Abstract
The Prader-Willi syndrome (PWS)/Angelman syndrome (AS) region, on human chromosome 15q11-q13, exemplifies coordinate control of imprinted gene expression over a large chromosomal domain. Establishment of the paternal state of the region requires the PWS imprinting center (PWS-IC); establishment of the maternal state requires the AS-IC. Cytosine methylation of the PWS-IC, which occurs during oogenesis in mice, occurs only after fertilization in humans, so this modification cannot be the gametic imprint for the PWS/AS region in humans. Here, we demonstrate that the PWS-IC shows parent-specific complementary patterns of H3 lysine 9 (Lys9) and H3 lysine 4 (Lys4) methylation. H3 Lys9 is methylated on the maternal copy of the PWS-IC, and H3 Lys4 is methylated on the paternal copy. We suggest that H3 Lys9 methylation is a candidate maternal gametic imprint for this region, and we show how changes in chromatin packaging during the life cycle of mammals provide a means of erasing such an imprint in the male germline.
The Prader-Willi syndrome (PWS [MIM 176270])/Angelman syndrome (AS [MIM 105830]) region, on human chromosome 15q11-q13, provides a dramatic example of the role that genetic imprinting plays in the pathogenesis of human disease (Nicholls et al. 1998). Deletions of an ∼4-Mb region from the paternal chromosome cause PWS, which is characterized by infantile hypotonia, mild developmental delay, and later-onset hyperphagia and obesity; deletions of the same region from the maternal chromosome 15 homolog cause AS, which is characterized by severe mental retardation, lack of speech, seizures, and easily provoked laughter. The PWS/AS region contains at least seven imprinted genes; five of these genes are expressed exclusively from the paternal chromosome, and loss of the active paternal alleles of these genes—through deletion, uniparental disomy, or imprinting defects—causes PWS. Two genes in this region show tissue-limited maternal-specific expression. One of these genes, UBE3A (MIM 601623), is imprinted only in certain brain regions (Albrecht et al. 1997), and loss of the maternal allele of UBE3A causes AS (Kishino et al. 1997; Matsuura et al. 1997). The PWS/AS region can exist in either of two mutually exclusive epigenetic states, the paternal state and the maternal state. Establishment and maintenance of the paternal state requires a DNA segment in cis, referred to as the “PWS imprinting center” (PWS-IC). Establishment of the maternal state requires a DNA segment ∼30 kb centromeric of the PWS-IC, referred to as the “AS-IC” (Nicholls et al. 1998). The functions that the PWS-IC and AS-IC have in establishing epigenetic states for this region are not known. Although the 4.3-kb PWS-IC, defined as the shortest region of IC-deletion overlap in human patients with PWS, includes the SNRPN (MIM 182279) promoter, a recent report indicates that targeted replacement of a 0.9-kb fragment containing the mouse Snrpn promoter by a 181-bp polylinker/LoxP fragment has no discernible effect on IC function (Bressler et al. 2001).
Although the mechanisms by which the PWS-IC and the AS-IC regulate gene expression over long distances in the PWS/AS region are not known, significant progress has been made in defining epigenetic marks that differ between the maternal and the paternal copies of the region. These marks, including cytosine methylation and histone acetylation, may play roles in establishing or maintaining differential gene expression between the maternal and the paternal alleles of imprinted genes in the region. These differential modifications (summarized in table 1) include hypermethylation of CpG dinucleotides on the maternal (i.e., silent) alleles in the promoter regions of SNRPN, of ZNF127 (MIM 603856), and of NDN (MIM 602117), as well as hypermethylation of CpGs on the paternal allele of SNRPN in intron 7. No differential methylation of the 5′ region of UBE3A has been found in either human lymphocyte DNA or mouse brain DNA (T. Kishino and J. Wagstaff, unpublished data). Hyperacetylation of the N-terminal tails of histones H3 and H4 has recently been reported in the promoter region of the SNRPN paternal (i.e., active) allele (Saitoh and Wada 2000; Fulmer-Smentek and Francke 2001).
