Abstract
Despite recent progress in anti-human immunodeficiency virus (HIV) therapy, drug toxicity and emergence of drug-resistant isolates during long-term treatment of HIV-infected patients necessitate the search for new targets that can be used to develop novel antiviral agents. One such target is the process of nuclear translocation of the HIV preintegration complex. Previously we described a class of arylene bis(methylketone) compounds that inhibit HIV-1 nuclear import by targeting the nuclear localization signal (NLS) in the matrix protein (MA). Here we report a different class of MA NLS-targeting compounds that was selected using computer-assisted drug design. The leading compound from this group, ITI-367, showed potent anti-HIV activity in cultures of T lymphocytes and macrophages and also inhibited HIV-1 replication in ex vivo cultured lymphoid tissue. The virus carrying inactivating mutations in MA NLS was resistant to ITI-367. Analysis by real-time PCR demonstrated that the compound specifically inhibited nuclear import of viral DNA, measured by two-long terminal repeat circle formation. Evidence of the existence of this mechanism was provided by immunofluorescent microscopy, using fluorescently labeled HIV-1, which demonstrated retention of the viral DNA in the cytoplasm of drug-treated macrophages. Compounds inhibiting HIV-1 nuclear import may be attractive candidates for further development.
The step of nuclear translocation of the human immunodeficiency virus type 1 (HIV-1) preintegration complex (PIC) is essential for viral replication, as it allows the PIC to get into contact with the cellular chromatin. A characteristic feature of HIV-1 is its ability to replicate in nondividing cells (14, 35, 60). Replication in nonproliferating cells depends on the aptitude of the virus to transport its PIC into the cell's nucleus through an intact nuclear membrane by an active, energy-dependent mechanism (8). This feature is not shared by oncogenic retroviruses such as murine leukemia virus, which does not use the active nuclear translocation mechanism but relies on the breakage of the nuclear membrane during mitosis to gain access to the nuclear compartment (35, 51). Initially it was presumed that this unusual feature of HIV-1 would manifest itself only in nondividing cells such as macrophages (29, 58). However, recent experiments demonstrated that HIV-1 employs its active nuclear import mechanism for infection of proliferating cells as well (3, 34). Since only a small proportion of T cells in the body divide, the capability to enter the cell nucleus during interphase would be extremely beneficial for the virus and may account for its very high replication rate observed in HIV-infected patients (31, 59).
There is still no consensus about the mechanisms that govern nuclear import of the HIV-1 PIC (43). Three HIV-1 proteins, matrix protein (MA), integrase (IN), and viral protein R (Vpr), have been proposed as karyophilic agents that recruit the cellular nuclear import machinery to the PIC. Another factor that may regulate HIV nuclear import is the specific structure of the viral cDNA intermediate termed “central DNA flap” (62). However, none of these factors appears to be essential for HIV-1 nuclear import, which seems to be regulated by a plethora of redundant determinants (46). While elimination of each individual determinant might have only a partial negative effect on viral nuclear entry, together they ensure the efficiency of this extremely important step for the virus. This redundancy in the regulation of the HIV nuclear import process explains the controversy surrounding this issue. The story of MA is a good example of this controversy. Historically, MA was the first protein implicated in HIV-1 nuclear import (7, 58). However, several later reports questioned the role of MA in HIV-1 nuclear import (18, 19). The main argument against MA being the principal HIV nuclear-targeting protein was the finding that the virus lacking most of the MA was still capable of infecting nondividing cells, albeit with a greatly reduced efficiency (49). As with other HIV proteins involved in PIC nuclear import, MA, while clearly required for efficient nuclear import, appears to be only one of several factors regulating this process (6).
