Abstract
The adenovirus (AdV) hexon constitutes the major virus capsid protein. The epitopes located on the hexon protein are targets of neutralizing antibodies in vivo, serve in the recognition by cytotoxic T cells, and provide the basis for the classification of adenoviruses into the 51 serotypes known to date. We have sequenced the entire hexon gene from human serotypes with incomplete or no sequence information available (n = 34) and performed a comparative analysis of all sequences. The overall sequence divergence between the 51 human serotypes ranged from 0.7 to 25.4% at the protein level. The sequence information has been exploited to assess the phylogeny of the adenovirus family, and protein distances between the six AdV species (A to F) and among individual serotypes within each species were calculated. The analysis revealed that the differences among serotypes within individual species range from 0.3 to 5.4% in the conserved regions (765 amino acids [aa]) and from 1.5 to 59.6% in the variable regions (154 to 221 aa). Serotypes of different species showed an expectedly greater divergence both in the conserved (5.9 to 12.3%) and variable (49.0 to 74.7%) regions. Construction of a phylogenetic tree revealed three major clades comprising the species B+D+E, A+F, and C, respectively. For serotypes 50 and 51, the original assignment to species B and D, respectively, is not in accordance with the hexon DNA and protein sequence data, which placed serotype 50 within species D and serotype 51 within species B. Moreover, the hexon gene of serotype 16, a member of species B, was identified as the product of recombination between sequences of species B and E. In addition to providing a basis for improved molecular diagnostics and classification, the elucidation of the complete hexon gene sequence in all AdV serotypes yields information on putative epitopes for virus recognition, which may have important implications for future treatment strategies permitting efficient targeting of any AdV serotype.
Human adenoviruses (AdV) represent a large family, currently comprising 51 different serotypes, which are divided into six species (designated A to F) based on different oncogenic, hemagglutinating, morphological, and DNA sequence properties. The prevalence of AdV infections is high, as revealed by serological studies (27). The most common sites of infection in immunocompetent individuals include the gastrointestinal tract, the upper respiratory tract, and the eyes (1, 9, 22). In the presence of a functional immune system, AdV infections are not associated with life-threatening disease, but latent infections with persistence of the viral genome, involving particularly species C, have been found in about 80% of the individuals investigated (12). Adenoviruses were described to enter human cells either by the coxsackievirus and adenovirus receptor CAR (3) or by the cell surface protein CD46 (11), which are present on most human cell types (23). Since adenovirus-based vectors are not regarded as harmful to healthy individuals, they are intensively studied for possible applications in gene therapy. For this purpose, AdV serotypes 2 and 5, belonging to species C, are predominantly being exploited (32). Hence, these two serotypes and serotype 12, which belongs to species A and has been described to have oncogenic properties (14), are well characterized on the genomic level. However, for many other human AdV serotypes only limited or no sequence information has been available.
Adenoviruses are nonenveloped and display an icosahedral capsid structure surrounding the viral genome, which consists of linear double-stranded DNA displaying a length ranging between 35 to 38 kb. The virus capsid is composed of three different proteins: 12 fiber attachment proteins associated with 12 penton base proteins, which are involved in the recognition and the interaction with cellular receptors, and 240 hexon proteins comprising 919 to 968 amino acids (aa), which form the main capsid component (27). Electron microscopy and X-ray crystallography of human AdV serotype 2 revealed that the hexon protein structure contains two pedestal regions and four loops. Loop 1 (aa 131 to 331) and loop 2 (aa 423 to 477) of the hexon protein are projected away from the virus surface (2, 25) and contain serotype-specific epitopes (33) encoded by the so-called hypervariable regions (HVRs) of the hexon gene (5). Owing to its hypervariable regions, the hexon protein is the most important part of the adenovirus proteome for the classification and recognition of individual serotypes. The remaining parts of the hexon protein (765 aa) elucidated to date show relatively little variability among different AdV serotypes (5), thus indicating that the hexon gene might be the most highly conserved component of the adenoviral genome. However, the number of AdV serotypes with complete hexon gene sequence information has been very limited. In particular, very little sequence information has been available on serotypes of species D, the largest group among human adenoviruses. The high variability of certain hexon DNA and protein sequences may imply distinct properties of different serotypes. Indeed, certain AdV types are known to cause specific diseases in immunocompetent individuals, such as the serotypes E04 and B07, which cause acute respiratory infections (6), or serotype D08, which is known to cause highly contagious keratoconjunctivitis (1). By contrast, relatively little is known about the relevance of many other AdV serotypes with regard to their infectious potential and the possible oncogenic properties in humans (15, 17).
Complete sequence information on the hexon gene, particularly with regard to the HVRs (26), could significantly expand our understanding of serotype-related characteristics at the molecular level. Additionally, the comprehensive sequence information would permit the establishment of highly specific detection assays covering all of the human adenovirus serotypes. Such diagnostic tests are of great importance in the surveillance of infections in immunosuppressed patients, where adenoviruses play an important role as pathogens, causing a life-threatening disease (4, 20, 29, 30). Moreover, detailed characterization of the adenovirus hexon gene might be helpful in the context of antiviral therapy. Currently available virustatic agents, such as cidofovir or ribavirin, are known to display limited efficacy in the treatment of invasive infections in immunosuppressed patients (17, 18, 21). There is, hence, growing interest in the implementation of specific cytotoxic T-cells in anti-adenoviral treatment. A recent study revealed that the AdV epitopes required for T-cell recognition reside within the hexon gene (19). Complete sequence information of the hexon gene in all human AdV serotypes may therefore be of importance for the prediction of epitopes relevant for efficient interaction with cytotoxic T cells and thus for the expected response to cell therapy.
