Abstract
The entry of human immunodeficiency virus type 1 (HIV-1) into the cell is initiated by the interaction of the viral surface envelope protein with two cell surface components of the target cell, CD4 and a chemokine coreceptor, usually CXCR4 or CCR5. The natural ligand of CXCR4 is stromal cell-derived factor 1α (SDF-1α). Whereas the overlap between HIV-1 and SDF-1α functional sites on the extracellular domains of CXCR4 has been well documented, it has yet to be determined whether there are sites in the transmembrane (TM) helices of CXCR4 important for HIV-1 and/or SDF-1α functions, and if such sites do exist, whether they are overlapping or distinctive for the separate functions of CXCR4. For this study, by employing alanine-scanning mutagenesis, 125I-SDF-1α competition binding, Ca2+ mobilization, and cell-cell fusion assays, we found that the mutation of many CXCR4 TM residues, including Tyr45, His79, Asp97, Pro163, Trp252, Tyr255, Asp262, Glu288, His294, and Asn298, could selectively decrease HIV-1-mediated cell fusion but not the binding activity of SDF-1α. Phe87 and Phe292, which were involved in SDF-1α binding, did not play a significant role in the coreceptor activity of CXCR4, further demonstrating the disconnection between physiological and pathological activities of CXCR4 TM domains. Our data also show that four mutations of the second extracellular loop, D182A, D187A, F189A, and P191A, could reduce HIV-1 entry without impairing either ligand binding or signaling. Taken together, our first detailed characterization of the different functional roles of CXCR4 TM domains may suggest a mechanistic basis for the discovery of new selective anti-HIV agents.
Chemokines are small soluble proteins of about 70 amino acid residues with a molecular mass of 8 to 10 kDa. They play prominent roles in leukocyte activation and trafficking to sites of inflammation by interacting with chemokine receptors. All known human chemokines are categorized based on the positions of two conserved cysteine residues in their amino (N)-terminal domains. The two major classes are the CXC and CC chemokines (1, 21). The chemokine receptor CXCR4, a member of the superfamily of G-protein-coupled receptors (GPCRs) possessing seven transmembrane (TM) helical domains, specifically binds the CXC chemokine stromal cell-derived factor 1α (SDF-1α), triggering multiple intracellular signals (1, 3, 24). Human immunodeficiency virus type 1 (HIV-1) requires a coreceptor, either CXCR4 or CCR5, for entry into target cells in addition to CD4, the primary receptor on the target cell surface. CXCR4 is the principal coreceptor for the T-cell-line-tropic HIV-1 isolate which is involved in the onset of AIDS-defining symptoms (1, 11, 20).
The HIV-1 envelope (Env) consists of gp120 and gp41. While gp120 contains the CD4 binding site, gp41 contains a hydrophobic fusion peptide directly involved in membrane fusion. In a plausible model, CD4 binding induces conformational changes in gp120 that expose the coreceptor binding determinants. The gp120 interaction with the coreceptor then induces a further conformational change in Env that results in insertion of the fusion peptide into the target cell membrane (1, 29, 30). This suggests an important role for CXCR4 as a potential target to combat the AIDS epidemic. Small-molecule inhibitors of CXCR4 have been described (8, 10, 12, 26, 31, 32). However, there is cause for concern regarding undesired side effects of blocking the normal CXCR4-SDF-1α function, since knockout mice lacking either CXCR4 (28, 34) or SDF-1α (22) die during embryogenesis, with evidence of hematopoietic, cardiac, vascular, and cerebellar defects. As a result, it would be desirable to develop compounds that can target specific regions of CXCR4 that are selective for HIV-1 coreceptor function only and not for the normal function of SDF-1α.
To develop such selective inhibitors of CXCR4, it is essential to first understand the mechanisms by which CXCR4 interacts with HIV-1 gp120 versus the normal ligand SDF-1α. Previous structure-function studies of CXCR4 have shown that there is significant overlap between HIV-1 and chemokine functional sites on the extracellular regions of CXCR4 (4-6, 9, 17, 33). However, it has yet to be determined whether residues and sites located near or within the TM helices of CXCR4 might play any structural and functional roles in ligand interactions, and if they do, whether such roles are different for different ligands (i.e., HIV-1 gp120 and SDF-1α). To address these questions, we constructed a panel of mutations at residues near or within the TM helices based on the following considerations: (i) charged residues such as D97, D171, and E288 might be involved in interactions with the positively charged residues of SDF-1α; (ii) residues such as H79, Y121, W161, Y219, N298, and Y302 are either highly conserved among chemokine receptors or analogous to corresponding sites in other GPCRs found to be functionally important (27); and (iii) residues such as P163 might play a role in affecting the helical conformation of CXCR4. In addition to these TM mutants, several mutants of the second extracellular loop (ECL2) residues were constructed to investigate the role of ECL2 in HIV-1 entry versus natural ligand binding and signaling. Through these studies, we hope to identify potentially different determinants for CXCR4 interactions with HIV-1 gp120 and SDF-1α and eventually use such information for the design of novel inhibitory molecules specifically targeting the CXCR4-HIV-1 gp120 interaction only.
MATERIALS AND METHODS
Materials.