Table 1.
Parent-Specific Epigenetic Modifications in the PWS/AS Region[Note]
| Gene/Region | Modification | Hypermodified on | Reference(s) |
| SNRPN promoter (human) | CpG methylation | Maternal chromosome (postfertilization) | Glenn et al. (1996), El-Maarri et al. (2001) |
| H3, H4 acetylation | Paternal chromosome | Saitoh and Wada (2000), Fulmer-Smentek and Francke (2001) | |
| Snrpn promoter (mouse) | CpG methylation | Maternal chromosome (prefertilization) | Shemer et al. (1997) |
| SNRPN intron 7 (human) | CpG methylation | Paternal chromosome | Glenn et al. (1993) |
| Snrpn exons 7–10 (mouse) | CpG methylation | Paternal chromosome (prefertilization) | Shemer et al. (1997) |
| NDN promoter (human) | CpG methylation | Maternal chromosome (postfertilization) | Jay et al. (1997), El-Maarri et al. (2001) |
| ZNF127 promoter (human) | CpG methylation | Maternal chromosome | Jong et al. (1999) |
| D15S63 (human) | CpG methylation | Maternal chromosome | Dittrich et al. (1992) |
Note.— In cases in which modifications have been assayed in gametes, timing of parent-specific modification is indicated either as “postfertilization,” when differential modification is not present in gametes, or as “prefertilization,” when differential modification is present in gametes.
The causal relationships between these epigenetic modifications and imprinted gene expression in 15q11-q13 are not clear. Studies involving treatment of cultured cells with inhibitors of DNA methylation or with inhibitors of histone deacetylases (HDACs) have yielded mixed results. Saitoh and Wada (2000) treated human lymphoblastoid cell lines lacking an active paternal SNRPN allele with the DNA methyltransferase (DNMT) inhibitor 5-azadeoxycytidine (5-aza-dC) and found partial reactivation of the inactive maternal SNRPN allele. Similar partial reactivation by 5-aza-dC was reported by Fulmer-Smentek and Francke (2001). El Kharroubi et al. (2001), on the other hand, found no reactivation of maternal Snrpn in parthenogenetic mouse embryonic fibroblasts treated with 5-aza-dC, under conditions that led to reactivation of other imprinted loci. All of these reports indicated that treatment with the HDAC inhibitor trichostatin A produced no detectable SNRPN reactivation. Shemer et al. (1997) examined Snrpn transcription in E9.5 mouse embryos homozygous or heterozygous for Dnmt1 mutations; −/− embryos showed increased Snrpn transcription compared to +/+ or +/− embryos, although derepression of the maternal allele was not directly demonstrated.
Shemer et al. (1997) showed that methylation of the maternal Snrpn-promoter/intron 1 region and of the paternal exons 7–10 region, occurs during gametogenesis in mice, so the differential methylation patterns observed in somatic tissues can be explained by the different patterns of methylation of the region inherited from oocyte and from sperm. Unexpectedly, El-Maarri et al. (2001) found that the timing of cytosine methylation of the maternal SNRPN-promoter region is quite different in mice and in humans: human oocytes lack methylation of the SNRPN-promoter region, although this region is heavily methylated on the maternal chromosome in human somatic cells.
