their HUGO nomenclature symbols. Microsatellite markers are indicated by arrows (↓) pointing to their exact position on the genomic bar map. Black denotes the boundaries of the linkage interval defined elsewhere (Lahat et al. 2001), and green indicates markers within the linkage interval defined in the present study. b, Expanded view of a 75-kb segment, showing the genomic organization of the CASQ2 interval. The interval is covered by two genomic clones that have 5-kb overlap. AL449264 contains the first four exons, and AL450389 contains the other seven exons, depicted by blue boxes. The lengths of the exons are shown below the boxes, and those of the introns are shown above the line. The translation-initiation site and the stop codon are indicated.
The recent implication of RYR2 in the autosomal dominant form of catecholamine-induced PVT (Laitinen et al. 2001; Priori et al. 2001) focused our attention on another gene involved in the same pathway of SR calcium control, located in the linkage interval calsequestrin 2 (CASQ2). CASQ2 (MIM 114251) is composed of 11 exons and encodes a 399–amino acid protein. This gene is known to be the only calsequestrin expressed in cardiac muscle, whereas skeletal muscle expresses CASQ2 as well as its homologue (91% identity), calsequestrin 1 (CASQ1 [MIM 114250]) (Yano and Zarain-Herzberg 1994). CASQ2 was previously mapped, by fluorescence in situ hybridization, to human chromosome 1p11-p13.3 (Otsu et al. 1993), but its accurate genomic disposition remained unknown. Searching the most recent version of genomic databases, we were able to identify two genomic clones that contained all 11 exons that constitute the CASQ2 cDNA (fig. 2b) (Ensembl Genome Server).
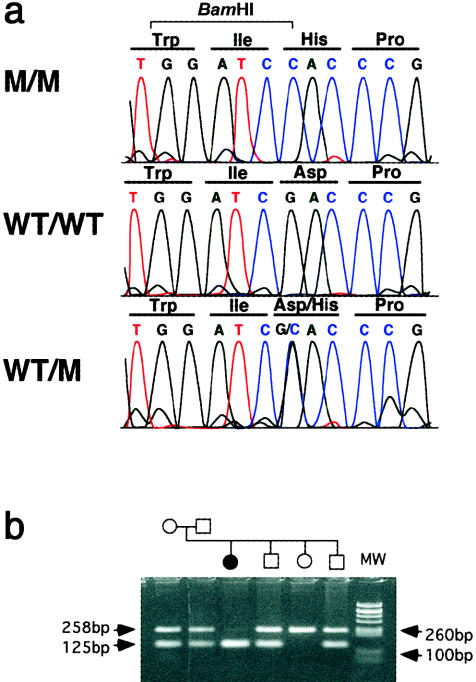
Sequencing of the CASQ2 exons revealed a G→C nonsynonymous substitution (fig. 3a) at nucleotide 1038 (exon 9), resulting in a change from aspartic acid to histidine at position 307 of the protein. No other sequence variations were noted in the coding region of this gene. The D307H mutation creates a BamHI restriction site in the mutated sequence. A full segregation of the mutation was observed in all of the families, and an example of one family is shown in figure 3b. Screening of 350 control subjects, including 250 Jewish, 50 Bedouin, and 50 Israeli Arabs, identified only the wild-type form. Alignment of sequences of two cardiac and three muscle calsequestrin genes from different vertebrates, as well as two calsequestrin genes from invertebrates, shows that the sequence variation that we have found occurs in a highly conserved amino acid, both in vertebrates and in invertebrates (fig. 4a).
Figure 3.
a, Sequence chromatograms from a patient (M/M), a noncarrier (WT/WT), and a carrier (WT/M). There is a G→C substitution at nucleotide 1038 in exon 9, converting aspartic acid to histidine at codon 307. b, BamHI restriction analysis of the D307H mutation in one of the Bedouin families. The 258-bp PCR product of exon 9 is cleaved in the carrier chromosome to yield 131-bp and 127-bp products that comigrate on the agarose gel. MW = molecular weight marker.