Whereas most of the MA in the virion localizes outside the core and forms the layer between the viral capsid and the envelope (hence its name), some MA molecules are found in tight association with the HIV core and the PIC (9, 41). The MA protein contains two well-defined basic-type nuclear localization signals (NLSs) with the core amino acid sequences of 26KKKYK (NLS-1) and 110KSKKK (NLS-2) (25, 42). Substitutions of selected lysine residues in the MA NLSs resulted in significant reduction in replication of mutated virus (25). NLS-1, 26KKKYK, exhibited less tolerance for such mutations. Import of cellular proteins containing such NLSs across the nuclear pore complex is mediated by special transport proteins called importins or karyopherins, which bind NLS-containing proteins in the cytosol and target them to the nucleus (37). Importin α is an adaptor that ensures binding of a basic NLS substrate to importin β, which is then responsible for docking of importin-NLS protein complexes to nucleoporins (a collective term for nuclear pore complex proteins) and translocation through the pore (50).
Targeting MA NLS-dependent nuclear import by using excess NLS mimetics was shown to inhibit HIV replication (7, 20, 24). The NLS mimetics presumably worked by competing with the MA NLS for binding to importin α. A class of small molecule inhibitors of HIV nuclear import, arylene bis(methylketone) compounds, which directly target the MA NLS, has also been described (12). The lead compound from this group, ITI-002, inactivated the NLS by forming Schiff bases with lysine residues (1), thus neutralizing the positive charges critical for NLS activity. ITI-002 demonstrated potent anti-HIV activity in primary macrophage and peripheral blood mononuclear cell cultures (26).
In this report, we describe another class of small molecule compounds targeting MA NLS. These compounds were selected using a computer-assisted drug design (CADD) approach and are predicted to bind to MA within a structural groove in NLS-1 generated by the inward orientation of the tyrosine 29 residue. The compounds show potent anti-HIV activity and may be good candidates for further preclinical development.
MATERIALS AND METHODS
Computer assisted drug design.
Computational docking of ∼145,000 Available Chemical Directory (ACD) compounds (Available Chemical Directory, 2000 release, MDL Information Systems, San Leandro, CA) was conducted against a solution structure of HIV-1 matrix protein (39) and determined with the aid of crystallographic data (36). Docking calculations were carried out using version 4.0.1 of the DOCK program (15). For these calculations, the putative binding site was converted into an arrangement of six docking spheres and a docking grid was generated from a 23-Å cube covering the site. The ACD compounds were flexibly docked into this site using anchored search (15) and were initially scored using a scheme combining contributions from molecular contacts and force field energy terms. The putative complexes of the top-scoring 1,000 compounds were stored and subjected to visual inspection. On the basis of this analysis, 259 plausible complexes were selected. The 259 compounds were sorted according to their contact scores, and from this list a total of 87 compounds were chosen for acquisition.
HIV-1 strains.
The primary isolate HIV-1M1 was described previously (33). Tumor necrosis factor alpha (TNF-α)-stimulated OM10.1 cells produce HIV-1LAI (10), which is a T-cell line-adapted virus isolate that uses CXCR4 as an entry coreceptor (X4 strain). HIV-1ADA and HIV-1SF162 are macrophage-tropic CCR5-using (R5) strains. NHLXADA is a recombinant R5 strain constructed using components of NL4-3, HLXB2, and ADA viruses (61).
Cells and infection.
CEM CD4+ T cells were infected with human T-lymphotropic virus 1 (HTLV-1) or HIV-1 by cocultivation with gamma-irradiated donor MT-2 cells (28) or TNF-stimulated OM10.1 cells (10), respectively, as previously described (40). The initial ratio of donor to CEM cells was 1:10.
Monocyte-derived macrophages were prepared from peripheral blood mononuclear cells, using adherence to plastic and differentiation in the presence of macrophage colony-stimulating factor essentially as previously described (52). T lymphocytes were isolated from tonsil tissue received through the National Disease Research Interchange (NDRI; Philadelphia, PA) 20 to 28 h postremoval from patients. The tissue was minced and ground between two frosted glass slides with just enough pressure to break the tissue structure and release the cells from the tonsils. The cell homogenate was sequentially filtered through a 100-μm and 40-μm cell strainer to remove any large clumps or aggregates of tissue and cells. The cells were pelleted by centrifugation and resuspended in serum-free media for cryogenic storage until use.
