Abstract
Spinal neurofibromatosis (SNF) is considered to be an alternative form of neurofibromatosis, showing multiple spinal tumors and café-au-lait macules. Involvement of the neurofibromatosis type 1 (NF1) locus has been demonstrated, by linkage analysis, for three families with SNF. In one of them, a cosegregating frameshift mutation in exon 46 of the NF1 gene was identified. In the present study, we report four individuals from two families who carry NF1 null mutations that would be expected to cause NF1. Three patients have multiple spinal tumors and no café-au-lait macules, and the fourth has no clinical signs of NF1. In the first family, a missense mutation (Leu2067Pro) in NF1 exon 33 was found, and, in the second, a splice-site mutation (IVS31-5A→G) enlarging exon 32 by 4 bp at the 5′ end was found. The latter mutation has also been observed in an unrelated patient with classical NF1. Both NF1 mutations cause a reduction in neurofibromin of ∼50%, with no truncated protein present in the cells. This demonstrates that typical NF1 null mutations can result in a phenotype that is distinct from classical NF1, showing only a small spectrum of the NF1 symptoms, such as multiple spinal tumors, but not completely fitting the current clinical criteria for SNF. We speculate that this phenotype is caused by an unknown modifying gene that compensates for some, but not all, of the effects caused by neurofibromin deficiency.
Paraspinal nerve-sheath tumors occur solitarily and sporadically. Several paraspinal tumors have been observed in a minority of patients with neurofibromatosis type 1 (NF1 [MIM 162200]) (Huson et al. 1988; McGaughran et al. 1999; Thakkar et al. 1999). These tumors occur as a main symptom in familial spinal neurofibromatosis (SNF [MIM 162210]). Four families with SNF have been reported to date (Pulst et al. 1991 [two families]; Poyhonen et al. 1997; Ars et al. 1998); in three of them, involvement of the NF1 locus has been demonstrated by linkage analysis. In one family, Ars et al. (1998) identified a cosegregating frameshift mutation (8042insA), in exon 46 of the NF1 gene, which is also suggested to result in a truncated neurofibromin in vivo. All affected family members fit the current clinical criteria for SNF, showing multiple spinal tumors and café-au-lait macules. In addition, one had cutaneous neurofibromas, whereas the others showed different signs of NF1. In the present study, we describe two families with multiple spinal tumors. The patients show spinal tumors but no café-au-lait macules. Two of the patients show a few dermal tumors, histologically identified as neurofibroma or schwannoma. Other signs of NF1 or NF2 (MIM 101000) are missing in both families. The patients were examined clinically in the Interdisciplinary Neurofibromatosis Clinic of the University of Ulm, and the appropriate informed consent was obtained from the patients. Our investigations determined that two members of each family are heterozygous for NF1 null mutations.
In the first family (NF134), the propositus was a female aged 32 years (NF134-3). An intrathoracic tumor was detected in her upper mediastinum, at age 17 years. The biopsy of the tumor was histologically identified as neurinoma. In addition, multiple tumors of the cervical and lumbar spine and of the psoas muscle were detected by magnetic resonance imaging (MRI) scan. At ages 23 and 27 years, tumors were excised in C6/7 and L2. These were identified as schwannoma and neurofibroma, respectively. At ages 29 and 32 years, a subcutaneous neurofibroma and a subcutaneous schwannoma, respectively, were excised. During the present investigations, multiple intradural and intramedullar tumor masses were detected by MRI scan in all segments of the spine. Tumors in the central nervous system (CNS) that are typical of NF2 were not found. The patient did not show café-au-lait macules, intertrigenous freckling, or Lisch nodules. The patient, a student, shows no signs of mental retardation or scoliosis. Her parents’ (NF134-1 and NF134-2, aged 60 and 61 years, respectively) medical history revealed no brain tumors, spinal cord tumors, neurofibromas, café-au-lait macules, unexplained deafness, neurological disability, or lumbago. Additional clinical investigations were not possible.