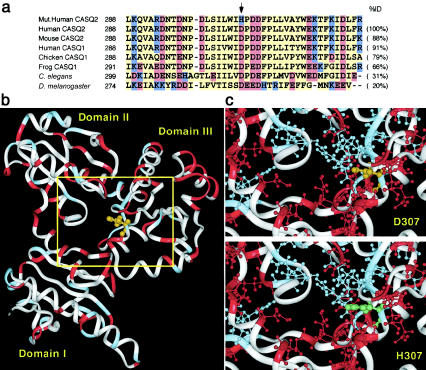
Figure 4.
a, Multiple alignment of a segment of the human CASQ2 protein with the corresponding segments in some of its homologues. The sequences were selected using a BLASTP (Altschul et al. 1997; NCBI BLAST Home Page) search of human CASQ2 against the nonredundant protein database. Multiple alignment of the entire protein length was performed by the ClustalW program, using the default parameters (Higgins et al. 1996). Acidic amino acids are shown in red, basic amino acids in blue, and yellow denotes all other amino acids. The top sequence is for the mutated human CASQ2, and the arrow (↓) indicates the D307H mutated position. The GenBank accession numbers of the aligned proteins are as follows: human cardiac calsequestrin (CASQ2), O14958; mouse cardiac calsequestrin (CASQ2), O09161; human skeletal muscle calsequestrin (CASQ1), P31415; chicken skeletal muscle calsequestrin (CASQ1), P19204; frog skeletal muscle calsequestrin (CASQ1), P31231; Caenorhabditis elegans calsequestrin, Q20203; and Drosophila melanogaster disulfide isomerase, gi|1709616. This is the Drosophila protein showing highest protein similarity to human CASQ2, and both share a thiorodoxin-like structural motif. b, Ribbon drawing of human CASQ2 three-dimensional homology model. The D307 amino acid is shown in orange. The model was calculated using “homology” and “discovery” modules of Biosym/MSI. The protein termini were omitted for the modeling calculation, leaving a G22-to-L366 amino acid core. The default parameters were used to generate a loop (E345 to P353). Energy minimization was performed using steepest-descent (100 iterations) and conjugate-gradient (1,000 iterations) algorithms. c, A closer look at the mutant position domain. The basic and acidic amino acids are shown, in blue and red, respectively, as “balls and sticks.” The top model is of the wild-type protein (D307), and the bottom model is of the mutant protein (H307). The substitute position is shown in orange (for D307) and green (for H307). The histidine in the mutant form appears to disrupt a band of negative amino acids and could thus upset the protein’s charge balance.
The calsequestrin 2 protein, which serves as the major Ca2+ reservoir within the SR of cardiac myocytes, has an ability to bind extremely large numbers of Ca2+ cations. The rabbit CASQ1 can bind and release 40–50 Ca2+ ions per protein molecule at each contraction-relaxation cycle (Yano and Zarain-Herzberg 1994). This is explained by the existence of >60 highly conserved anionic Asp and Glu residues in a protein of ∼400 amino acids (Yano and Zarain-Herzberg 1994). Figure 4b shows a three-dimensional model of human CASQ2 that we have constructed using rabbit skeletal muscle calsequestrin structure as a template (Wang et al. 1998). The predicted structure of CASQ2, like that determined experimentally for CASQ1, is composed of three thioredoxin-like domains (MacLennan and Reithmeier 1998; Wang et al. 1998), which enclose an interdomain space with a highly hydrophilic surface lining and a strong net negative charge. This acidic core has been proposed to be stabilized by binding of Ca2+ cations (Wang et al. 1998) and is likely to play an important role in the chelation and sequestration of Ca2+ ions. Residue 307, which harbors the mutation, protrudes into the interdomain space (fig. 4c), and its undergoing a change from a negatively charged aspartic acid to a positively charged histidine may disrupt the normal chelation function.