The task of the current work was the identification of the complete hexon gene sequences of all human adenoviruses, including primarily the 34 serotypes with limited or no sequence information available. The implications of comparative analysis of hexon gene sequences across the entire spectrum of human adenoviruses are discussed with regard to (i) the elucidation of the phylogeny of the adenovirus family, (ii) the taxonomy of AdV serotypes, and (iii) clinical aspects including diagnosis and therapy.
MATERIALS AND METHODS
Virus strains and isolation of DNA.
Adenovirus reference strains from all 51 serotypes were kindly provided by H. Niesters (Department of Virology, University of Rotterdam, The Netherlands). Additionally, reference strains for the virus serotypes B16, B50, and D51 were provided by A. Heim (Department of Virology, Medical University Hannover, Germany). All strains were obtained as supernatants of culture cells infected and lysed by the respective virus type. Adenovirus DNA was extracted using the QIAmp DNA Blood Mini kit (QIAGEN, Hilden, Germany) according to the manufacturer's recommendations.
Sequencing strategy and PCR.
A number of primers, including degenerate sequences to account for mismatched nucleotides between individual serotypes, were designed for each AdV species on the basis of sequence data available from the National Center for Biotechnology Information (NCBI) database. The length of the primers targeting selected regions of the hexon gene ranged from 20 to 25 bp. Four fragments of approximately 1 kb in size spanning the entire hexon gene were amplified. The amplicons contained overlapping regions of about 300 bp, and in some instances additional overlapping fragments have been sequenced to provide reliable sequence data. In total, 45 different primer pairs were employed to amplify overlapping fragments of the complete hexon genes for sequence analysis of 34 different serotypes. PCRs were set up in a total volume of 100 μl, including 2 μl virus DNA solution, 2 U QIAGEN HotStarTaq DNA Polymerase (QIAGEN, Hilden, Germany), the appropriate buffer (QIAGEN, Hilden, Germany), deoxynucleotide triphosphates (Invitrogen GmbH), and primers at 10 pmol per reaction. The amplification profile included an initial denaturation step at 96°C for 10 min, followed by 35 cycles with denaturation at 95°C for 30 s, annealing at 45°C for 45 s, and extension at 72°C for 2 min. Direct sequencing of the purified PCR products was done in a specialized commercial laboratory (Vienna Bio Center, Vienna, Austria). All fragments were sequenced in both directions. In total, more than 200 overlapping hexon gene fragments representing a total of >300,000 nucleotides (nt) were subjected to sequence analysis to establish a reliable sequence database for the complete hexon gene in all human AdV serotypes, in which only partial or no sequence information has been available (n = 34).
Alignments and phylogenetic analysis.
A DNA alignment was performed with newly identified hexon gene sequences from 34 human adenovirus serotypes, together with the available sequence data from the NCBI nucleotide database covering the serotypes C01, C02, E04, C05, B03, B07, D09, B11, C12, B16, D17, B21, B34, B35, F40, F41, and D48 using the BioEdit software package, version 5.0.9 (13). In the variable sections, sequence divergence proved to be too high to obtain a reasonable alignment. The DNA sequences were therefore translated into protein sequences, which were used for further phylogenetic analysis. Neighbor-joining (NJ) and maximum parsimony (MP) dendrograms were calculated using the PAUP software package, version 4.0b10 (29). Bootstrap values were calculated from 100 replicates. NCBI GenBank accession numbers of previously analyzed hexon gene sequences of human adenoviruses are AF534906, AJ293905, X76549, X84646, J01966, AF515814, AJ854486, AF532578, NC_001460, X74662, BK000406, AY008279, AB052911, AB052912, X51782, D13781, and U20821. In addition, bovine (Bos taurus; AAF82136) and equine (Equus caballus; AAB88060) adenovirus protein sequences were used as outgroups to root the phylogenetic tree. In addition to the hexon, fiber and penton sequences of all adenovirus serotypes available in public databases were used for phylogenetic analysis. The sequence data were derived from NCBI GenBank under the following accession numbers: fiber gene, X73487, X76548, X01998, AY921622, AB162822, AB065116, AB073632, U06107, AB073168, AY271307, AF534906, AJ278923, AY224419, AB125751, X74660, X74659, X76706, Y14241, X94485, Y14242, AF447393, X94484, X76547, X16583, and L19443; penton gene, X73487, Z29487, AD001675, AJ293911, M22141, AJ249343, AJ854486, AJ296009, AF217410, AJ296010, AJ296012, AY458656, and AF105145.
The DNA and protein alignments of all adenovirus serotypes are displayed on the CCRI webpage (http://www.ccri.at/frameset4.html).
Nucleotide sequence accession numbers.
The sequences determined in the course of the present study are registered under the following GenBank accession numbers: DQ149610 to DQ149643.
RESULTS
AdV hexon protein sequence analysis.