In the second family (NF138), the female propositus (NF138-3, aged 31 years), observed multiple painful intradermal tumors on the extremities and the trunk, at age 17 years. One of these tumors was excised and was histologically identified as a neurofibroma. Another tumor was excised from the thoracic spine at age 29 years and was histologically identified as schwannoma. Multiple spinal tumors currently have been found, through use of MRI scans, in all segments of the spine, especially in C5/6. Other symptoms typical for NF1, such as café-au-lait macules, freckles, Lisch nodules, scoliosis, or tumors of the CNS, were not found. Clinical investigation of her mother (NF138-2, aged 57 years) revealed two lumbar hyperpigmentations that were 2 cm in diameter, without sharp borders. Signs of NF1 or NF2 were not found. She presented with acute lumbago. The MRI scan revealed enlarged spinal nerves from L3/4 to L5/S1. An additional MRI scan showed these alterations in all segments of the spine. The anamnesis of her husband (NF138-1), her son (NF138-4), and her second daughter (NF138-5) did not reveal symptoms of NF1 or NF2.
NF1 mutation screening, of complementary DNA (cDNA) as well as of genomic DNA (gDNA), was performed in these families through use of the enzymatic mutation detection (EMD) method, for all exons (Eisenbarth et al. 2000), with subsequent genomic sequencing of the altered fragments (Fahsold et al. 2000). With this method we are able to recognize >70% of NF1 mutations (data not shown). The screening for NF2 mutations was performed by means of genomic sequencing (Kehrer-Sawatzki et al. 1997). Fibroblasts were isolated from the skin of the index patients and were cultured as described by Kaufmann et al. (1999). In NF134-3, the cells were obtained from the right upper arm, and in NF138-3 they were obtained from the shoulder. In addition, peripheral blood cells from members of the families (NF134-1/2/3 and NF138-2/3/4/5) were investigated.
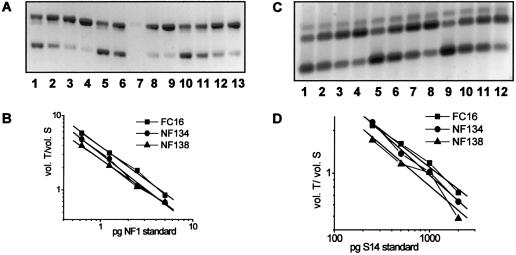
In both families, NF1 mutations could be characterized. In the first family, in NF134-3, only one altered fragment was detected by EMD. Through sequencing of gDNA, a base exchange was found in NF1 exon 33 (T6200C). Investigation of <1,000 other patients with NF1 did not reveal this base exchange in any of the patients, indicating that it is not a polymorphism. Screening for the NF2 mutation showed no alterations in NF134-3. The NF1 mutation has no impact on the amount of NF1 mRNA in cultured cells of NF134-3. The amount of NF1 mRNA was measured in three independent experiments by quantitative competitive reverse-transcriptase PCR (RT-PCR) using internal standard NF1 RNA, as described in detail by Kaufmann et al. (1999). The resulting measurement was 87.4%±8.2% of that found in the fibroblasts of a healthy donor, as shown in a typical experiment (fig. 1A and B). To rule out significant variation of the amount of RNA, the expression of S14 (encoding ribosomal subunit protein) was measured using the same method. The mRNA amount of S14 in NF134-3 was 78.7% of that found in the control, FC16 (fig. 1C and D). Correction of the NF134-3 NF1 mRNA amount for the S14 mRNA amount results in 111%. This indicates that the mRNA transcribed from the mutated NF1 allele is stable. Interestingly, the change of leucine to proline at position 2067 of neurofibromin reduces the stability of the protein. This was shown by investigating the amount of total neurofibromin in cultured cells of NF134-3 in comparison with controls, as described by Griesser et al. (1997) and Klose et al. (1998). In three independent experiments, neurofibromin was found to be reduced to 51.9%±5.4% of the amount in control cells. A typical experiment is shown in figure 2A. As a control, the amount of p120GAP was measured (fig. 2B). Truncated neurofibromin was found neither in immunoprecipitation of lysates from 35S-cysteine-methionin–labelled fibroblasts, using an antibody against the amino-terminal domain of neurofibromin (data not shown), nor in immunoprecipitation and western blotting using this antibody (fig. 2C). This shows that the NF1 germline mutation of NF134-3 is a null mutation at the protein level of the investigated cells, suggesting a functional haploinsufficiency. The parents were investigated for this mutation, by restriction analysis of PCR products from gDNA (fig. 3A). Unexpectedly, her father (NF134-1), clinically unaffected and aged 61 years, also showed this NF1 mutation in his blood cells. Additional molecular investigations of other tissues, to exclude a mosaicism, were not feasible, and additional clinical investigations through MRI scans could not be performed. Therefore, nonpenetrance of this NF1 mutation cannot be ruled out.