Radioiodinated SDF-1α was purchased from Perkin-Elmer Life Sciences (Boston, MA). Antibodies 12G5 and 44708.111, plasmid pcDNA-CXCR4, HEK 293 cells, murine 3T3-T4 cells, and HeLa cells were obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, National Institutes of Health, Bethesda, MD). Cell culture media and G418 were purchased from CAMBREX (Walkersville, MD). Dulbecco's modified Eagle's medium (DMEM) plus 10% fetal bovine serum and 5% penicillin-streptomycin (DMEM-10) was used to maintain 293, 3T3-T4, and HeLa cells. Recombinant vaccinia viruses encoding three types of envelopes of HIV-1, i.e., vSC60 (IIIB), vBD3 (89.6), and vCB28 (JR-FL), and vTF1.1 encoding T7 RNA polymerase were obtained from Robert W. Doms (University of Pennsylvania, Philadelphia, PA).
Total chemical synthesis of SDF-1α.
Automated stepwise incorporations of protected amino acids were performed in an Applied Biosystems 433A peptide synthesizer (Foster City, CA) using a CLEAR amide resin (Peptides International, Louisville, KY) as the solid support. 9-Fluorenylmethoxy carbonyl chemistry was employed for synthesis. 2-(1H-Benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) and N-hydroxybenzotriazole were used as coupling reagents in the presence of diisopropylethylamine. In certain coupling steps with potentially slow reaction rates, a double coupling followed by capping of the unreacted amino functional groups was performed. After the incorporation of the 50th residue, 2% (vol/vol) dimethyl sulfoxide was introduced to the coupling solution to enhance the coupling reaction. After removal of the N-terminal 9-fluorenylmethoxy carbonyl protection, the protein was cleaved from the resin support by adding a cleavage cocktail of phenol (4% [wt/vol]), thioanisole (5% [vol/vol]), water (5% [vol/vol]), ethanedithiol (2.5% [vol/vol]), triisopropylsilane (1.5% [vol/vol]), and trifluoroacetic acid (TFA; 82% [vol/vol]). The protein was precipitated by adding ice-cold tert-butyl methyl ether and then washed repeatedly by adding cold ether. The crude protein was dissolved in 25% CH3CN in water containing 0.1% TFA before being lyophilized, and it was then dissolved in water and purified using semipreparative reverse-phase high-performance liquid chromatography. The folding of the purified protein was performed in 1 M guanidinium hydrochloride and 0.1 M Trisma base, pH 8.5 (1 mg protein/ml folding buffer), and it was monitored by analytical reverse-phase high-performance liquid chromatography using a Vydac C18 column (0.46 × 15 cm, 5 μm) with a flow rate of 1 ml/min with solvent A (water with 0.1% TFA) and solvent B (20% water in CH3CN with 0.1% TFA), with a linear gradient of 30 to 70% solvent B over 30 min. Protein desalting and purification were performed. The purified protein was characterized by matrix-assisted laser desorption ionization-time of flight mass spectrometry.
Site-directed mutagenesis.
Wild-type CXCR4 was inserted into the pcDNA3 vector as previously described (33). All mutants of CXCR4 were prepared with a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. Most of the mutations were individual substitutions of alanine for the original residues. The mutations were confirmed by sequencing.
Transfection of adherent 293 cells.
Wild-type or mutant CXCR4 was transfected into 293 cells using Tfx-50 reagents (Promega, Madison, WI) according to the manufacturer's instructions. Selective medium containing G418 (800 μg/ml) was used to isolate stably transfected cells. Each stably transfected cell was cloned from a single colony to isolate a colony that displayed a comparable expression level to that of wild-type CXCR4.
Flow cytometry.
Transfected 293 cells (5 × 105 cells/well) were washed with fluorescence-activated cell sorting (FACS) buffer (0.5% bovine serum albumin, 0.05% sodium azide in phosphate-buffered saline [PBS]) twice and incubated with the anti-CXCR4 monoclonal antibody (MAb) 12G5 or 44708.111 (10 μg/ml) for 30 min at 4°C. After being washed twice with FACS buffer, cells were incubated with 10 μg of fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (IgG; Sigma, St. Louis, MO) for 30 min at 4°C. After being washed twice with FACS buffer, cells were fixed in fixing buffer (2% paraformaldehyde in PBS) for 30 min at 4°C before being analyzed on a FACScan flow cytometer (33). At least three independent experiments were performed.
125I-SDF-1α competition binding assay.
Transfected 293 cells were harvested in PBS and washed twice with PBS. Ligand binding experiments were performed using a single concentration (0.2 nM) of 125I-SDF-1α in a final volume of 100 μl binding buffer (50 mM HEPES, pH 7.4, 1 mM CaCl2, 5 mM MgCl2, 0.1% bovine serum albumin) containing 5 × 105 cells/well in the presence of various concentrations of unlabeled SDF-1α. Nonspecific binding was determined by adding 150 nM of unlabeled SDF-1α. Samples were incubated for 60 min at room temperature. After 60 min of incubation, cells were washed twice with 200 μl binding buffer. Bound ligands were determined by counting gamma emissions. At least three independent experiments were performed. The binding data were analyzed using the PRISM program (GraphPad Inc., San Diego, CA).