Macrophage cultures were inoculated with HIV-1 (4 × 105 cpm of reverse transcriptase (RT) activity per 106 cells) in the presence of the drug. After an overnight incubation, cells were washed and cultured in the presence of the drug in Dulbecco modified Eagle medium supplemented with 10% heat-inactivated human serum. Every 3 days, half the medium was replenished, and the drug concentration was maintained. Tonsil-derived T lymphocytes were thawed and cultured without exogenous stimulation in Iscove's media with 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 8% heat-inactivated human serum. Cells were treated with ITI-367 for 2 h prior to challenge with the patient HIV-1 isolate M1. Four hours after infection, the cells were washed to remove the virus inoculum and cultured in the presence of ITI-367.
Analysis of HIV-1 infection in ex vivo cultured lymphoid tissue.
Human tonsils were obtained from patients undergoing tonsillectomy. Tissues were dissected into 2-mm blocks and incubated on the top of collagen gels at the air-liquid interface (histoculture) as previously described in detail (21-23). The drugs were added to the medium 3 h prior to infection and were replenished with each medium change. Tissue blocks were infected by slowly applying 3 μl of clarified virus-containing medium on the top of each tissue block. For each experiment, supernatants from 20 tissue blocks (this number was found empirically to produce statistically reliable data [21]) obtained from the same donor were pooled and used for p24 analysis (measured by enzyme-linked immunosorbent assay [ELISA]). All experiments involving ex vivo cultured lymphoid tissue were performed in the laboratory of Leonid Margolis at the NIH/NICHD.
Analysis of PIC-importin α interaction.
Preparation of cytoplasmic PICs and binding to glutathione S-transferase (GST)-importin α fusion protein immobilized on glutathione-coated beads were performed essentially as previously described (26). The beads were then assayed for the presence of HIV-1-specific DNA by real-time PCR, using FOR-LATE (5′-TGTGTGCCCGTCTGTTGT GT-3′) and REV-LATE (5′-GAGTCCTGCGTCGAGAGAGC-3′) primers specific for the late reverse transcription products (11) and the DyNAmo SYBR Green qPCR kit (Finnzymes, Espoo, Finland).
Analysis of HIV-1 reverse transcription and nuclear import.
Macrophages were infected with HIV-1 ADA and cultured in the presence of anti-CD4 monoclonal antibody (MAb) (to block secondary infection) with or without ITI-367 (1μM) or zidovudine (AZT) (1 μM). Seventy-two hours after infection, total DNA was extracted and analyzed by real-time PCR using the TaqMan method with primers and probes specific for late reverse transcripts (57) or two-long terminal repeat (2-LTR) circle forms (32).
Fluorescent labeling of HIV-1 DNA.
HIV-1 DNA was labeled using endogenous reverse transcription as previously described (2). Briefly, HIV-1 ADA was resuspended at an RT activity of 90,000 cpm/μl in 200 μl of 100 mM Tris-HCl (pH 7.5), 10 mM NaCl, 2.5 mM MgCl2, 100 μM deoxynucleoside triphosphate (dA, dG, and dC), 100 μM ChromaTide Alexa Fluor 488-5-dUTP (Molecular Probes, Eugene, OR), 3 mM Spermine, 0.1 mM Spermidine, and 50 μM Melittin in phosphate-buffered saline (PBS) and incubated for 4 h at 37°C. The virus was then centrifuged at 60,000 × g for 2 h at 4°C, resuspended in 200 μl of PBS, and used for infection and analysis.
Fluorescent microscopy.
An aliquot (5 μl) of fluorescently labeled virus was spotted on Poly-L-Lysine-coated coverslips (Becton Dickinson), air dried, fixed in 4% Formalin, treated with 0.2% Triton-PBS for 15 min at room temperature, washed in PBS, and incubated in 4% goat serum-PBS for 1 h at 4°C. p24 was stained by incubation with mouse anti-p24 MAb (18, 54) diluted 1:250 in 4% goat serum-PBS for 1 h at room temperature, followed by Cy5-labeled goat anti-mouse secondary antibody (Jackson Immunoresearch Lab) in 1% goat serum-PBS for 1 h at room temperature. After being washed and air dried, the slips were mounted on a glass slide, using a ProLong Antifade kit (Molecular Probes), and analyzed on a BX-60 Olympus Epi-fluorescence microscope (Melville, NY) with an Evolution MP digital camera using Image-Pro Plus version 4.5 acquisition software (Media Cybernetics, Silver Spring, MD).