An alignment of hexon proteins of the 34 AdV serotypes sequenced in our laboratory was performed together with the 17 already known hexon DNA sequences, which had been derived from public databases and translated into putative protein sequences. Two nonhuman hexon proteins derived from bovine and equine adenoviruses were included in the comparative analysis. The sequence divergence across the complete hexon protein among all 51 human adenovirus serotypes ranged from 0.7 to 25.4%. The newly sequenced AdV serotypes confirmed the general pattern of conserved and highly variable regions in the hexon protein described previously (5, 21). Based on the sequence analysis, we divided the protein into four conserved (C1 to C4) and three variable regions (V1 to V3). The conserved regions (765 aa) have a constant length, and the protein sequences differ by less than 15%. Taking the hexon of serotype 2 (C02) as a positional reference, C1 extends from 1 to 137 aa, C2 from 222 to 248 aa, C3 from 317 to 419 aa, and C4 from 461 to 961 aa. The variable regions, which are interspersed between the constant regions (Fig. 1), vary considerably in length (range, 154 to 203 aa) and sequence, revealing a divergence of up to 59.6% within and up to 74.7% between species. When using the C02 sequence as a reference, V1 extends from 138 to 221 aa, V2 from 249 to 316 aa, and V3 from 420 to 460 aa. This classification differs somewhat from earlier descriptions of variable hexon regions (5, 23) but fits in with the three-dimensional structure of the hexon gene, according to which the variable regions V1 and V2, as described herein, are located on loop 1, while the variable region V3 resides on loop 2 of the hexon protein. Hence, the variable regions V1 to V3 are located on outwards-oriented positions of the hexon protein on the virus surface and represent potential epitopes for recognition by neutralizing antibodies (31).
FIG. 1.
Schematic illustration of the hexon protein structure. Variable regions (V1 to V3) are located in the loops designated 1 and 2 and are therefore outwardly oriented on the surface of the virus particle. The indicated amino acid positions are based on the reference serotype C02.
Phylogenetic analysis.
Based on the protein alignment, distances between individual sequences were calculated separately for the conserved (Table 1) and the variable regions (Table 2). In the conserved regions C1 to C4, which extend over 765 aa of the hexon protein, the highest interspecies similarity observed was between AdV serotypes of the species B, E, and D (average divergence ranged from 6.8% to 7.3%). Species A and F also appear to be closely related (divergence, 7.4%), whereas species C is separated from the rest (11.2 to 12.0% divergence). The divergence between species C and all others is about half of that between human and bovine or equine adenoviruses, indicating a rather early phylogenetic differentiation of the human adenovirus species. Not surprisingly, the distances among serotypes within individual species (Table 3) are rather low in the conserved regions C1 to C4. The lowest average divergence was observed between serotypes of species D (range, 0.3 to 2.7%), which is remarkable because this species comprises the highest number of serotypes by far. The greatest divergence was detected between the serotypes of species B, which ranged from 0.5 to 5.4%. By contrast, the variable regions V1 to V3 showed a relatively high average divergence even within individual species. The most pronounced divergence was present between the two serotypes of species F (52.8%), whereas the smallest divergence was observed within species A (38.1%). The divergence of protein sequences within the variable regions between AdV serotypes of different species was in the range of 49.0 to 74.7% (Table 2). The highest value is close to the divergence between human and bovine or equine adenoviruses. A significant part of the divergence is attributable to insertions or deletions, which account for 14.4% to 42.2% of the total difference.
TABLE 1.
Average protein divergence (%) in conserved sections (765 aa) between the five major clades and the two outgroup sequences (Bos and Equus)
| Clade or sequence | Ba | E | Da | F | A | C | Bos | Equus |
|---|---|---|---|---|---|---|---|---|
| B | 6.81 | 7.20 | 10.40 | 11.11 | 11.55 | 21.18 | 24.02 | |
| E | 5.9-7.5 | 7.30 | 10.59 | 11.02 | 11.57 | 21.57 | 23.92 | |
| D | 6.1-8.6 | 6.8-8.1 | 9.10 | 9.25 | 11.17 | 20.89 | 23.73 | |
| F | 9.9-11.2 | 10.2-11.0 | 8.5-9.7 | 7.36 | 11.98 | 20.39 | 24.05 | |
| A | 10.4-11.8 | 10.7-11.2 | 8.4-10.2 | 7.1-7.8 | 11.64 | 21.44 | 23.49 | |
| C | 11.1-12.2 | 11.4-11.6 | 10.6-12.0 | 11.6-12.3 | 11.2-12.0 | 21.31 | 24.51 | |
| Bos | 20.7-21.6 | 20.4-21.2 | 20.1-20.7 | 21.3-21.6 | 21.2-21.6 | 24.41 | ||
| Equus | 23.5-24.4 | 23.3-24.2 | 23.9-24.2 | 23.1-24.1 | 24.2-24.7 |
Sequence B50 is included in clade D, and sequence D51 is in clade B.
TABLE 2.