In cardiac excitation-contraction coupling, the SR plays an essential role in the regulation of the cytosolic free Ca2+ concentrations. Three major stages have been identified: (1) Ca2+ uptake from the cytosol into the SR lumen, resulting in muscle relaxation; (2) Ca2+ storage in the SR lumen; and (3) Ca2+ release from the SR into the cytosol, resulting in muscle contraction (Carafoli 1987). Stimulation of voltage-sensitive L-type calcium channels (dihydropyridine) located in the sarcolemma permits the entry of extra small amounts of Ca2+, which activate the ryanodine receptors. This leads to the release of large amounts of Ca2+ from calsequestrin into the cytosol, initiating contractility (Puglisi et al. 1996). Two additional proteins involved in the Ca2+ cascade release, junctin and triadin, interact directly in the junctional SR membrane and stabilize a quaternary protein complex that anchors calsequestrin to the ryanodine receptor and that may be required for normal operation of Ca2+ release (Zhang et al. 1997). Studies of isolated human myocardial preparations suggested that impaired Ca2+ handling by the SR is an important subcellular mechanism contributing to the depressed contractility in heart failure (Hasenfuss et al. 1997). In these studies, CASQ2-expression levels were unaltered, suggesting specific and rigid regulation of this protein in cardiac tissue.
Recently, transgenic mice were formed that exhibited a 10-fold overexpression of CASQ2. These mice developed severe cardiac hypertrophy and heart failure, with a twofold increase in heart mass and cell size, in addition to a resting bradycardia, a prolonged electrocardiographic QTc interval, and an increased incidence of sudden death attributed to arrhythmia (Jones et al. 1998; Knollmann et al. 2000). The last three features are similar to those found in our patients, whereas cardiac hypertrophy and heart failure are absent. Electrophysiological studies of these mice suggested that at least some of the pathological manifestations are caused by an impairment in the Ca2+-release process.
The detailed mechanism whereby the D307H mutation induces PVT remains to be elucidated. Calcium release from the SR is regulated by at least three factors: the magnitude of the triggering Ca2+ influx, the state of the ryanodine receptors, and the Ca2+ content within the SR (Volders et al. 2000). Increased intracellular Ca2+ (“Ca2+ overload”) can trigger both early and delayed afterdepolarizations, which are oscillations of the membrane potential that occur during the plateau/repolarization phase of the action potential or after its completion, respectively (Rubart and Zipes 2001). Afterdepolarizations have been implicated as the pathophysiological basis for different clinical arrhythmias, including bidirectional ventricular tachycardia and PVT (Priori and Corr 1990). Thus, these experiments may suggest that an intracellular “Ca2+ overload” is the triggering force initiating the arrhythmias. The mutated CASQ2 may directly increase the Ca2+ content within the SR, alter the function of the ryanodine receptor to which it is connected, or impair the Ca2+ release process in a way analogous to the transgenic mice model. Catecholamines induce intracellular overload of Ca2+ by a variety of mechanisms (Rona 1985; Marban and O'Rourke 1995). In the patients with PVT, catecholamines may further increase intracellular Ca2+ levels and trigger PVT.
In a setting in which all of the carrier chromosomes are derived from a common ancestor, any genetic variant linked to the actual disease-causing gene would be expected to segregate with the disease in all of the studied families. An amino acid substitution like the one we have found can represent a silent polymorphism; however, several lines of evidence presented above implicate the D307H substitution as the cause of catecholamine-induced PVT in our families. First, this amino acid change was not found in any of 700 chromosomes from normal control individuals. Second, this change occurred in an amino acid that is highly conserved throughout evolution. Third, molecular modeling shows that this mutation is associated with a change in the electrostatic properties of the protein that may affect its Ca2+-binding capacity. Fourth, mutations in RYR2, which takes part in the same biochemical pathway as CASQ2 and forms a protein complex with it, causes a phenotype very similar to the phenotype presented in this study. Future identification of more mutations in CASQ2 in other families with this disorder, as well as the creation of a mouse knock-in model, will provide additional evidence that this is indeed the disease-causing gene.