For analysis of HIV-infected cells, monocyte-derived macrophages were plated on Poly-L-Lysine-coated coverslips, preincubated with 10 mM ITI-367 for 2 h, and infected with fluorescently labeled HIV-1 in the presence of the drug. Twenty-four hours after infection, the cells were washed and either analyzed or incubated for an additional 48 h with the drug. For immunofluorescent analysis, cells were fixed with 4% Formalin for 20 min, washed, and permeabilized by incubation in 0.1% Triton X-100 for 15 min at room temperature. Nuclei were stained by incubation with TOPRO-3 iodide (Molecular Probe) diluted 4,000 times in PBS for 15 min. After being washed and air dried, the slips were mounted on a glass slide and analyzed as described above.
RESULTS
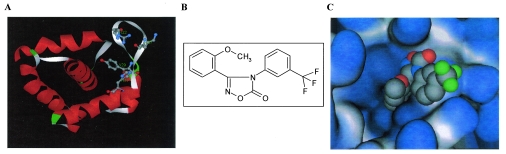
The oxadiazol class of compounds was selected using CADD by modeling a 145,000-compound library on the crystal structure of the HIV-1 matrix protein (30, 38). The oxadiazols were predicted to bind to MA within a structural groove in NLS-1 generated by the inward orientation of the tyrosine residue in position 29 and involving the amino acids arginine 22, lysine 27, and histidine 33 (Fig. 1A). The chemical structure of the lead compound from this class, 3-(2-methoxyphenyl)-4-[3- (trifluoromethyl)phenyl-1,2,4-oxadiazol-5(4H)-1] (ITI-367), is shown in Fig. 1B. The hypothetical orientation of ITI-367 within the structural groove generated by tyrosine 29 is displayed in Fig. 1C.
FIG. 1.
CADD of ITI-367. (A) Hypothetical structural pocket within the N-terminal NLS of HIV-1 MA protein. The amino acids shown (R22, K27, Y29, and H33) participate in the formation of the hypothetical binding pocket for ITI-367. (B) The chemical structure of ITI-367. (C) ITI-367 in its docked (hypothetical) conformation and orientation within the “tyrosine pocket” of HIV-1 MA. In this molecular model, the protein is rendered as a solid molecular surface (light gray/light blue) and ITI-367 is shown in space-filling representation (standard atom coloring: gray, carbon; blue, nitrogen; red, oxygen; green, fluorine).
Inhibition of HIV replication by ITI-367.
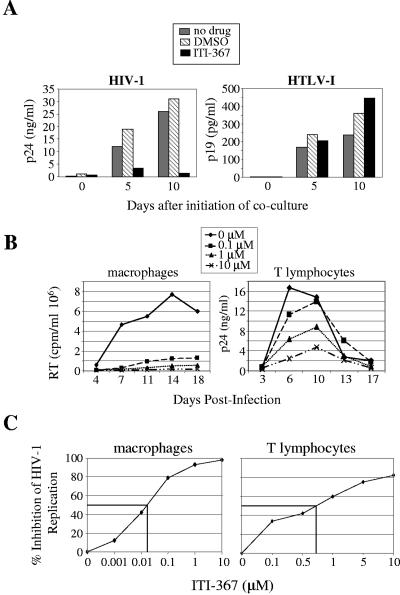
We first tested the effect of ITI-367 on spreading infection of CEM cells by HIV-1 and HTLV-I. CEM cells were cocultured at a ratio of 1:10 with gamma-irradiated MT-2 cells (a source of HTLV-I) or TNF-α-stimulated OM10.1 cells (a source of HIV-1), and viral replication was followed by measuring p19 (for HTLV-I) or p24 (for HIV-1) in the culture supernatant. ITI-367 at a concentration of 10 μM potently inhibited infection by HIV-1, but not by HTLV-I, as measured by ELISA specific for viral proteins p24 (HIV-1) or p19 (HTLV-I) (Fig. 2A). This result indicates that ITI-367 specifically targets HIV-1 replication.