Average protein divergence (%) in variable sections (154 to 221 aa) between the five major clades and the two outgroup sequences (Bos and Equus)
| Clade or sequence | Ba | E | Da | F | A | C | Bos | Equus |
|---|---|---|---|---|---|---|---|---|
| B | 55.30 | 59.07 | 61.31 | 65.48 | 69.33 | 73.08 | 63.93 | |
| E | 49.0-61.0 | 53.06 | 60.06 | 64.02 | 67.56 | 72.83 | 68.39 | |
| D | 50.2-66.0 | 49.0-58.5 | 63.13 | 63.82 | 68.56 | 73.73 | 68.31 | |
| F | 54.8-65.7 | 58.6-61.5 | 56.5-68.5 | 56.59 | 63.37 | 68.86 | 71.08 | |
| A | 59.5-70.5 | 62.5-65.9 | 55.8-69.3 | 53.5-61.0 | 64.72 | 64.49 | 69.73 | |
| C | 65.2-74.2 | 66.5-69.3 | 63.6-74.7 | 61.1-65.3 | 60.3-70.6 | 73.94 | 73.04 | |
| Bos | 67.8-77.7 | 71.1-77.7 | 67.1-70.7 | 67.1-67.7 | 73.1-75.5 | 68.18 | ||
| Equus | 57.6-68.1 | 63.3-73.9 | 69.9-72.3 | 68.7-70.6 | 70.5-76.5 |
Sequence B50 is included in clade D, and sequence D51 is in clade B.
TABLE 3.
Average protein divergence (% diff) and ranges (%) within clades
| Clade | Protein (conserved sections)
|
Protein (variable sections)
|
||
|---|---|---|---|---|
| % Diff | Range | % Diff | Range | |
| A | 2.92 | 2.1-3.5 | 38.10 | 19.9-48.4 |
| Ba,b | 3.37 | 0.5-5.4 | 39.82 | 17.3-59.6 |
| C | 1.46 | 0.9-2.0 | 47.69 | 41.6-54.5 |
| Da | 1.45 | 0.3-2.7 | 40.80 | 1.5-56.9 |
| F | 2.61 | 52.76 | ||
Sequence B50 is included in clade D, and sequence D51 is in clade B.
The recombined sequence B16 has been excluded from these comparisons.
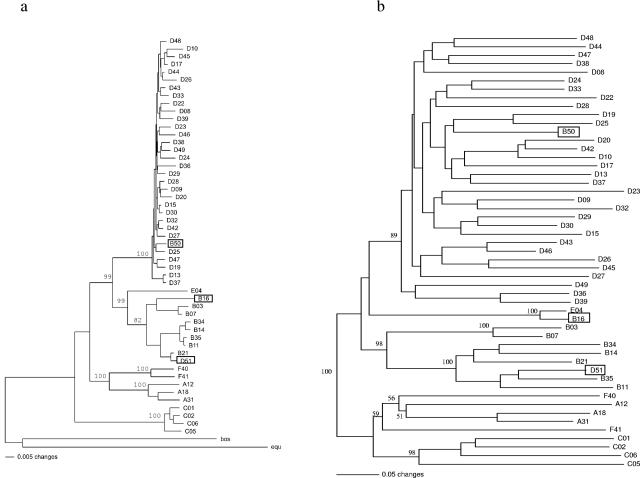
Based on the protein sequence data, phylogenetic trees were constructed. Neighbor-joining (NJ) trees were calculated separately for the conserved regions (length of alignment, 765 aa; Fig. 2a) and for the variable regions (length of alignment, 229 aa; Fig. 2b). For the tree based on the conserved regions, the bovine and equine hexon sequences were used as outgroups to root the tree. For the variable regions, alignment of animal and human adenovirus sequences was not feasible, and midpoint rooting was therefore employed.
FIG. 2.
Phylogenetic analysis of AdV hexon. (a) NJ tree based on the conserved regions (765 aa) of hexon protein sequences from all 51 human serotypes as well as bovine and equine adenoviruses. Bootstrap values are given for the basal nodes of each species (A to F). (b) NJ tree based on the variable regions (length of the alignment, 229 aa) from all 51 human serotypes. The positions of the framed serotypes (D51 and B50 in both trees and B16 in tree b) are not in accordance with their previous species assignments.
The NJ tree derived from the conserved regions shows six well-defined clades, which are supported by high bootstrap values (99 to 100%) (Fig. 2a). With the exception of serotypes B50 and D51, which will be addressed separately in the following section, the clades correspond to the six species A to F. The basal node separates species C from the rest, which is further divided into the groups A+F and B+E+D. Species E, which is represented by a single serotype (E04), is closer to species B than to D. The D clade (which includes B50) forms a homogeneous cluster of closely related sequences. Due to the high sequence similarity, the number of phylogenetically informative positions is limited and the relationships within the clade are thus not resolved. Therefore, NJ and MP trees of the D clade were also calculated from the DNA sequences (data not shown), where the p-distances ranged from 1.0% to 7.1%. However, since the conserved sections of the hexon protein are apparently under strong functional constraints, most nucleotide changes are synonymous substitutions and the variable sites therefore have reached saturation for the deeper splits already. Although there is no resolution of the basal branching points within the D clade, some groupings are consistently found both in NJ and MP analyses. The groups D13+D37 (bootstrap values: NJ, 99; MP, 99; p-distance, 2.6%), D15+D29+D30 (NJ, 92; MP, 90; p-distance, 1.6%), and D44+D48 (NJ, 86; MP, 96; p-distance, 2.4%) are well supported, whereas only weak support was obtained for the groups D9+D10 (NJ, 58; MP, 63; p-distance, 3.4%) and D39+D43 (NJ, <50; MP, 57; p-distance, 3.5%). The branching pattern within the D clade is characterized by short distances between nodes compared to the lengths of the terminal branches. This finding suggests that over a rather short period of time a radiation burst has occurred within species D giving rise to the high number of serotypes observed today. In contrast to the D clade, the B clade is divided into two clearly separated subclades consisting of the serotypes B03+B07+B16 and B14+B34+B11+B35+B21+B51. According to the sequence divergence (about 5% difference at the protein level), the split between these two subclades must be considerably older than the radiation within the D clade.