Acknowledgments
We thank Tangiz Bahan, Ph.D. for his assistance. This study was supported by Israel Science Foundation grant 385/00-1 (to E.P.), as well as by the Crown Human Genome Center, a Ministry of Science grant to the National Laboratory for Genome Infrastructure, the Alfried Krupp Foundation, the German-Israeli Foundation for Scientific Research and Development, and the Weizmann Institute Glasberg, Levy, Nathan Brunschwig, and Levine Funds (support to D.L.). This work was performed in partial fulfillment of the requirements for a Ph.D. degree of H.L., Sackler Faculty of Medicine, Tel Aviv University, Israel. D.L. holds the Ralph and Lois Silver Chair in Human Genomics.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Ensembl Genome Server, http://www.ensembl.org/perl/geneview?gene=ENSG00000118729 (for CASQ2 location, gene sequence, and primer design)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human cardiac calsequestrin [CASQ2; accession number O14958], mouse cardiac calsequestrin [CASQ2; accession number O09161], human skeletal muscle calsequestrin [CASQ1; accession number P31415], chicken skeletal muscle calsequestrin [CASQ1; accession number P19204], frog skeletal muscle calsequestrin [CASQ1; accession number P31231]; Caenorhabditis elegans calsequestrin [accession number Q20203], and Drosophila melanogaster disulfide isomerase [accession number gi|1709616])
- Human Genome Working Draft, http://genome.ucsc.edu/goldenPath/hgTracks.html (December 2000 freeze; for genes within the linkage interval)
- NCBI BLAST Home Page, http://www.ncbi.nlm.nih.gov/BLAST/ (for CASQ2 homologues and orthologues, as well as genetic markers)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PVT [MIM 604772], CASQ1 [MIM 114250], CASQ2 [MIM 114251])
- Weizmann Institute of Science Crown Human Genome Center, The, http://bioinformatics.weizmann.ac.il/genome_center/PVT/genes.html (for list of 46 genes located in the PVT genomic linkage interval)
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafoli E (1987) Intracellular calcium homeostasis. Annu Rev Biochem 56:395–433 [DOI] [PubMed] [Google Scholar]
- Hasenfuss G, Meyer M, Schillinger W, Preuss M, Pieske B, Just H (1997) Calcium handling proteins in the failing human heart. Basic Res Cardiol 92 Suppl 1:87–93 [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266:383–402 [DOI] [PubMed] [Google Scholar]
- Jones LR, Suzuki YJ, Wang W, Kobayashi YM, Ramesh V, Franzini-Armstrong C, Cleemann L, Morad M (1998) Regulation of Ca2+ signaling in transgenic mouse cardiac myocytes over expressing calsequestrin. J Clin Invest 101:1385–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knollmann BC, Knollmann-Ritschel BE, Weissman NJ, Jones LR, Morad M (2000) Remodeling of ionic currents in hypertrophied and failing hearts of transgenic mice overexpressing calsequestrin. J Physiol 525:483–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahat H, Eldar M, Levy-Nissenbaum E, Bahan T, Friedman E, Khoury A, Lorber A, Kastner DL, Goldman B, Pras E (2001) Autosomal recessive catecholamine- or exercise-induced polymorphic ventricular tachycardia: clinical features and assignment of the disease gene to chromosome 1p13-21. Circulation 103:2822–2827 [DOI] [PubMed] [Google Scholar]
- Laitinen PJ, Brown KM, Piippo K, Swan H, Devaney JM, Brahmbhatt B, Donarum EA, Marino M, Tiso N, Viitasalo M, Toivonen L, Stephan DA, Kontula K (2001) Mutations of the cardiac ryanodine receptor (RyR2) gene in familial polymorphic ventricular tachycardia. Circulation 103:485–490 [DOI] [PubMed] [Google Scholar]