Intracellular calcium measurement.
Transfected cells (5 × 106 cells/ml) were loaded with 2 μM fura-2/AM (Molecular Probes, Eugene, OR) and 0.01% Pluronic F-127 (Sigma) in Hanks' balanced salt saline (140 mM NaCl, 5 mM KCl, 10 mM HEPES, pH 7.4, 1 mM CaCl2, 1 mM MgCl2, 1 mg/ml glucose, and 0.025% bovine serum albumin) for 20 min at room temperature. The cells were washed twice and resuspended in the same buffer to 106 cells/ml. Fura-2 fluorescence was measured at room temperature with a fluorescence spectrophotometer (ISA SPEX FluoroMax-2) using excitation wavelengths of 340 nm and 380 nm and an emission wavelength of 510 nm. Calibration was performed using 0.2% Triton X-100 for total fluorophore release and 10 mM EGTA to chelate free Ca2+. [Ca2+]i was calculated using the Grynkiewicz equation (13) with a Kd of 135 nM.
Cell-cell fusion assay.
Following a modified procedure published by others (23, 25), HIV-1 Env proteins and T7 RNA polymerase were introduced into HeLa cells by infection with a recombinant vaccinia virus at a multiplicity of infection of 10. Infected HeLa cells were trypsinized, washed with PBS, resuspended in DMEM-10, and incubated overnight at 32°C in the presence of rifampin (100 μg/ml; Sigma). 3T3-T4 cells stably expressing CD4 were cotransfected with 1.5 μg of plasmid encoding wild-type or mutant CXCR4 and luciferase under the control of the T7 promoter, using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). The cells were incubated at 37°C in a CO2 incubator for 24 h. To initiate fusion, HeLa cells were washed at least four times with PBS and resuspended at 2 × 106 cells/ml in DMEM-10 supplemented with rifampin (100 μg/ml) and araC (cytosine β-d-arabinofuranoside, 10 μM; Sigma). The medium for 3T3-T4 cells was replaced with 0.5 ml DMEM-10 supplemented with rifampin and araC per well. HeLa cells (105) were added to each well and incubated at 37°C. After 5 h of fusion, cells were lysed in 160 μl luciferase reporter lysis buffer (Promega) and assayed for luciferase activity using luciferase assay reagent (Promega). At least three independent experiments were performed.
RESULTS AND DISCUSSION
Cell surface expression of CXCR4.
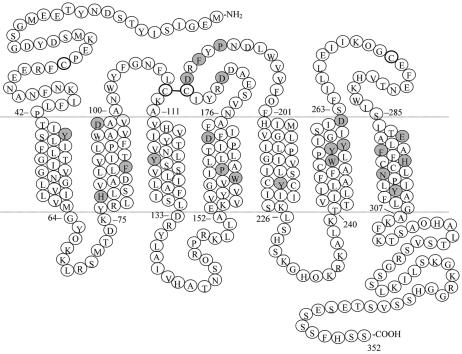
Based on the published structural model of the CXCR4-SDF-1α complex by our laboratory (33), several TM and ECL2 residues were selected for further site-directed mutagenesis studies (Fig. 1). Wild-type or mutant CXCR4 was either stably or transiently transfected into 293 cells. An analysis of cell surface expression by flow cytometry indicated that all the TM and ECL2 mutants displayed expression levels comparable to that of wild-type CXCR4, whether 12G5 or 44708.111 was used (Table 1). One interesting observation was that 12G5 was not able to bind the D182A mutant, which is in agreement with the previous finding that the epitope for 12G5 is contained in the second and third extracellular loops (5, 18). However, when another CXCR4 MAb, 44708.111, was used, the expression level of D182A was comparable to that of wild-type CXCR4.
FIG. 1.
Mutation sites on CXCR4. The sequence and predicted topology of the extracellular, TM, and cytoplasmic regions are shown, with shaded residues indicating the sites of mutations.
TABLE 1.