That cytosine methylation of the PWS-IC—previously thought to be crucial in distinguishing the maternal copy from the paternal copy of the PWS/AS region and in producing differential gene expression on homologous chromosomes—does not appear in humans until after fertilization raises the question: What is the structural difference between the copy of this region inherited from human sperm and that inherited from the oocyte that leads to differential function after fertilization? In principle, this structural difference may be (a) a heritable covalent DNA modification other than cytosine methylation; (b) a DNA-associated protein that remains stably associated with either the maternal or the paternal chromosome from the gamete through somatic cell divisions; or (c) a covalent modification of a DNA-associated protein that is inherited in a parent-specific fashion. We have examined an evolutionarily conserved histone modification—the methylation of H3 on Lys9—that has recently been associated with the formation of stably silenced chromatin regions in Drosophila and in fission yeast (Rea et al. 2000; Bannister et al. 2001; Lachner et al. 2001; Nakayama et al. 2001). Histone modifications, especially acetylation, previously have been shown to mediate effects of a number of transcriptional regulatory proteins—presumably, by changing chromatin structure to increase accessibility to other transcriptional factors (Strahl and Allis 2000). Unlike acetylated histones, which are quite labile, methyl groups attached to histones show a very low level of turnover, making histone methylation a good candidate modification in epigenetic processes such as imprinting (Byvoet et al. 1972). We have also examined the methylation of H3 on Lys4, which has been correlated with transcriptional activity in Tetrahymena (Strahl et al. 1999).
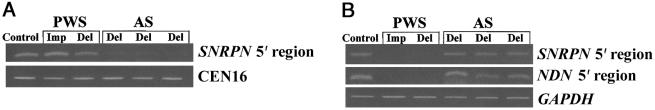
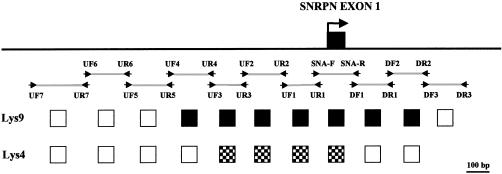
Chromatin prepared from stimulated lymphocytes from controls, from individuals with PWS (lacking a paternal copy of 15q11-q13, because of deletion or imprinting defect), and from individuals with AS (lacking a maternal copy of 15q11-q13) was immunoprecipitated with antibodies specific to either H3 methylated on Lys9 or H3 methylated on Lys4. The patient with a PWS imprinting defect had no detectable deletion of the PWS-IC (authors' unpublished data). DNA recovered from the immunoprecipitations was assayed by PCR for sequences in the PWS-IC, including the SNRPN promoter, and for other sequences in the region, as well as for a control single-copy sequence near the centromere of chromosome 16 (positive control for methyl Lys9 H3 antibody; Horvath et al. 2000) and for GAPDH (positive control for methyl Lys4 H3 antibody). The maternal copy of the SNRPN-promoter region, within the PWS-IC (present in PWS chromatin), was immunoprecipitated by anti–methyl Lys9 H3 antibody, whereas there was dramatically reduced precipitation of this sequence on the paternal copy (present in AS chromatin) (fig. 1). This result correlates well with the observation that maintenance of silenced heterochromatin in both Drosophila and fission yeast requires the function of H3 Lys9 methyltransferases (Rea et al. 2000; Nakayama et al. 2001). The region of maternal-specific H3 Lys9 methylation extends ∼0.6 kb 5′ and ∼0.5 kb 3′ around SNRPN exon 1 (fig. 2). Conversely, the paternal copy of the SNRPN promoter was immunoprecipitated by anti–methyl Lys4 H3 antibody. This sequence was not precipitated on the maternal copy (fig. 1). Previous reports that this modification is associated with active chromatin (Strahl et al. 1999) are consistent with these findings.
Figure 1.