Although the alignment was problematic in some sections and the sequence is rather short, the general structure of the phylogenetic tree obtained from the variable regions (Fig. 2b) differs only slightly from that based on the conserved regions. The clades D, B, and C are well supported. Clade A is only marginally supported and clade F is disrupted, but the bootstrap values for this rearrangement are low. The relationships within clades are not in accordance with those in Fig. 2a. Nevertheless, within the D clade the groups D13+D37, D15+D29+D30, and D44+D48 can still be recognized, and within the B clade the separation into two subclades is confirmed. The positions of B50 in clade D and D51 in clade B are consistent with the topology in Fig. 2a. Interestingly, B16 clusters with E04.
Phylogenetic analysis of two other capsid proteins, fiber and penton.
Fiber gene sequences of 25 different serotypes and penton gene sequences of 13 serotypes derived from public databases were translated into putative protein sequences, and phylogenetic trees (NJ and MP) were calculated (data not shown). Both data sets included representatives of all six species. Unfortunately, no penton sequences are available for serotypes B50 and D51, where the hexon data indicate affiliation to a different species. With the penton sequences, a reasonable alignment of the 13 proteins was obtained over the entire length (588 aa). Sequence divergence varied from 1.7 to 32.8% and thus proved to be somewhat higher than that among the hexon proteins. Nevertheless, the results confirmed the classification into the three major clades A+F, C, and B+D+E and the assignment of the serotypes to their respective species. In contrast to the penton proteins, the 25 fiber proteins differed considerably in length (319 aa for B3, 582 aa for C2) as well as in sequence. Reasonable alignments were only achieved when the groups A+F+C and B+D+E were treated separately. Even then, only partial sequences could be used for the calculation of phylogenetic trees. The results corroborate the species assignment of the various serotypes. The fiber protein of serotype B16 (partial sequence of 111 aa available only), which was not unequivocally assigned in the hexon trees (see also the discussion below), clearly belongs to species B. The results also show that the hexon protein is more appropriate for phylogenetic analyses. Among the three AdV protein sequences compared, conserved sections of the hexon protein permitting unequivocal alignment extend over 765 aa. This sequence is considerably longer than that in the penton or fiber protein and therefore provides a much higher number of phylogenetically informative sites. Moreover, sequence similarity within the hexon is higher than in the other two proteins, where the deeper nodes in the trees are more affected by saturation of amino acid replacements. Finally, because of the pronounced length variation among AdV species with available sequence information, the fiber proteins cannot be employed for phylogenetic analyses.
Reclassification of the serotypes B50 and D51 on the basis of the hexon protein sequence.
The phylogenetic analysis presented has confirmed earlier classifications of most adenovirus serotypes. Two AdV serotypes, B50 and D51, however, clustered with serotypes of species different from the original assignment: serotype 50, previously described to belong to species B (8), clustered with serotypes of species D, and serotype 51, described earlier as a representative of species D (8), clustered together with serotypes of species B. These results are indicated in Fig. 2a and b, demonstrating that these two serotypes have greater similarity to species other than those originally described within both conserved and variable regions of the hexon protein. Table 4 gives an overview of the closest relatives to the B50 and D51 protein sequences based on conserved and variable regions, respectively. To exclude the possibility of erroneous numbering of the two serotypes, either in the laboratory of origin or in our center, we have obtained the AdV serotypes B50 and D51 from two independent sources and sequenced the variable regions for close comparison. The appropriate DNA alignments revealed that no inadvertent exchange of serotype numbers had occurred. Careful comparison of the DNA sequences showed, however, that the reference strains obtained from a center in The Netherlands and the second set provided to us by a center in Germany (see Materials and Methods) differed by a small number of mutations (data not shown). These observations indicate that the sets of reference strains of the two serotypes represent different substrains of the respective viruses and suggest that our assignment of the serotypes 50 and 51 to other AdV species was not attributable to an error in numbering.
TABLE 4.
Assignment of the sequences B50, D51, and B16 according to similarities at the protein level
| Sequence | Conserved sections
|
Variable sections
|
||||||
|---|---|---|---|---|---|---|---|---|
| Closest relative | % Diffa | Original assignment (reference) | Mean % diffb | Closest relative | % Diff | Original assignment (reference) | Mean % diff | |
| B50 | D15 | 0.78 | B clade (8) | 7.01 | D15, D29, D30 | 30.48 | B clade (8) | 56.69 |
| D51 | B21 | 0.52 | D clade (32) | 6.65 | B21 | 36.56 | D clade (32) | 54.46 |
| B16 | B07, B11 | 3.53 | B clade (8) | 3.77 | E04 | 10.29 | B clade (8) | 53.90 |
Difference from the most similar sequence.
Mean difference from the clade to which the respective sequence has been previously assigned.
Adenovirus B16: a recombinant serotype?