HIV-1 coreceptor and SDF-1α binding and signaling activities of CXCR4 mutantsa
| CXCR4 domain | CXCR4 mutant | HIV-1 coreceptor activity | SDF-1α binding activity (nM)b | SDF-1α signaling activity | CXCR4 expression level (%)c |
|---|---|---|---|---|---|
| 293 | NA | NA | − | 10.0 ± 0.6d | |
| WT | +++ | 2.2 ± 1.4 | +++ | 100 | |
| TM1 | Y45A | − | 1.0 ± 0.1 | +++ | 139.6 ± 5.8 |
| TM2 | H79A | + | 1.6 ± 0.1 | +++ | 121.9 ± 5.2 |
| F87A | ++ | NAe | NAf | 107.0 ± 4.5 | |
| D97A | − | 2.1 ± 1.1 | +++ | 223.5 ± 7.6 | |
| D97E | + | 5.4 ± 1.2 | +++ | 103.9 ± 6.7 | |
| TM3 | Y121A | +++ | 2.3 ± 0.04 | +++ | 87.5 ± 6.7 |
| TM4 | W161A | ++ | 12.5 ± 3.6 | +++ | 259.0 ± 10.8 |
| P163A | + | 8.6 ± 0.8 | +++ | 112.4 ± 7.3 | |
| D171A | − | NAe | NAf | 123.1 ± 8.0 | |
| TM5 | Y219A | ++ | 4.7 ± 1.3 | +++ | 87.3 ± 6.7 |
| TM6 | W252A | + | 4.6 ± 0.7 | +++ | 84.6 ± 3.5 |
| Y255A | − | 7.9 ± 0.05 | +++ | 64 ± 2.9 | |
| Y256A | +++ | 3.7 ± 0.9 | +++ | 174.2 ± 8.0 | |
| D262A | ++ | 4.4 ± 0.2 | +++ | 137.9 ± 5.8 | |
| D262E | + | 4.2 ± 1.5 | +++ | 177 ± 7.4 | |
| TM7 | E288A | − | 4.4 ± 1.4 | +/++ | 101.2 ± 6.5 |
| E288D | + | 5.9 ± 0.7 | +/++ | 127.5 ± 5.8 | |
| F292A | ++ | NAe | NAf | 63.3 ± 2.9 | |
| H294A | + | 2.8 ± 0.8 | +++ | 112.7 ± 4.7 | |
| N298A | + | 1.3 ± 0.1 | +++ | 139.7 ± 6.4 | |
| Y302A | +++ | 11.5 ± 0.3 | +++ | 141.7 ± 6.5 | |
| ECL2 | D182A | + | 3.4 ± 1.5 | +++ | 98.2 ± 21.8g |
| D187A | − | 4.7 ± 2.1 | +++ | 111.9 ± 23.1 | |
| F189A | − | 8.0 ± 2.1 | +++ | 98.5 ± 23.3 | |
| P191A | + | 2.5 ± 0.4 | +++ | 114.2 ± 26.0 |
Activities that are similar to those of wild-type CXCR4 are denoted “+++,” whereas “++,” “+,” and “−” indicate varying degrees of reduction, as follows: ++, 10 to 30% reduction; +, 30 to 60% reduction; and −, 60 to 100% reduction. Data are means ± standard deviations. NA, not available.
The binding activity of each CXCR4 mutant was determined by its 50% inhibitory concentration.
Mean fluorescence intensity (± standard deviation) for cells expressing mutant CXCR4 relative to cells expressing wild-type CXCR4.
Negative control.
The competition binding curve did not reach a plateau of nonspecific binding.
Ca2+ mobilization assays were not performed on these mutants.
The expression level was determined with another CXCR4 MAb, 44708.111.
Distinct functional sites for SDF-1α on CXCR4 TM domains.
As demonstrated by 125I-SDF-1α competition binding assays (Fig. 2; Table 1), most of the TM mutants had no effect on SDF-1α binding. Only three TM mutants, F87A, D171A, and F292A, drastically reduced the binding activity of SDF-1α, as their binding curves did not reach a plateau of nonspecific binding, even with 300 nM unlabeled SDF-1α. As for nontransfected 293 cells, a negative control, the competition binding curve was unobtainable. According to the previously published structural model of the CXCR4-SDF-1α complex by our laboratory (33), Phe87 is located very close to Phe292. These two residues, possibly with two additional residues, Val88 and Tyr116, may form a hydrophobic site that can accommodate a residue at the N terminus of SDF-1α. Consequently, the mutation of either Phe87 or Phe292 may diminish the hydrophobic interaction between CXCR4 and SDF-1α. As for Asp171, it is located next to a proline residue, which usually forms a kink in the helical structure. The presence of the proline residue may bend Asp171 towards SDF-1α, possibly forming a favorable interaction with SDF-1α.
FIG. 2.
SDF-1α ligand binding curve of a representative CXCR4 mutant, D97E. The 50% inhibitory concentration (IC50) was determined for each mutant by using radioisotope competition binding assays with a single concentration of 125I-SDF-1α (0.2 nM) in the presence of various concentrations of unlabeled SDF-1α. The data shown here are representative of at least three independent experiments.
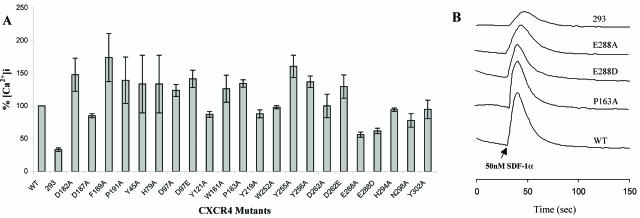
In accordance with the binding data, almost all of the TM mutants showed Ca2+ signaling activities comparable to or higher than that of wild-type CXCR4, indicating that these TM mutants are fully functional for SDF-1α binding (Fig. 3; Table 1). Note, however, that Ca2+ mobilization assays were not performed with the F87A, D171A, and F292A mutants, as they impaired the binding activity of SDF-1α. Among the TM residues examined, only the mutation of Glu288 impaired the signaling activity of CXCR4 to some extent. The difference in signaling activity between the wild type and the E288A or E288D mutant was not due to different levels of CXCR4 expression because E288A and E288D had similar expression levels to that of the wild type (Table 1). Highly conserved residues among many GPCRs, such as Tyr219, Asn298, and Tyr302, which were previously suggested to play a key role in the signal transduction of other receptors (15, 16, 19, 27), did not show any sign of involvement in the activation of CXCR4.
