To the Editor:
Mutations in the gene encoding spastin, an ATPase of unknown function, cause the most common form of autosomal dominant hereditary spastic paraplegia (SPG4 [MIM 182601]; Hazan et al. 1999), a neurodegenerative disorder characterized by progressive spasticity of the lower limbs. Recently, we described 11 mutations in this gene, 7 of which cause missplicing of the spastin transcript (Svenson et al. 2001). Because of the presence of a rare spastin gene–coding region polymorphism in the proband of one family with an IVS9+4a→g mutation in the spastin gene, we were able to determine that this mutation is only partially penetrant—that is, both normally and aberrantly spliced (skipped exon 9) transcripts are produced from the mutant allele. This “leakiness” of the mutation was not recapitulated in in vitro splicing experiments with minigene constructs containing only exon 9 and flanking intronic sequence. However, when we included additional sequence (exons 8–12, with flanking intronic sequence), the leakiness of the mutation was recapitulated. These findings demonstrate the importance of genomic-sequence context in the determination of splice-site selection and the extent of missplicing caused by this mutation.
Because of the absence of any other spastin gene–coding region polymorphisms, we were unable to test several other splice-site mutations for leakiness by reverse-transcriptase (RT)–PCR analysis of patient-derived RNA. None of these other mutations appeared leaky in our in vitro splicing experiments with minigene constructs, even in the larger genomic-sequence context that included flanking exons.
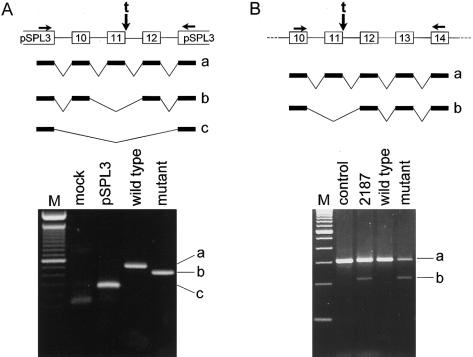
We have since found that at least one of these other splice-site mutations is, in fact, leaky in a much larger genomic-sequence context—that is, in the complete human chromosome 2 that harbors the mutation. This mutation, an IVS11+2t insertion, causes skipping of exon 11, as determined by RT-PCR analysis of patient-derived RNA (see the report by Svenson et al. [2001], for pedigree information and experimental data). In our in vitro splicing experiments with a minigene construct that carries this mutation as well as the complete sequence of both flanking introns and exons on each side of the skipped exon, we detected only aberrantly spliced (skipped exon 11) transcript (fig. 1A). To determine whether the presence of additional genomic-sequence context would affect the change in splice-site selection caused by this mutation, we performed RT-PCR analysis of RNA extracted from mouse-human hybrid cell lines containing either the wild-type or the mutant chromosome 2 of a patient with SPG who is heterozygous for this mutation. As shown in figure 1B, we detected both normally and aberrantly spliced transcripts in the RNA extracted from the hybrid cell line containing the mutant—but not the wild-type—chromosome 2. We confirmed through sequencing that the transcripts were derived from the human and not the mouse spastin gene. We were unable to test the other splice-site mutations for leakiness by this method, because of the lack of availability of appropriate patient-derived cell lines.
Figure 1.
A, The pSPL3 exon-trapping vector was used to assay splice-site selection. COS1 cells were transfected with vector alone (pSPL3), with wild-type exons 10–12 and flanking intronic sequence (wild type), or with the same construct but containing the IVS11+2t insertion (mutant). Top panel, Schematic showing construct design, vector-specific primers used for amplification (horizontal arrows), site of mutation (vertical arrow), and splice variants a, b, and c. Bottom panel, Transcripts identified on the basis of sequencing of the correspondingly labeled RT-PCR products (a, b, and c). B, RT-PCR analysis of RNA extracted from a lymphoblastoid cell line from a normal control individual (control), from a patient heterozygous for the IVS11+2t insertion (2187), or from a mouse E2-human hybrid cell line hemizygous for human chromosome 2, without (wild type) or with (mutant) the IVS11+2t insertion. Top panel, Schematic showing positions of primers used for amplification (horizontal arrows), site of mutation (vertical arrow), and splice variants a and b. Bottom panel, Transcripts identified on the basis of sequencing of the correspondingly labeled RT-PCR products (a and b).
These findings provide further evidence of the dependence of splice-site selection on genomic-sequence context. The exon-definition model of splice-site selection proposes that an exon is defined by the binding of splicing factors to the 3′ and 5′ splice sites of the same exon (Berget 1995). Pairing of U1 small nuclear RNA (snRNA) to the 5′ splice junction includes at least six nucleotides beyond the invariant gt. The IVS11+2t insertion mutation would shift the base pairing by one nucleotide, resulting in a net loss of four base pairs relative to the pairing with the wild-type sequence. Despite this drastic alteration in U1 snRNA:hnRNA (i.e., heterogeneous nuclear RNA) pairing, this mutation, in its full genomic-sequence context, is only partially penetrant.
Given these observations, we suggest that, in the case of splicing mutations outside the invariant gt–ag splice-site sequences, splice-site selection might also depend on the definition of exons and the strength of intron-exon junction sequences not directly adjacent to the mutant splice junction. In addition, splice-site selection might be influenced, in part, by intronic sequences distant from the intron-exon junctions.
Although the function and normal range of expression level of spastin are still unknown, our findings reported here provide additional support for the hypothesis that the function of spastin is highly concentration dependent. Our finding that normally spliced transcript is produced from at least two different mutant alleles is consistent with the threshold of spastin required for transition from normal function to disease state lying within a narrower interval than the 50% decrease predicted by a disease model of haploinsufficiency. Thus, as we have speculated elsewhere (Svenson et al. 2001), even slight variation in spastin expression level might have dramatic phenotypic consequences, accounting, at least in part, for the wide variability in age at onset, symptom severity, and rate of symptom progression reported to occur in SPG caused by spastin gene mutations.
Acknowledgments
We thank the patients and their families for their participation. This work was supported, in part, by National Institutes of Health program project grant 2P01-NS26630-11A1.
Electronic-Database Information
The accession number and URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SPG4 [MIM 182601])
References
- Berget SM (1995) Exon recognition in vertebrate splicing. J Biol Chem 270:2411–2414 [DOI] [PubMed] [Google Scholar]
- Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, Davoine C-S, Cruaud D, Dürr A, Wincker P, Brottier P, Cattolico L, Barbe V, Burgunder J-M, Prud’homme J-F, Brice A, Fontaine B, Heilig R, Weissenbach J (1999) Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet 23:296–303 [DOI] [PubMed] [Google Scholar]
- Svenson IK, Ashley-Koch AE, Gaskell PC, Riney TJ, Cumming WJK, Kingston HM, Hogan EL, Boustany R-MN, Vance JM, Nance MA, Pericak-Vance MA, Marchuk DA (2001) Identification and expression analysis of spastin gene mutations in hereditary spastic paraplegia. Am J Hum Genet 68:1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]