The complete hexon gene sequence information and the phylogenetic trees revealed that, when the conserved regions were analyzed, AdV serotype 16, a representative of species B (24), clustered together with other B serotypes (Fig. 2a). On the basis of the variable regions, however, it was found to cluster with serotype E04, the only representative of species E (Fig. 2b). A high similarity between the serotypes B16 and E04 within loops 1 and 2 has been described previously (24). Table 4 gives details on the composite character of the B16 protein sequence. A protein alignment of hexon sequences derived from all B serotypes (including serotype 51 and excluding serotype 50) together with the E serotype E04 shows high similarity between B16 and the other B serotypes inside the C1 region. At the C-terminal end of C1, exactly at amino acid position 133 displaying a cysteine residue, there is a shift in homology. The adjacent regions V1, C2, V2, and C3 display higher similarity of B16 to the serotype E04. At the N-terminal end of C4, at position 489 displaying a serine residue, the similarity to serotype E04 is over and the downstream sequences again display high similarity to other serotypes of species B. This observation suggests that serotype B16 could be a recombinant adenovirus, composed of sequences derived from species B and E, with breakpoints located close to positions 132 and 488. Based on the DNA alignment of this set of serotypes (data not shown), we were able to determine the breakpoint locations more precisely: nucleotide positions 300 to 308 within C1 and 25 to 41 in C4 (Table 5).
TABLE 5.
Shared sequence positions of B16a
| Hexon region | Nucleotide or amino acid positionb
|
DNAc
|
Proteinc
|
|||
|---|---|---|---|---|---|---|
| nt | aa | Species E | Species B | Species E | Species B | |
| C1 (5′ end) | 1-300 | 1-100 | 0 | 14 | 0 | 2 |
| C1 | 301-411 | 101-137 | 14 | 0 | 2 | 0 |
| V1 | 412-615 | 138-205 | 55 | 1 | 24 | 1 |
| C2 | 616-696 | 206-232 | 1 | 1 | 4 | 0 |
| V2 | 697-891 | 233-297 | 34 | 1 | 19 | 0 |
| C3 | 892-1200 | 298-400 | 11 | 0 | 0 | 0 |
| V3 | 1201-1326 | 401-442 | 20 | 3 | 7 | 2 |
| C4 | 1327-1365 | 443-455 | 2 | 0 | 0 | 0 |
| Recombinant region | 301-1365 | 101-455 | 137 | 6 | 56 | 3 |
| C4 (3′-end) | 1366-2820 | 456-940 | 3 | 156 | 1 | 27 |
Based on an alignment of hexon sequence E04 and sequences of the serotypes of species B (including D51).
Positions relate to the DNA and protein sequences of B16.
Numbers indicate how many nucleotide or amino acid positions in B16 were identical to either E or B serotypes.
To verify this notion and to investigate whether this recombination is found in other representatives of serotype B16, we sequenced the regions adjacent to the putative breakpoints in a reference strain of serotype B16 obtained from a second independent source (see Materials and Methods). Sequence analysis revealed the same positions of the putative breakpoints encompassing a fragment with characteristics of variable regions of species E, confined by the constant regions C1 and C4, displaying high homology to other serotypes of species B (data not shown). Nevertheless, a few point mutations (including two missense mutations and one silent mutation) were observed in the C4 region, indicating that a reference substrain of serotype B16 different from the one described earlier has been investigated. The presence of the putative breakpoints at identical positions within the hexon sequence confirmed, however, that AdV serotype B16 may represent a recombinant virus between serotypes of species B and E.
Presence of species-specific domains in the variable regions.
The variable domains representing approximately one-fifth of the hexon protein length show very limited similarity among AdV serotypes belonging to the same or to different species. Despite this fact, the phylogenetic trees (Fig. 2a and b) revealed a nearly identical cluster distribution, regardless of whether the calculations were based on the conserved (C1 to C4) or the variable (V1 to V3) regions, serotype 16 being the only exception. This implies the existence of species-specific domains within the variable regions. A good candidate for such a domain is a short stretch of 5 to 9 amino acids in region V1 (positions 173 to 194 in the reference sequence AdV C02). Flanked by the two motifs THTFG and GLQIG, which are common among all serotypes, this putative species-specific domain seems to be well conserved within species or serotype groups but differs considerably between species (Table 6). In addition, several motifs can be identified which are located in the variable regions of only one species. The most prominent feature of serotypes from species C is the greater length of the hexon protein sequence by 13 or more amino acid residues compared to all other AdV species. This difference is mainly due to a variable stretch of up to 16 acidic glutamate and aspartate residues at the beginning of the V1 region. Serotypes of species D contain the conserved motif QNQICK at the C-terminal end of V3 and the motif DID, which is shared with species B, in the V2 region. Another motif (NEIGV) that is characteristic for species C but absent in the other serotypes is found at the C-terminal end of the V3 region.
TABLE 6.
Putative species-specific sequence motifs in the variable region V1
| Serotype(s) (aa positions in AdVC02) | 5′ Flanking sequence (aa 173-177) | Species specific (aa 178-182) | 3′ Flanking sequencea (aa 183-194) | |
|---|---|---|---|---|
| D | THTFG | VAAMG | GENIT++GLQIG | |
| E04 plus B16 | MHTFG | VAAMPGVT | GKKIEADGLPIG | |
| B (03, 07) | TNTFG | IASMK | G+NITKEGL+IG | |
| B (11, 14, 21, 34, 51) | TYTFG | NAPVKAEAE | ITK+GLPIG | |
| A plus F | T+TFA | QAPYI | G++ITKDGIQVG | |
| C | THVYA | QAPLS | G++ITKEGLQIG |
+, No prevalent amino acid within the respective serotype groups at that position.