FIG. 3.
SDF-1α signaling activities of CXCR4 mutants. The intracellular Ca2+ influx in 293 cells was measured in response to 50 nM SDF-1α. (A) Changes in [Ca2+]i for all mutants, expressed as percentages of wild-type CXCR4 value. (B) Ca2+ signals of representative mutants. The data shown are representative of at least three independent experiments.
Distinct functional sites for HIV-1 on CXCR4 TM domains.
In contrast to the normal function of SDF-1α binding and signaling activities retained by many TM mutants, the mutations significantly reduced the coreceptor activity of CXCR4, as measured by cell-cell fusion assays (Fig. 4A; Table 1). 3T3-T4 cells cotransfected with plasmids encoding luciferase but not CXCR4 were used as a negative control. Although binding to CD4 clearly triggers conformational changes in Env, only in the presence of an appropriate coreceptor, generally CXCR4 or CCR5, does fusion occur (1). In order to confirm that fusion activity was solely due to the presence of CXCR4 and not CCR5, 3T3-T4 cells cotransfected with plasmids encoding wild-type CXCR4 and luciferase were allowed to undergo fusion with HeLa cells infected with the M-tropic (CCR5-peferring) JR-FL virus. No fusion activity was detected.
FIG. 4.
HIV-1 coreceptor activities of CXCR4 mutants. (A) Coreceptor activities of TM mutants, as determined by cell-cell fusion assays with the 89.6 dual-tropic isolate. (B) Coreceptor activities of ECL2 mutants, as determined by cell-cell fusion assays with the IIIB T-tropic and 89.6 dual-tropic isolates. The results are presented as average percentages of the luciferase activity of wild-type CXCR4. All data shown are means ± standard deviations from at least three independent experiments.
The finding that the coreceptor activities of many TM mutants were significantly impaired for the dual-tropic 89.6 HIV-1 envelope clearly illustrates that HIV-1 gp120 and SDF-1α functions of CXCR4 are separable and mediated by distinct TM sites. The observed sites that are selectively important for HIV-1 gp120 function include negatively charged residues such as Asp97, Asp262, and Glu288. In particular, D97A, E288A, and E288D mutants only retained 20 to 40% of the wild-type coreceptor activity. The only negatively charged residue that was involved in both coreceptor and chemokine receptor activities was Asp171. Our data from cell-cell fusion assays agree with previous reports by Chabot et al. (6) and Brelot et al. (4), in which the importance of Asp97, Asp171, Asp262, and Glu288 in HIV-1 coreceptor activity was demonstrated, although Asp262 showed less importance in our experiments. It is not surprising that these negatively charged residues play an important role in gp120 binding considering that these residues are generally located near the extracellular surfaces of TM domains (Fig. 1) and that the coreceptor binding loops of gp120, particularly the V3 loop, are rich in basic residues. However, despite the fact that both Asp97 and Glu288 showed their involvement in HIV-1 coreceptor activity, we noted that the two mutations of residue 97 (D97A and D97E) showed a significant difference in their effects on coreceptor activity (30% and 70% of the wild-type coreceptor activity, respectively), whereas the difference between the effects of the two mutations of residue 288 (E288A and E288D) was much smaller (30% versus 40%). These results suggest that the presence of a negatively charged group at position 97 is more important for the coreceptor activity than the exact spatial distance of this charged group, while both the negatively charged group and its appropriate spatial distance are required at position 288 for HIV-1 gp120 function.
In addition to the negatively charged TM residues, cell-cell fusion assays showed that some noncharged or positively charged TM residues of CXCR4 play a selective role in HIV-1 coreceptor activity without being involved in SDF-1α binding and signaling (Fig. 4A; Table 1). Two prominent examples include Tyr45 and Tyr255. Alanine substitution of these residues resulted in a significant reduction (>60%) in the coreceptor activity. In addition, H79A, P163A, W252A, H294A, and N298A mutants showed different degrees of reduced coreceptor activity. It is not clear whether these residues are directly involved in gp120 binding, as some, such as His79, Pro163, and Asn298, are located deep inside the TM “barrel,” as shown in Fig. 5, and are unlikely to contact gp120 directly. It is certainly possible that their main role is to help maintain the proper conformations of CXCR4 for gp120 binding. Consequently, the mutations may indirectly affect the conformations of CXCR4 and its ability to mediate HIV-1 entry. Importantly, even the potential conformational changes caused by mutations of His79, Pro163, and Asn298, if they do occur, seem to be quite selective in hindering the gp120 interaction, as SDF-1α binding and signaling were not affected. If this notion of conformational changes is indeed true, then it would strongly suggest that in addition to the distinct residues discussed above, different conformations of CXCR4 are responsible for HIV-1 and SDF-1α activities. The disconnection between the physiological and pathological functions of CXCR4 in the TM domains, as mediated by different residues and even possibly by conformations, is further shown by the finding that F87A and F292A mutations impaired the binding activity of SDF-1α but did not significantly reduce the coreceptor activity of CXCR4.