A, PCR analysis of chromatin prepared from lymphocytes from controls, from individuals with PWS, and from individuals with AS, immunoprecipitated with methyl H3 Lys9 antibody. Primary lymphocytes from controls (Control), from individuals with PWS deletion (Del), from individuals with PWS imprinting defect (Imp), and from individuals with AS deletion (Del) were stimulated with phytohemagglutinin and IL-2; then, chromatin was prepared, was sonicated to average size ∼0.5 kb, and was immunoprecipitated, as described by Kuo and Allis (1999), with rabbit antibody to a keyhole-limpet-hemocyanin–conjugated peptide containing amino acids 6–13 of H3 with dimethyl modification of Lys9 (Nakayama et al. 2001). DNA from the immunoprecipitated material was amplified by PCR with the following primers: SNRPN 5′ region (SNA-F, 5′-GATGCTCAGGCGGGGATGTGTGCG-3′; and SNA-R, 5′-GCTCCCCAGGCTGTCTCTTGAGAG-3′) (Saitoh and Wada 2000) and CEN16 (16CEN-F, 5′-GTCTCTTTCTTGTTTTTAAGCTGGG-5′; and 16CEN-R, 5′-TGAGCTCATTGAGACATTTGG-3′) (sequence from GenBank [accession number AC002307]). CEN16 was used as the control PCR for DNA immunoprecipitated with methyl H3 Lys9 antibody. Sample volumes were adjusted so that, for a given antibody, all samples yielded equal amounts of product with the control PCR primers. CEN16 PCR cycling conditions were 94°C for 3 min; 30 cycles at 94°C for 1 min, 54°C for 1 min, and 72°C for 1 min; and 1 cycle at 94°C for 1 min, 54°C for 1 min, and 72°C for 5 min. PCR products were separated on agarose gels and were visualized by ethidium bromide staining. Product sizes were 173 bp, for SNRPN 5′ region, and 198 bp, for CEN16. B, PCR analysis of chromatin prepared from lymphocytes from controls, from individuals with PWS, and from individuals with AS, immunoprecipitated with methyl H3 Lys4 antibody. The same chromatin preparations used in panel A were immunoprecipitated with rabbit antibody to a BSA-conjugated peptide containing amino acids 1–8 of H3 with dimethyl modification of Lys4. DNA from the immunoprecipitated material was amplified by PCR with primers used in panel A, as well as primers for the NDN promoter and for the positive control sequence GAPDH: GAPDH (GAPDH-F, 5′-GCATCACCCGGAGGAGAAAATCGG-3′; and GAPDH-R, 5′-GTCACGTGTCGCAGAGGAGC-3′) (Saitoh and Wada 2000) and NDN 5′ region (NCD-A, 5′-ATGGCGAGGCTTCACCTG-3′; and NCD-B, 5′-AACTGGCCCCTTCTCCAGTA-3′) (sequence from GenBank [accession number AF001013]). NDN PCR was performed in the presence of 10% dimethyl sulfoxide; cycling conditions were 94°C for 3 min; 30 cycles at 94°C for 1 min, 59°C for 1 min, and 72°C for 1 min; and 1 cycle at 94°C for 1 min, 59°C for 1 min, and 72°C for 5 min. Product sizes were 111 bp, for NDN 5′ region, and 269 bp, for GAPDH.
Figure 2.
Extent of methyl modifications of H3 around SNRPN exon 1. DNA extracted from chromatin immunoprecipitated with methyl Lys9 H3 and methyl Lys4 H3 antibodies was amplified using primer pairs as shown. Primer pairs that showed preferential amplification of the maternal chromosome after immunoprecipitation with methyl H3 Lys9 antibody are shown as blackened squares; primer pairs that showed preferential amplification of the paternal chromosome after immunoprecipitation are shown as checkered squares; and primer pairs that showed no parent-specific preferential amplification are shown as unblackened squares. Primer sequences are as follows: UF1 (5′-CTAGAGGCCCCCTCTCATTGCAAC-3′), UR1 (5′-CTTCGCACACATCCCCGCCTGAGC-3′), UF2 (5′-TAGGAAGACCTGAGGGTGAG-3′), UR2 (5′-CACAGCACTGTTGCAATGAG-3′), UF3 (5′-TTTTCACAATTTACCCCCTC-3′), UR3 (5′-GGTCTTCCTATGTGCGGTAC-3′), UF4 (5′-ATTATCTCCCCCAAAATCAC-3′), UR4 (5′-TGTATCCAATTCTAATAGGC-3′), UF5 (5′-GTCTTTTCCCAAGCTACATC-3′), UR5 (5′-TGCCTATTAGAATTGGATAC-3′), UF6 (5′-GATCATTGGGAACTGAGCAG-3′), UR6 (5′-GATGTAGCTTGGGAAAAGAC-3′), UF7 (5′-TCCATCAGCCATATTGCATG-3′), UR7 (5′-TGCTCAGTTCCCAATGATCC-3′), DF1 (5′-CTCTCAAGAGACAGCCTGGGGAGC-3′), DR1 (5′-CCCCAATGCGAGCGGACAGGATAC-3′), DF2 (5′-ATCCTGTCCGCTCGCATTGG-3′), DR2 (5′-ACACGGAACTGCAATCACCC-3′), DF3 (5′-TCAGGGTGATTGCAGTTCCG-3′), and DR3 (5′-TACCCTGCTCCACCACGCAG-3′).