Figure 1.
Mutations of NF134-3 and of NF138-3, causing no alteration in the amount of NF1 mRNA (A and B) in relation to the expression of S14 (C and D). Fibroblasts from NF134-3, NF138-3, and a healthy donor (FC16) were seeded at a density of 4,000 cells/cm2, were cultured for 2 d, and were total-RNA isolated. For each cell culture, 4× 1 μg RNA was reverse transcribed with random hexamers and different amounts of standard RNAs for NF1 and S14 and then were PCR amplified with primer pair NF1-H/NF1-R and S14-H/S14-R. Measurement of the band intensities of the PCR products was performed as described by Kaufmann et al. (1999), with the following modification: the volume of the heteroduplex band was additionally measured, with half of its amount being added to both the volume of the transcript and the standard band. A, RT-PCR products for measurement of NF1 mRNA amounts, 1 μg of total RNA each: FC16 (lanes 1–4), NF134-3 (lanes 5–8), and NF138-3 (lanes 9–12). The products were separated on a 1.5% agarose gel stained with ethidium bromide. Lower band (345 bp), NF1 standard; middle band, heteroduplexes; upper band (395 bp), NF1 transcript. Amounts of NF1 standard RNA added: lanes 1, 5, and 9, 5 pg; lanes 2, 6, and 10, 2.5 pg; lanes 3, 7, and 11, 1.25 pg; lanes 4, 8, and 12, 0.625 pg. B, Graphical representation with linear regression for each of the NF1 mRNA measurements (vol. = integration of the volume of the band areas; T = transcript; S = standard). C, RT-PCR products for measurement of S14 mRNA amounts, 1 μg of total RNA each: FC16 (lanes 1–4), NF134-3 (lanes 5–8), and NF138-3 (lanes 9–12). The products were separated on a 1.5% agarose gel stained with SYBR-Gold. Lower band (148 bp), S14 standard; middle band (197 bp), S14 transcript; upper band, heteroduplexes. Amounts of S14 standard RNA added: lanes 1, 5, and 9, 2,000 pg; lanes 2, 6, and 10, 1,000 pg; lanes 3, 7, and 11, 500 pg; lanes 4, 8, and 12, 250 pg. D, Graphical representation with linear regression for each of the S14 mRNA measurements. Abbreviations are the same as those listed for figure 1B.
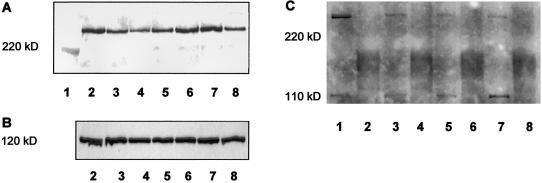
Figure 2.
Reduction in the amount of neurofibromin in fibroblasts of NF134-3 and NF138-3 (A and B). No additional shortened neurofibromin band is detectable in either cell culture (C). A, Detection of neurofibromin in lysates, by western blotting of immunoprecipitated neurofibromin. Cells were cultured in parallel, were seeded at a density of 4,000 cells/cm2, were cultured for 3 d, and were lysed as described elsewhere (Griesser et al. 1995). Neurofibromin was immunoprecipitated from equal amounts of total protein (lanes 2–7, 600 μg; lane 8, 300 μg) by incubation with an antibody against the amino-terminal domain of neurofibromin (sc-68; Santa Cruz Biotechnology) for 1.5 h, followed by incubation with protein-A-Sepharose (Amersham Pharmacia Biotech). The samples were boiled in SDS-sample buffer and were separated with SDS-PAGE. After blotting on polyvinylidene fluoride (PVDF) membrane, immunodetection was performed using an antibody against the carboxy-terminal domain of neurofibromin (sc-67; Santa Cruz Biotechnology) and the ECL system. The resulting bands were evaluated using a video-densitometric system that allows neurofibromin quantification (Griesser et al. 1997; Klose et al. 1998). Lane 