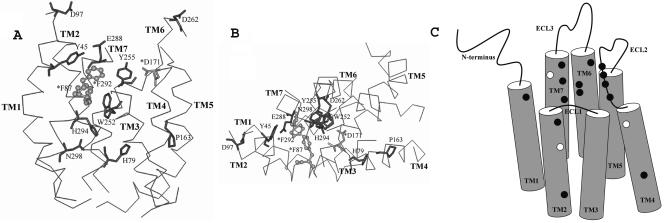
FIG. 5.
Distinct functional sites for HIV-1 gp120 and SDF-1α on a hypothetical structural model of CXCR4. The TM residues Phe87 and Phe292, required for SDF-1α binding only, are highlighted in a lighter color and represented as ball-and-stick figures with asterisks. Asp171, the only residue found to be involved in both HIV-1 gp120 and SDF-1α interactions, is highlighted in a lighter color with an asterisk. The TM residues that are involved in HIV-1 coreceptor activity only are highlighted in the darker color. Only the TM domains of CXCR4, with side (A) and top (B) views, are shown for simplicity. (C) Schematic illustration of the locations of residues found to be important for HIV-1 or SDF-1α on CXCR4 TM and ECL2 domains. The residues required for SDF-1α binding are shown as white spots, while those that are involved in HIV-1 coreceptor activity are shown as black spots. Asp171 is highlighted as a white spot.
The important functional roles of TM residues in CXCR4 are consistent with other lines of evidence previously reported for other GPCRs. For instance, studies of human neurokinin peptide receptor (NK1R) (16) and melanin-concentrating hormone receptor (19) suggested that some residues in the TM regions contribute to ligand binding and activation. A recent study of a CCR5 chimera also suggested that the N termini of macrophage inflammatory protein 1α and RANTES (regulated on activation, normal T cell expressed and secreted) mediate receptor activation by interacting with the TM helix bundle (2). Furthermore, according to a two-site model of the CXCR4-SDF-1α interaction, the motif consisting of amino acids 12 to 17 of SDF-1α first docks on the N-terminal domain of CXCR4, which allows its flexible N terminus to interact with the receptor groove formed by TM domains and/or extracellular loops of CXCR4, thus triggering the receptor function (7, 14). When these results and models published by others are combined with the findings from the present study, one can hypothesize that the N terminus of SDF-1α or certain flexible functional determinants of gp120 may readily interact with some residues in the TM domains of CXCR4. The interaction of the TM domains with HIV-1 gp120 should involve an extensive set of TM residues that are different from those residues required for the interaction with SDF-1α.
Selective role of CXCR4 ECL2 in HIV-1 entry.
We previously reported that three residues in ECL2, i.e., Arg183, Tyr184, and Tyr190, are not involved in SDF-1α binding but play an important role in HIV-1 coreceptor activity (33). This finding suggested, as demonstrated for the TM domains, that the CXCR4-gp120 interaction might be potentially disrupted without impairing its natural chemokine receptor activity. For this report, by studying four additional ECL2 mutants, D182A, D187A, F189A, and P191A, we obtained more extensive evidence demonstrating the selective involvement of ECL2 in HIV-1 coreceptor function. In fact, alterations of ECL2 by these mutations had little effect on SDF-1α binding but significantly diminished the ability of CXCR4 to support HIV-1 entry (Fig. 4B; Table 1). For instance, an alanine substitution for Asp187 or Pro191 resulted in a significant reduction (>60%) in HIV-1 coreceptor activity for both T-cell-tropic IIIB and dual-tropic 89.6 HIV-1 envelope proteins. The mutants D182A and F189A exhibited 40 to 60% of the wild-type coreceptor activity. As expected, the experiments using either the T-cell-tropic IIIB or dual-tropic 89.6 HIV-1 envelope showed similar results for the ECL2 mutants. Thus, only a vaccinia virus encoding the dual-tropic 89.6 HIV-1 envelope was used to test the coreceptor activities of TM mutants as discussed above. The results of this study also provide new insights into the importance of electrostatic interactions between CXCR4 and HIV-1 Env, as charged residues, such as Asp182 and Asp187, in ECL2 contributed to the coreceptor activity. Yet the observation that other mutants, such as F189A and P191A, affected HIV-1 coreceptor activity indicates that the negative charge may not be the only requirement for the ECL2 interaction with HIV-1.
In agreement with the binding data, none of the ECL2 mutants impaired Ca2+ signaling (Fig. 3; Table 1). This is consistent with data on other critical ECL2 residues studied previously (4, 9, 33). Although Brelot et al. reported that Asp187 in ECL2 is required for binding and signaling (4), the D187A mutant was involved in neither binding nor signaling according to our experiments. This discrepancy may be due to the fact that we used stably expressing cell lines for both Ca2+ mobilization and SDF-1α competition binding assays, whereas Brelot et al. employed transiently expressing cells for their assays. Zhou et al. also reported that two ECL2 mutants, R183A and Y184A, have little effect on signaling, although a Y190A mutation in ECL2 impairs the signaling activity of CXCR4 somewhat (33). Moreover, our Ca2+ mobilization and cell-cell fusion assays demonstrate that signaling and coreceptor functions are independent activities of CXCR4.