Parent-specific differential association of methyl Lys9 H3 was not detected with primers from the promoter regions of other imprinted genes in 15q11-q13—including ZNF127, NDN, MAGEL2 (MIM 605283), and IPW (MIM 601491), which are paternally expressed, as well as UBE3A and ATP10C (MIM 605855), which show tissue-specific maternal expression (data not shown). Also, methyl Lys9 H3 was not associated with the AS-IC. However, methyl Lys4 H3 was found to be specifically associated with the promoter region of the paternal allele of the paternally active gene NDN (fig. 1). Parent-specific Lys4 methylation in lymphocyte chromatin was not detected for ZNF127, MAGEL2, IPW, UBE3A, or ATP10C (data not shown). Loci that showed no parent-specific differential H3 methylation were hypomethylated on both chromosomes.
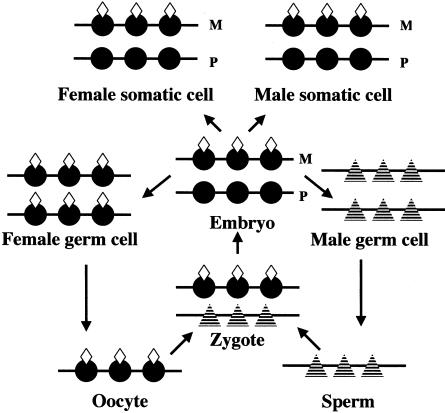
The preferential methylation of H3 Lys9 on the maternal allele of the human SNRPN-promoter region is the first maternal modification, other than cytosine methylation, to be reported in the PWS-IC. It is clear that the human PWS-IC lacks cytosine methylation in oocytes (El-Maarri et al. 2001); therefore, cytosine methylation of the PWS-IC cannot be the gametic imprint for the human PWS/AS region. Among the parent-specific histone modifications of the PWS-IC, acetylation of H3 and of H4, as well as H3 Lys4 methylation, cannot be the gametic imprint, because sperm lack histones (so a histone modification cannot function as a paternal gametic imprint). Methyl Lys9 H3, however, is a potential imprinting mark that could be imposed on histones in the PWS-IC during gametogenesis. A maternal histone-modification imprint would have the unique quality of having undergoing programmed erasure during spermatogenesis, when histones are removed from chromatin and replaced by protamines (fig. 3).
Figure 3.
Hypothetical model for histone-modification–based gametic imprint. Blackened circles represent histone, and striped triangles represent protamine; “M” denotes maternal chromosome, and “P” denotes paternal chromosome. Both in male somatic cells and in female somatic cells, maternal chromosome histone is modified (represented by unblackened diamonds), whereas paternal chromosome histone is unmodified. In female germ cell, histones on both chromosomes become modified; in male germ cell, histones are replaced by protamines (Braun 2001). After fertilization, protamine associated with paternal chromosome is replaced by unmodified histone. Replacement of histones by protamines, during spermatogenesis, would provide a mechanism to reverse the chemically stable histone-methylation mark.
How could a histone-modification imprint inherited from the oocyte be somatically propagated after fertilization, throughout the life span of an organism? Bannister et al. (2001) have suggested that silenced chromatin marked with methyl Lys9 H3 could be inherited during chromosome replication, by association of methyl Lys9 H3 and the heterochromatin protein HP1 and by association of HP1 and the mammalian H3 Lys9 methyltransferase SUVAR39H1. This multiprotein complex may allow H3 methyltransferase associated with an unreplicated chromosome region to methylate histones associated with newly synthesized DNA, at a nearby replication fork.