DISCUSSION
The comparative sequence analysis performed in this study provides information on the complete hexon protein in the entire spectrum of human adenoviruses. To date, complete hexon gene coding sequences have been available only for a small number of AdV serotypes (5, 7, 16). Partial hexon gene sequences of the conserved region of some AdV serotypes were published recently (28). Previous studies on the hexon gene, based on the limited sequence information available, divided the hexon protein into eight conserved domains and seven hypervariable regions (HVRs) (5, 23). Based on our hexon gene sequence analysis and the alignment of all 51 human serotypes, we propose a different designation of variable and conserved regions for practical reasons. The conserved regions (C1 to C4) appear to be appropriate for the calculation of phylogenetic relationships among all AdV serotypes and to differentiate between the species A to F. The variable regions (V1 to V3), on the other hand, are useful for the characterization of the serotypes within particular species. As these regions correspond quite well to the hexon loops protruding from the capsid surface, they may also contain the specific epitopes that trigger the immune response.
The complete hexon gene sequence data and the results of the comparative analysis have facilitated the development of a highly specific and economic pan-adenovirus molecular detection method (10) (patent pending). More accurate characterization of individual adenovirus types has permitted the reclassification of the serotypes 50 and 51, which had originally been assigned to other species, based on their hemagglutination capacity of rat and monkey erythrocytes and the pattern produced after restriction digestion (8). Sequence analysis of the entire hexon gene revealed that the serotypes 50 and 51 can be assigned to species D and B, respectively, on the basis of our molecular data. Additionally, serotype B16 has been identified as a recombinant virus containing sequences from two different AdV species, B and E, as revealed by comparative sequence analysis of the hexon gene. In addition to the new insights into adenovirus taxonomy, the elucidation of the complete hexon sequence in all human adenoviruses may have an impact on the establishment of specific approaches to adenovirus treatment. In a recently published study, a number of predicted AdV recognition epitopes for cytotoxic T cells have been identified (19). The study revealed the limitations in predicting the recognition of a broad range of adenoviruses by specific lymphocyte subsets generated in vitro if the protein sequence of putative AdV surface epitopes is not known in sufficient detail. In view of the growing importance of cytotoxic T-cell treatment in immunocompromised patients suffering from life-threatening adenovirus infection, improved possibilities of surface epitope prediction could have an important impact on the development of efficient immune therapy approaches. Taken together, the availability of the complete hexon sequences in all human adenoviruses offers new possibilities in adenovirus classification, diagnosis, and characterization of surface epitopes, which may contribute to more successful anti-adenoviral therapy.
Acknowledgments
This work was supported by the Jubiläumsfonds of the National Bank of Austria (grant no. 11168).
REFERENCES
- 1.Adhikary, A. K., T. Inada, U. Banik, A. Mukouyama, Y. Ikeda, M. Noda, T. Ogino, E. Suzuki, T. Kaburaki, J. Numaga, and N. Okabe. 2004. Serological and genetic characterisation of a unique strain of adenovirus involved in an outbreak of epidemic keratoconjunctivitis. J. Clin. Pathol. 57:411-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Athappilly, F. K., R. Murali, J. J. Rux, Z. Cai, and R. M. Burnett. 1994. The refined crystal structure of hexon, the major coat protein of adenovirus type 2, at 2.9 A resolution. J. Mol. Biol. 242:430-455. [DOI] [PubMed] [Google Scholar]
- 3.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 4.Blanke, C., C. Clark, E. R. Broun, G. Tricot, I. Cunningham, K. Cornetta, A. Hedderman, and R. Hromas. 1995. Evolving pathogens in allogeneic bone marrow transplantation: increased fatal adenoviral infections. Am. J. Med. 99:326-328. [DOI] [PubMed] [Google Scholar]
- 5.Crawford-Miksza, L., and D. P. Schnurr. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70:1836-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford-Miksza, L. K., R. N. Nang, and D. P. Schnurr. 1999. Strain variation in adenovirus serotypes 4 and 7a causing acute respiratory disease. J. Clin. Microbiol. 37:1107-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davison, A. J., M. Benko, and B. Harrach. 2003. Genetic content and evolution of adenoviruses. J. Gen. Virol. 84:2895-2908. [DOI] [PubMed] [Google Scholar]
- 8.De Jong, J. C., A. G. Wermenbol, M. W. Verweij-Uijterwaal, K. W. Slaterus, P. Wertheim-Van Dillen, G. J. Van Doornum, S. H. Khoo, and J. C. Hierholzer. 1999. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 37:3940-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durepaire, N., S. Ranger-Rogez, and F. Denis. 1996. Evaluation of rapid culture centrifugation method for adenovirus detection in stools. Diagn. Microbiol. Infect. Dis. 24:25-29. [DOI] [PubMed] [Google Scholar]