1, molecular weight marker, myosin (220 kD); lanes 2 and 6, fibroblasts from a healthy donor (FC7); lane 3, fibroblasts of NF71 carrying a stop mutation (Stark et al. 1992); lane 4, fibroblasts of NF138-3; lane 5, fibroblasts of NF134-3; lane 7, fibroblasts from a healthy donor (FC4); lane 8, fibroblasts from a peripheral nerve of a healthy donor (N21). The densitometric units (100% = median of the values of the four controls) are as follows—FC7: lane 2, 240 (108%), and lane 6, 218 (98%); FC4: 223 (100%); NF71: 126 (57%); NF138-3: 86 (37%); NF134-3: 126 (57%); peripheral nerve cells (300 μg protein): 104 (47%, corresponding to 94% for 600 μg protein). B, Detection of p120GAP by western blotting. Equal amounts of total protein (20 μg) were separated with SDS-PAGE and were blotted on PVDF membrane. Immunodetection was performed by incubation with anti-p120GAP antibody (G12920; Transduction Laboratories) for 2 h, followed by detection with the ECL system. The densitometric units are as follows—FC7: lane 2, 263 and lane 6, 212; FC4: 234; NF71: 268; NF138-3: 204; NF134-3: 202; peripheral nerve cells: 220. C, Investigation of shortened neurofibromin by western blotting of immunoprecipitated neurofibromin, using sc-68. Lane 1, FC7; lane 3, NF134-3; lane 5, NF138-3; lane 7, NF71. To evaluate unspecific signals, sc-68 was preincubated with the corresponding peptide (lanes 2, 4, 6, and 8), and the same detection was performed. 240 kD: neurofibromin; 110 kD: unspecific product. The expected neurofibromin should be detected at ∼225 kD.
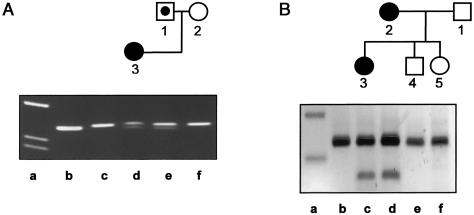
Figure 3.
NF1 mutations in family NF134 (A) and family NF138 (B). In the pedigrees, blackened symbols denote affected individuals, unblackened symbols denote unaffected indivuals, and unblackened symbols with a blackened circle denote carriers. A, Detection of the T→C mutation in NF1 exon 33 in family NF134. gDNA was amplified using PCR primers ex33mut (5′-TTT TAG CAC GCT ACA TGC TGG TGC) and ex33r (5′-TTT TGG TAA TAT TTC ATG TCA TTA CTG). Primer ex33mut was designed such that an additional NlaIV restriction site is created if the T→C mutation is present. Lanes b and c show the PCR product derived from gDNA of a healthy donor, both uncut (335 bp) and restricted with NlaIV, respectively. Lane d shows PCR products derived from the index patient (NF134-3), lane e from her father (NF134-1), and lane f from her mother (NF134-2), cut with NlaIV. The lower band (313 bp) in lanes d and e demonstrates the presence of the T→C mutation in the index patient and in her father. Lane a shows a size marker. B, Detection of A→G exchange in NF1 intron 31 in family NF138. gDNA PCR products including NF1 exon 32, using the primers ex32h (5′-TGA CAG GCC TGT AAA TAA AAT CTA G) and ex32r (5′-TTT CCA GAA GCC AAA ACT ACA G) are shown, from a healthy control (lane b), the index patient (NF138-3, lane c), the mother (NF138-2, lane d), her brother (NF138-4, lane e), and her sister (NF138-5, lane f), all restricted with AluI; lane a shows a 100-bp ladder (200-bp and 300-bp bands). The upper band corresponds to the wild-type PCR product of 225 bp. The lower band (144 bp) seen in lanes c and d results from the restriction at an additional AluI site created by the mutation in intron 31. The index patient and her mother carry this NF1 mutation.