A primary requirement for a gametic imprint is that it be present in the gamete—and absent from the gamete arising from the other sex. Testing for histone modifications in oocytes will require development of techniques of chromatin immunoprecipitation that are more sensitive than those that are currently available, to test for association of specific DNA sequences and modified histones in small numbers of cells. If H3 Lys9 is methylated in the oocytes, a rigorous test of the role that H3 Lys9 methylation plays in PWS/AS imprinting will require both identification of the histone methyltransferase responsible for this methylation in the female germline and targeted inactivation of this methyltransferase in the female germline. An alternative or additional gametic imprint based on histone modification, rather than on DNA methylation, may provide a resolution to several paradoxes—such as (a) that a number of imprinted genes (including mouse Mash2 [Tanaka et al. 1999] and human CDKN1C [MIM 600856] [Chung et al. 1996]) show no regions of parent-specific DNA methylation, (b) that Mash2 imprinting is not disturbed in Dnmt1 −/− embryos (Caspary et al. 1998), and (c) that some genes show evolutionary conservation of imprinting without showing conservation of differential DNA methylation (Hatada and Mukai 1995; Chung et al. 1996; Killian et al. 2000).
Acknowledgments
This work was supported by grants from the National Institutes of Health (to J.W. and C.D.A.) and from the Ward Family Foundation (to J.W.). We thank Burak Gezen for technical assistance.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for CEN16 [accession number AC002307] and NDN 5′ [accession number AF001013])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PWS [MIM 176270], AS [MIM 105830], UBE3A [MIM 601623], SNRPN [MIM 182279], ZNF127 [MIM 603856], NDN [MIM 602117], MAGEL2 [MIM 605283], IPW [MIM 601491], ATP10C [MIM 605855], and CDKN1C [MIM 600856])
References
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL (1997) Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet 17:75–78 [DOI] [PubMed] [Google Scholar]
- Bannister A, Zegerman P, Partridge J, Miska E, Thomas J, Allshire R, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120–124 [DOI] [PubMed] [Google Scholar]
- Braun R (2001) Packaging paternal chromosomes with protamine. Nat Genet 28:10–12 [DOI] [PubMed] [Google Scholar]
- Bressler J, Tsai T-F, Wu M-Y, Tsai S-F, Ramirez MA, Armstrong D, Beaudet AL (2001) The SNRPN promoter is not required for genomic imprinting of the Prader-Willi/Angelman domain in mice. Nat Genet 28:232–240 [DOI] [PubMed] [Google Scholar]
- Byvoet P, Shepherd G, Hardin J, Noland B (1972) The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys 148:558–567 [DOI] [PubMed] [Google Scholar]
- Caspary T, Cleary M, Baker C, Guan X-J, Tilghman SM (1998) Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol Cell Biol 18:3466–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W-Y, Yuan L, Feng L, Hensle T, Tycko B (1996) Chromosome 11p15.5 regional imprinting: comparative analysis of KIP2 and H19 in human tissues and Wilms' tumors. Hum Mol Genet 5:1101–1108 [DOI] [PubMed] [Google Scholar]
- Dittrich B, Robinson WP, Knoblauch H, Buiting K, Schmidt K, Gillessen-Kaesbach G, Horsthemke B (1992) Molecular diagnosis of the Prader-Willi and Angelman syndromes by detection of parent-of-origin specific DNA methylation in 15q11-13. Hum Genet 90:313–315 [DOI] [PubMed] [Google Scholar]
- El Kharroubi A, Piras G, Stewart CL (2001) DNA demethylation reactivates a subset of imprinted genes in uniparental mouse fibroblasts. J Biol Chem 276:8674–8680 [DOI] [PubMed] [Google Scholar]
- El-Maarri O, Buiting K, Peery E, Kroise P, Balaban B, Wagner K, Urman B, Heyd J, Lich C, Brannan C, Walter J, Horsthemke B (2001) Methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet 27:341–344 [DOI] [PubMed] [Google Scholar]