- 10.Ebner, K., M. Suda, F. Watzinger, and T. Lion. 2005. Molecular detection and quantitative analysis of the entire spectrum of human adenoviruses by a two-reaction real-time PCR assay. J. Clin. Microbiol. 43:3049-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 12.Garnett, C. T., D. Erdman, W. Xu, and L. R. Gooding. 2002. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J. Virol. 76:10608-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 14.Hilger-Eversheim, K., and W. Doerfler. 1997. Clonal origin of adenovirus type 12-induced hamster tumors: nonspecific chromosomal integration sites of viral DNA. Cancer Res. 57:3001-3009. [PubMed] [Google Scholar]
- 15.Javier, R. T. 1994. Adenovirus type 9 E4 open reading frame 1 encodes a transforming protein required for the production of mammary tumors in rats. J. Virol. 68:3917-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovacs, G. M., A. J. Davison, A. N. Zakhartchouk, and B. Harrach. 2004. Analysis of the first complete genome sequence of an Old World monkey adenovirus reveals a lineage distinct from the six human adenovirus species. J. Gen. Virol. 85:2799-2807. [DOI] [PubMed] [Google Scholar]
- 17.Kuwano, K., M. Kawasaki, R. Kunitake, N. Hagimoto, Y. Nomoto, T. Matsuba, Y. Nakanishi, and N. Hara. 1997. Detection of group C adenovirus DNA in small-cell lung cancer with the nested polymerase chain reaction. J. Cancer Res. Clin. Oncol. 123:377-382. [DOI] [PubMed] [Google Scholar]
- 18.Lankester, A. C., B. Heemskerk, E. C. Claas, M. W. Schilham, M. F. Beersma, R. G. Bredius, M. J. van Tol, and A. C. Kroes. 2004. Effect of ribavirin on the plasma viral DNA load in patients with disseminating adenovirus infection. Clin. Infect. Dis. 38:1521-1525. [DOI] [PubMed] [Google Scholar]
- 19.Leen, A. M., U. Sili, E. F. Vanin, A. M. Jewell, W. Xie, D. Vignali, P. A. Piedra, M. K. Brenner, and C. M. Rooney. 2004. Conserved CTL epitopes on the adenovirus hexon protein expand subgroup cross-reactive and subgroup-specific CD8+ T cells. Blood 104:2432-2440. [DOI] [PubMed] [Google Scholar]
- 20.Lion, T., R. Baumgartinger, F. Watzinger, S. Matthes-Martin, M. Suda, S. Preuner, B. Futterknecht, A. Lawitschka, C. Peters, U. Potschger, and H. Gadner. 2003. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood 102:1114-1120. [DOI] [PubMed] [Google Scholar]
- 21.Ljungman, P. 2004. Treatment of adenovirus infections in the immunocompromised host. Eur. J. Clin. Microbiol. Infect. Dis. 23:583-588. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell, L. S., B. Taylor, W. Reimels, F. F. Barrett, and J. P. Devincenzo. 2000. Adenovirus 7a: a community-acquired outbreak in a children's hospital. Pediatr. Infect. Dis. J. 19:996-1000. [DOI] [PubMed] [Google Scholar]
- 23.Philipson, L., and R. F. Pettersson. 2004. The coxsackie-adenovirus receptor-a new receptor in the immunoglobulin family involved in cell adhesion. Curr. Top. Microbiol. Immunol. 273:87-111. [DOI] [PubMed] [Google Scholar]
- 24.Pring-Akerblom, P., F. E. Trijssenaar, and T. Adrian. 1995. Sequence characterization and comparison of human adenovirus subgenus B and E hexons. Virology 212:232-236. [DOI] [PubMed] [Google Scholar]
- 25.Roberts, M. M., J. L. White, M. G. Grutter, and R. M. Burnett. 1986. Three-dimensional structure of the adenovirus major coat protein hexon. Science 232:1148-1151. [DOI] [PubMed] [Google Scholar]
- 26.Rux, J. J., P. R. Kuser, and R. M. Burnett. 2003. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol. 77:9553-9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shenk, T., and M. S. Horwitz. 2001. Adenoviridae: the viruses and their replication, adenoviruses, p. 2265-2326. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 28.Shimada, Y., T. Ariga, Y. Tagawa, K. Aoki, S. Ohno, and H. Ishiko. 2004. Molecular diagnosis of human adenoviruses D and E by a phylogeny-based classification method using a partial hexon sequence. J. Clin. Microbiol. 42:1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swofford, D. L. 2002. PAUP* -Phylogenetic analysis using parsimony (*and other methods), version 4.0b-10. Sinauer, Sunderland, Mass.
- 30.Teramura, T., M. Naya, T. Yoshihara, G. Kanoh, A. Morimoto, and S. Imashuku. 2004. Adenoviral infection in hematopoietic stem cell transplantation: early diagnosis with quantitative detection of the viral genome in serum and urine. Bone Marrow Transplant. 33:87-92. [DOI] [PubMed] [Google Scholar]
- 31.Varghese, R., Y. Mikyas, P. L. Stewart, and R. Ralston. 2004. Postentry neutralization of adenovirus type 5 by an antihexon antibody. J. Virol. 78:12320-12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wivel, N. A., G. P. Gao, and J. M. Wilson. 1999. Adenovirus vectors: the development of human gene therapy, p. 87-110. In T. Friedman (ed.), The development of human gene therapy. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Wohlfart, C. 1988. Neutralization of adenoviruses: kinetics, stoichiometry, and mechanisms. J. Virol. 62:2321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]