Conclusion.
We have demonstrated that many residues throughout the CXCR4 TM and ECL2 domains, including Tyr45, His79, Asp97, Pro163, Asp182, Asp187, Phe189, Pro191, Trp252, Tyr255, Asp262, Glu288, His294, and Asn298, are specifically involved in interaction with HIV-1 gp120, as most of these sites did not play a role in either SDF-1α binding or signaling. As shown in Fig. 5, many distinct sites for either HIV-1 or SDF-1α function are located on the upper part of the TM barrel close to the extracellular side or in ECL2. Their role is likely to be involved with direct interactions with different ligands. On the other hand, some residues predicted to be located deep inside the TM barrel and thus unlikely to contact gp120 directly were also able to reduce the coreceptor activity of CXCR4 without impairing the binding and signaling activities of SDF-1α. It is possible that these mutations change the conformations of CXCR4 that are important for HIV-1 but not SDF-1α function. Further work is needed to understand the mechanism of how the function of HIV-1 or SDF-1α is mediated differently by distinct sets of residues and possibly by conformational states. Nevertheless, the findings from the present study provide a basis for the development of new inhibitory agents that modulate the functional sites or conformations of CXCR4 for the purpose of reducing or avoiding the limitations and side effects caused by nonselective inhibitors of this important coreceptor.
Acknowledgments
We thank Chang-Zhi Dong and Santosh Kumar in our laboratory for synthesizing SDF-1α.
This work was supported by NIH grant GM57761 to Z. Huang. S. Tian was supported by NIH grant HL16059 to A. Jonas.
REFERENCES
- 1.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 2.Blanpain, C., B. J. Doranz, A. Bondue, C. Govaerts, A. De Leener, G. Vassart, R. W. Doms, A. Proudfoot, and M. Parmentier. 2003. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J. Biol. Chem. 278:5179-5187. [DOI] [PubMed] [Google Scholar]
- 3.Bleul, C. C., M. Farzan, H. Choe, C. Parolin, I. Clark-Lewis, and J. Sodroski. 1996. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 382:829-833. [DOI] [PubMed] [Google Scholar]
- 4.Brelot, A., N. Heveker, M. Montes, and M. Alizon. 2000. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J. Biol. Chem. 275:23736-23744. [DOI] [PubMed] [Google Scholar]
- 5.Brelot, A., N. Heveker, O. Pleskoff, N. Sol, and M. Alizon. 1997. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J. Virol. 71:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chabot, D. J., P. F. Zhang, G. V. Quinnan, and C. C. Broder. 1999. Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates. J. Virol. 73:6598-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crump, M. P., J. H. Gong, P. Loetscher, K. Rajarathnam, A. Amara, F. Arenzana-Seisdedos, J. L. Virelizier, M. Baggiolini, B. D. Sykes, and I. Clark-Lewis. 1997. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 16:6996-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donzella, G. A., D. Schols, S. W. Lin, J. A. Este, K. A. Nagashima, P. J. Maddon, G. P. Allaway, T. P. Sakmar, G. Henson, E. De Clercq, and J. P. Moore. 1998. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med. 4:72-77. [DOI] [PubMed] [Google Scholar]
- 9.Doranz, B., M. Orsini, J. Turner, T. Hoffman, J. Berson, J. Hoxie, S. Peiper, L. Brass, and R. Doms. 1999. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J. Virol. 73:2752-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doranz, B. J., K. Grovit-Ferbas, M. P. Sharron, S. H. Mao, M. B. Goetz, E. S. Daar, R. W. Doms, and W. A. O'Brien. 1997. A small-molecule inhibitor directed against the chemokine receptor CXCR4 prevents its use as an HIV-1 coreceptor. J. Exp. Med. 186:1395-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fauci, A. S. 1996. Host factors and the pathogenesis of HIV-induced disease. Nature 384:529-534. [DOI] [PubMed] [Google Scholar]
- 12.Fenard, D., G. Lambeau, T. Maurin, J. C. Lefebvre, and A. Doglio. 2001. A peptide derived from bee venom-secreted phospholipase A2 inhibits replication of T-cell tropic HIV-1 strains via interaction with the CXCR4 chemokine receptor. Mol. Pharmacol. 60:341-347. [DOI] [PubMed] [Google Scholar]
- 13.Grynkiewicz, G., M. Poenie, and R. Y. Tsien. 1985. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260:3440-3450. [PubMed] [Google Scholar]
- 14.Gupta, S. K., K. Pillarisetti, R. A. Thomas, and N. Aiyar. 2001. Pharmacological evidence for complex and multiple site interaction of CXCR4 with SDF-1alpha: implications for development of selective CXCR4 antagonists. Immunol. Lett. 78:29-34. [DOI] [PubMed] [Google Scholar]