- Fulmer-Smentek S, Francke U (2001) Association of acetylated histones with paternally expressed genes in the Prader-Willi deletion region. Hum Mol Genet 15:645–652 [DOI] [PubMed] [Google Scholar]
- Glenn CC, Porter KA, Jong MTC, Nicholls RD, Driscoll DJ (1993) Functional imprinting and epigenetic modification of the human SNRPN gene. Hum Mol Genet 2:2001–2005 [DOI] [PubMed] [Google Scholar]
- Glenn CC, Saitoh S, Jong MTC, Filbrandt MM, Surti U, Driscoll DJ, Nicholls RD (1996) Gene structure, DNA methylation, and imprinted expression of the human SNRPN gene. Am J Hum Genet 58:335–346 [PMC free article] [PubMed] [Google Scholar]
- Hatada I, Mukai T (1995) Genomic imprinting of p57Kip2, a cyclin-dependent kinase inhibitor, in mouse. Nat Genet 11:204–206 [DOI] [PubMed] [Google Scholar]
- Horvath JE, Viggiano L, Loftus BJ, Adams MD, Archidiacono N, Rocchi M, Eichler EE (2000) Molecular structure and evolution of an alpha satellite/non-alpha satellite junction at 16p11. Hum Mol Genet 9:113–123 [DOI] [PubMed] [Google Scholar]
- Jay P, Rougeulle C, Massacrier A, Moncla A, Mattei M-G, Malzac P, Roeckel N, Taviaux S, Lefranc J-LB, Cau P, Berta P, Lalande M, Muscatelli F (1997) The human necdin gene, NDN, is maternally imprinted and located in the Prader-Willi syndrome critical region. Nat Genet 17:357–360 [DOI] [PubMed] [Google Scholar]
- Jong MTC, Gray TA, Ji Y, Glenn CC, Saitoh S, Driscoll DJ, Nicholls RD (1999) A novel imprinted gene, encoding a RING zinc-finger protein, and overlapping antisense transcript in the Prader-Willi syndrome critical region. Hum Mol Genet 8:783–793 [DOI] [PubMed] [Google Scholar]
- Killian J, Byrd J, Jirtle J, Munday B, Stoskopf M, MacDonald R, Jirtle R (2000) M6P/IGF2R imprinting evolution in mammals. Mol Cell 5:707–716 [DOI] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J (1997) UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet 15:70–73 [DOI] [PubMed] [Google Scholar]
- Kuo M-H, Allis CD (1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425–433 [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120 [DOI] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard R-J, Jiang Y, Benton CS, Rommens JM, Beaudet AL (1997) De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet 15:74–77 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice J, Strahl BD, Allis CD, Grewal S (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110–113 [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Saitoh S, Horsthemke B (1998) Imprinting in Prader-Willi and Angelman syndromes. Trends Genet 14:194–200 [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun Z-W, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromosome structure by site-specific histone H3 methyltransferases. Nature 406:593–599 [DOI] [PubMed] [Google Scholar]
- Saitoh S, Wada T (2000) Parent-of-origin specific histone acetylation and reactivation of a key imprinted gene locus in Prader-Willi syndrome. Am J Hum Genet 66:1958–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer R, Birger Y, Riggs A, Razin A (1997) Structure of the mouse SNRPN gene and establishment of its parental-specific methylation pattern. Proc Natl Acad Sci USA 94:10267–10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook R, Allis CD (1999) Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci USA 96:14967–14972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Puchyr M, Gertsenstein M, Harpal K, Jaenisch R, Rossant J, Nagy A (1999) Parental origin-specific expression of Mash2 is established at the time of implantation with its imprinting mechanism highly resistant to genome-wide demethylation. Mech Dev 87:129–142 [DOI] [PubMed] [Google Scholar]