With regard to the second family (NF138), an NF1 alteration was found by EMD in the cDNA of the index patient (NF138-3). Genomic sequencing of the corresponding fragment showed a base exchange in the NF1 splice-acceptor site of exon 31 (intron 31 AACTAG/exon32). This mutation (IVS31-5A→G) creates a new splice-acceptor site (intron 31 AGCTAG/exon32 [base change underlined]). The new splice site is indeed used in cultured fibroblasts of NF138-3, as shown by sequencing of RT-PCR products. This results in the insertion of four bases in NF1 exon 32 (exon 31 GGGCAG/CTAG exon 32) and, subsequently, in a premature termination codon at amino acid (aa) 1995 (exon 32). The amount of the mutated NF1 mRNA was not reduced, as shown by quantitative RT-PCR. The amount of NF1 mRNA measured was 71% of that found in the control FC16 (fig. 1A and B). Standardized to the expression of S14 (50.4% of that in cells of the control; fig. 1C and D), the NF1 mRNA amount in cells of NF138-3 is 141%. At the protein level, this NF1 mutation also resulted in instability of the mutated neurofibromin; a reduction to 37% of the control level was measured in the cells of NF138-3 (fig. 2A and B). Again, a truncated neurofibromin was not detected (fig. 2C). To test for the presence of this mutation in other members of this family, gDNA was investigated by restriction analysis. The base change A→G creates a new restriction site for AluI. Restriction of PCR products obtained from gDNA of blood cells with this enzyme resulted in an additional shortened DNA fragment in the index patient (NF138-3) and in her mother (NF138-2) (fig. 3B). Since the mother's tumors involve all segments of the spine and the mutation was found in peripheral blood cells, we suggest that the NF1 mutation in NF138-2 occurred in the germline. In this case, the clinical differences between NF138-2 and her daughter (NF138-3) suggest a variable expressivity of this mutation.
It is assumed that most germline mutations in NF1 result in haploinsufficiency through the reduction of neurofibromin levels (Heim et al. 1995; Hoffmeyer et al. 1995; Fahsold et al. 2000), which then causes NF1. We have demonstrated that the NF1 mutations reported here also result in reduction of neurofibromin in our patients with spinal tumors. In both cases, the mutant NF1 mRNAs are stable, whereas the mutant neurofibromin is unstable. This is also known to occur for other NF1 mutations. For example, patient NF71 in the study by Stark et al. (1992) showed equal expression of the NF1 alleles and a normal amount of NF1 mRNA (Kaufmann et al. 1999) but a level of neurofibromin that was reduced by half (fig. 2A, lane 3).
Our patients do not fit the current strict clinical criteria for SNF, because they show multiple spinal tumors but no café-au-lait macules. We assume that the investigation of additional families that have spinal tumors and show NF1 mutations will help to define additional criteria for this disease that would permit the inclusion of our patients.
Our observation is not consistent with the assumption that the type or effect of the NF1 mutation can be invoked to explain the special phenotype of SNF. This is especially true with regard to the NF1 mutation IVS31-5A→G in family NF138, which we detected in an earlier study, in an unrelated patient showing classical NF1 (Fahsold et al. 2000). In the family reported by Ars et al. (1998), SNF was described to have been the result of a residual function of the truncated neurofibromin. However, it was not demonstrated that the truncated protein was, in fact, present in cells of the patients in vivo. Furthermore, among mutations found in patients with regular NF1, premature termination codons in the same region are quite frequent—for example, at aa 2604, 2640, 2643, and 2717 (Fahsold et al. 2000). In our two patients, the NF1 mutations result in a reduction of neurofibromin levels, with no phenotypic difference in comparison with other patients with NF1. To explain the specific phenotype of our index patients, which represents only a small portion of the spectrum of NF1 symptoms, we speculate that an unknown modifying gene may be having an effect. This modifying gene should compensate for the majority, but not all, of the effects caused by neurofibromin deficiency in the cells of these patients.
Acknowledgments
We wish to thank Anna Siegel, Birgit Schmoll, and Sigrid Wieland-Lange, for their excellent technical assistance. This work was supported by a grant from the Deutsche Krebshilfe (to B.B., B.S., and S.W.-L.).