- 15.Huang, R. R., D. Huang, C. D. Strader, and T. M. Fong. 1994. Conformational compatibility as a basis of differential affinities of tachykinins for the neurokinin-1 receptor. Biochemistry 34:16467-16472. [DOI] [PubMed] [Google Scholar]
- 16.Huang, R. R., P. P. Vicario, C. D. Strader, and T. M. Fong. 1995. Identification of residues involved in ligand binding to the neurokinin-2 receptor. Biochemistry 34:10048-10055. [DOI] [PubMed] [Google Scholar]
- 17.Kajumo, F., D. A. Thompson, Y. Guo, and T. Dragic. 2000. Entry of R5X4 and X4 human immunodeficiency virus type 1 strains is mediated by negatively charged and tyrosine residues in the amino-terminal domain and the second extracellular loop of CXCR4. Virology 271:240-247. [DOI] [PubMed] [Google Scholar]
- 18.Lu, Z., J. F. Berson, Y. Chen, J. D. Turner, T. Zhang, M. Sharron, M. H. Jenks, Z. Wang, J. Kim, J. Rucker, J. A. Hoxie, S. C. Peiper, and R. W. Doms. 1997. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc. Natl. Acad. Sci. USA 94:6426-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macdonald, D., N. Murgolo, R. Zhang, J. P. Durkin, X. Yao, C. D. Strader, and M. P. Graziano. 2000. Molecular characterization of the melanin-concentrating hormone/receptor complex: identification of critical residues involved in binding and activation. Mol. Pharmacol. 58:217-225. [DOI] [PubMed] [Google Scholar]
- 20.Moore, J. P., A. Trkola, and T. Dragic. 1997. Co-receptors for HIV-1 entry. Curr. Opin. Immunol. 4:551-562. [DOI] [PubMed] [Google Scholar]
- 21.Murphy, P. M. 1994. The molecular biology of leukocyte chemoattractant receptors. Annu. Rev. Immunol. 12:593-633. [DOI] [PubMed] [Google Scholar]
- 22.Nagasawa, T., S. Hirota, K. Tachibana, N. Takakura, S. Nishikawa, Y. Kitamura, N. Yoshida, H. Kikutani, and T. Kishimoto. 1996. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature 382:635-638. [DOI] [PubMed] [Google Scholar]
- 23.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oberlin, E., A. Amara, F. Bachelerie, C. Bessia, J. L. Virelizier, F. Arenzana-Seisdedos, O. Schwartz, J. M. Heard, I. Clark-Lewis, D. F. Legler, M. Loetscher, M. Baggiolini, and B. Moser. 1996. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature 382:833-835. [DOI] [PubMed] [Google Scholar]
- 25.Rucker, J., B. Doranz, A. Edinger, D. Long, J. Berson, and R. Doms. 1997. Cell-cell fusion assay to study role of chemokine receptors in human immunodeficiency virus type 1 entry. Methods Enzymol. 288:118-133. [DOI] [PubMed] [Google Scholar]
- 26.Sachpatzidis, A., B. K. Benton, J. P. Manfredi, H. Wang, A. Hamilton, H. G. Dohlman, and E. Lolis. 2003. Identification of allosteric peptide agonists of CXCR4. J. Biol. Chem. 278:896-907. [DOI] [PubMed] [Google Scholar]
- 27.Strader, C. D., T. M. Fong, M. R. Tota, and D. Underwood. 1994. Structure and function of G protein-coupled receptors. Annu. Rev. Biochem. 63:101-132. [DOI] [PubMed] [Google Scholar]
- 28.Tachibana, K., S. Hirota, H. Iizasa, H. Yoshida, K. Kawabata, Y. Kataoka, Y. Kitamura, K. Matsushima, N. Yoshida, S. Nishikawa, T. Kishimoto, and T. Nagasawa. 1998. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature 393:591-594. [DOI] [PubMed] [Google Scholar]
- 29.Wild, C., J. W. Dubay, T. Greenwell, T. J. Baird, T. G. Oas, C. McDanal, E. Hunter, and T. Matthews. 1994. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc. Natl. Acad. Sci. USA 91:12676-12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou, N., Z. Luo, J. Luo, X. Fan, M. Cayabyab, M. Hiraoka, D. Liu, X. Han, J. Pesavento, C. Z. Dong, Y. Wang, J. An, H. Kaji, J. G. Sodroski, and Z. Huang. 2002. Exploring the stereochemistry of CXCR4-peptide recognition and inhibiting HIV-1 entry with D-peptides derived from chemokines. J. Biol. Chem. 277:17476-17485. [DOI] [PubMed] [Google Scholar]
- 32.Zhou, N., Z. Luo, J. Luo, J. W. Hall, and Z. Huang. 2000. A novel peptide antagonist of CXCR4 derived from the N-terminus of the viral chemokine vMIP-II. Biochemistry 39:3782-3787. [DOI] [PubMed] [Google Scholar]
- 33.Zhou, N., Z. Luo, J. Luo, D. Liu, J. W. Hall, R. J. Pomerantz, and Z. Huang. 2001. Structural and functional characterization of human CXCR4 as a chemokine receptor and HIV-1 co-receptor by mutagenesis and molecular modeling studies. J. Biol. Chem. 276:42826-42833. [DOI] [PubMed] [Google Scholar]
- 34.Zou, Y., A. Kottmann, M. Kuroda, I. Taniuchi, and D. Littman. 1998. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature 393:595-599. [DOI] [PubMed] [Google Scholar]