- Leenhardt A, Lucet V, Denjoy I, Grau F, Ngoc DD, Coumel P (1995) Catecholaminergic polymorphic ventricular tachycardia in children: a 7 year follow up of 21 patients. Circulation 91:1512–1519 [DOI] [PubMed] [Google Scholar]
- MacLennan DH, Reithmeier RA (1998) Ion tamers. Nat Struct Biol 5:409–411 [DOI] [PubMed] [Google Scholar]
- Marban E, O’Rourke B (1995) Calcium channels: structure, function, and regulation. In: Zipes DP, Jaliffe J (eds) Cardiac electrophysiology: from cell to bedside. W. B. Saunders, Philadelphia, pp 11–21 [Google Scholar]
- Otsu K, Fujii J, Periasamy M, Difilippantonio M, Uppender M, Ward DC, MacLennan DH (1993) Chromosome mapping of five human cardiac and skeletal muscle sarcoplasmic reticulum protein genes. Genomics 17:507–509 [DOI] [PubMed] [Google Scholar]
- Priori SG, Corr PB (1990) Mechanisms underlying early and delayed afterdepolarizations induced by catecholamines. Am J Physiol 258:H1796–H1805 [DOI] [PubMed] [Google Scholar]
- Priori SG, Napolitano C, Tiso N, Memmi M, Vignati G, Bloise R, Sorrentino VV, Danieli GA (2001) Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation 103:196–200 [DOI] [PubMed] [Google Scholar]
- Puglisi JL, Bassani RA, Bassani JW, Amin JN, Bers DM (1996) Temperature and relative contributions of Ca transport systems in cardiac myocyte relaxation. Am J Physiol 270:H1772–H1778 [DOI] [PubMed] [Google Scholar]
- Rona G (1985) Catecholamine cardiotoxicity. J Mol Cell Cardiol 17:291–306 [DOI] [PubMed] [Google Scholar]
- Rubart M, Zipes DP (2001) Genesis of cardiac arrhythmias: electrophysiological considerations. In: Braunwald E, Zipes DP, Libby P (eds) Heart disease. W. B. Saunders, Philadelphia, pp 680-684 [Google Scholar]
- Stokes DL, Wagenknecht T (2000) Calcium transport across the sarcoplasmic reticulum: structure and function of Ca2+-ATPase and the ryanodine receptor. Eur J Biochem 267:5274–5279 [DOI] [PubMed] [Google Scholar]
- Swan H, Piippo K, Viitasalo M, Heikkila P, Paavonen T, Kainulainen K, Kere J, Keto P, Kontula K, Toivonen L (1999) Arrhythmic disorder mapped to chromosome 1q42-q43 causes malignant polymorphic ventricular tachycardia in structurally normal hearts. J Am Coll Cardiol 34:2035–2042 [DOI] [PubMed] [Google Scholar]
- Tiso N, Stephan DA, Nava A, Bagattin A, Devaney JM, Stanchi F, Larderet G (2001) Identification of mutations in the cardiac ryanodine receptor gene in families affected with arrhythmogenic right ventricular cardiomyopathy type 2 (ARVD2). Hum Mol Genet 10:189–194 [DOI] [PubMed] [Google Scholar]
- Viskin S, Belhassen B (1998) Polymorphic ventricular tachyarrhythmias in the absence of organic heart disease: classification, differential diagnosis, and implication for therapy. Prog Cardiovasc Dis 41:17–34 [DOI] [PubMed] [Google Scholar]
- Volders PGA, Vos MA, Szabo B, Sipido KR, de Groot SHM, Gorgels APM, Wellens HJJ, Lazzara R (2000) Progress in the understanding of cardiac early afterdepolarizations and torsades de pointes: time to revise current concepts. Cardiovasc Res 46:376–392 [DOI] [PubMed] [Google Scholar]
- Wang S, Trumble WR, Liao H, Wesson CR, Dunker AK, Kang CH (1998) Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum. Nat Struct Biol 5:476–483 [DOI] [PubMed] [Google Scholar]
- Yano K, Zarain-Herzberg A (1994) Sarcoplasmic reticulum calsequestrins: structural and functional properties. Mol Cell Biochem 135:61–70 [DOI] [PubMed] [Google Scholar]
- Zhang L, Kelley J, Schmeisser G, Kobayashi YM, Jones LR (1997) Complex formation between junctin, triadin, calsequestrin, and the ryanodine receptor: proteins of the cardiac junctional sarcoplasmic reticulum membrane. J Biol Chem 272:23389–23397 [DOI] [PubMed] [Google Scholar]