Electronic-Database Information
Accession numbers and the URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SNF [MIM 162210], NF1 [MIM 162200], and NF2 [MIM 101100])
References
- Ars E, Kruyer H, Gaona A, Casquero P, Rosell J, Volpini V, Serra E, Lázaro C, Estivill X (1998) A clinical variant of neurofibromatosis type 1: familial spinal neurofibromatosis with a frameshift mutation in the NF1 gene. Am J Hum Genet 62:834–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth I, Beyer K, Krone W, Assum G (2000) Toward a survey of somatic mutation of the NF1 gene in benign neurofibromas of patients with neurofibromatosis type 1. Am J Hum Genet 66:393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahsold R, Hoffmeyer S, Mischung C, Gille C, Ehlers C, Kücükceylan N, Abdel-Nour M, Gewies A, Peters H, Kaufmann D, Buske A, Tinschert S, Nürnberg P (2000) Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am J Hum Genet 66:790–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesser J, Kaufmann D, Eisenbarth I, Bauerle C, Krone W (1995) Ras-GTP regulation is not altered in cultured melanocytes with reduced levels of neurofibromin derived from patients with neurofibromatosis 1 (NF1). Biol Chem Hoppe Seyler 376:91–101 [DOI] [PubMed] [Google Scholar]
- Griesser J, Kaufmann D, Maier B, Mailhammer R, Kuehl P, Krone W (1997) Post-transcriptional regulation of neurofibromin level in cultured human melanocytes in response to growth factors. J Invest Dermatol 108:275–280 [DOI] [PubMed] [Google Scholar]
- Heim RA, Kam-Morgan LNW, Binnie CG, Corns DD, Cayouette MC, Farber RA, Aylsworth AS, Silverman LM, Luce MC (1995) Distribution of 13 truncating mutations in the neurofibromatosis 1 gene. Hum Mol Genet 4:975–981 [DOI] [PubMed] [Google Scholar]
- Hoffmeyer S, Assum G, Griesser J, Kaufmann D, Nurnberg P, Krone W (1995) On unequal allelic expression of the neurofibromin gene in neurofibromatosis type 1. Hum Mol Genet 4:1267–1272 [DOI] [PubMed] [Google Scholar]
- Huson SM, Harper PS, Compston DAS (1988) Von Recklinghausen neurofibromatosis: a clinical and population study in south-east Wales. Brain 111:1355–1381 [DOI] [PubMed] [Google Scholar]
- Kaufmann D, Gruener S, Braun F, Stark M, Griesser J, Hoffmeyer S, Bartelt B (1999) EVI2B, a gene lying in an intron of the neurofibromatosis type 1 (NF1) gene, is as the NF1 gene involved in differentiation of melanocytes and keratinocytes and is over-expressed in cells derived from NF1 neurofibromas. DNA Cell Biol 18:345–356 [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Udart M, Krone W, Baden R, Fahsold R, Thomas G, Schmucker B, Assum G (1997) Mutational analysis and expression studies of the neurofibromatosis type 2 (NF2) gene in a patient with a ring chromosome 22 and NF2. Hum Genet 100:67–74 [DOI] [PubMed] [Google Scholar]
- Klose A, Ahmadian MR, Schuelke M, Scheffzek K, Hoffmeyer S, Gewies A, Schmitz F, Kaufmann D, Peters H, Wittinghofer A, Nurnberg P (1998) Selective disactivation of neurofibromin GAP activity in neurofibromatosis type 1. Hum Mol Genet 7:1261–1268 [DOI] [PubMed] [Google Scholar]
- McGaughran JM, Harris DI, Donnai D, Teare D, MacLeod R, Westerbeek R, Kingston H, Super M, Harris R, Evans DG (1999) A clinical study of type 1 neurofibromatosis in north west England. J Med Genet 36:197–203 [PMC free article] [PubMed] [Google Scholar]
- Poyhonen M, Leisti EL, Kytölä S, Leisti J (1997) Hereditary spinal neurofibromatosis: a rare form of NF1? J Med Genet 34:184–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulst SM, Riccardi VM, Fain P, Korenberg JR (1991) Familial spinal neurofibromatosis: clinical and DNA linkage analysis. Neurology 41:1923–1925 [DOI] [PubMed] [Google Scholar]
- Stark M, Assum G, Kaufmann D, Kehrer H, Krone W (1992) Analysis of segregation and expression of an identified mutation at the neurofibromatosis type 1 locus. Hum Genet 90:356–359 [DOI] [PubMed] [Google Scholar]
- Thakkar SD, Feigen U, Mautner VF (1999) Spinal tumours in neurofibromatosis type 1: an MRI study of frequency, multiplicity and variety. Neuroradiology 41:625–629 [DOI] [PubMed] [Google Scholar]