FIG. 2.
ITI-367 inhibits HIV-1 replication. (A) HIV-1-producing OM10.1 cells (left panel) and HTLV-I-producing MT-2 cells (right panel) were gamma irradiated and cocultured with CEM cells without any drug or in the presence of 0.1% dimethyl sulfoxide (DMSO) or 10 μM ITI-367 (prepared from 10 mM stock in DMSO). Samples were collected for ELISA at the indicated time points after initiation of cocultures (p19 for HTLV-1 and p24 for HIV-1). Results for one representative experiment out of two performed are presented. (B) Primary monocyte-derived macrophages (left panel) or tonsil-derived T lymphocytes (right panel) were infected with R5 isolates ADA or M1, respectively, and cultured in the presence of the indicated concentrations of ITI-367. Analysis of virus replication, as measured by RT activity or p24 concentration in culture supernatant, is presented. Results are shown for one representative experiment out of three performed. (C) Dose-response analysis plotted from the results of three independent experiments.
We next tested the effect of ITI-367 on HIV-1 replication in its physiological targets. Replication of an R5 HIV-1 ADA strain in primary monocyte-derived macrophages was inhibited by ITI-367 in a dose-dependent manner (Fig. 2B, left panel), with a 50% inhibitory concentration (IC50) of about 0.05 μM (Fig. 2C, left panel). A similar experiment was performed with T-lymphocyte cultures. To approximate in vivo conditions, we used T lymphocytes isolated from tonsil tissue 24 h after surgical removal. Similar to their behavior in the tissue (21), such cells support HIV-1 replication without unnatural exogenous stimulation, due to the endogenously activated state at the time of isolation. Cells were infected with the patient-derived HIV-1 R5 isolate M1. Results presented in Fig. 2B (right panel) demonstrate a dose-dependent inhibition of virus replication with an IC50 of about 0.7 μM (Fig. 2C, right panel).
To determine whether ITI-367 inhibited a pre- or postintegration step in the HIV-1 life cycle, we tested the effect of the drug on HIV-1 production from the PMA-activated T-cell line ACH-2 (17) and the promonocytic cell line U1 (55), which carry latent integrated provirus. No inhibitory effects of the drug were observed in these experiments (results not shown), suggesting that inhibition occurred at a step preceding integration.
Due to solubility constraints for ITI-367 in aqueous solutions, the maximum concentration tested for cell toxicity by the Alamar Blue assay (27) was 50 μM. At this concentration, cell proliferation in T-cell cultures was inhibited by approximately 5% (data not shown). The same level of toxicity was measured in macrophage cultures, using the MTT assay. Therefore, although we were unable to calculate the exact selectivity index for ITI-367, it appears to be >100 in T-cell cultures and >1,000 in macrophages.
ITI-367 exhibits an additive effect when combined with drugs used in highly active antiretroviral therapy (HAART).
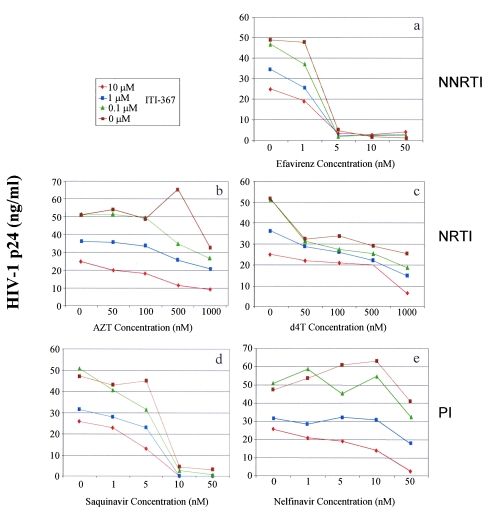
The inhibitory potential of ITI-367 was tested in an HIV-1 infectivity assay in combination with FDA-approved drugs. Tonsil-derived T lymphocytes were used as target cells for this experiment, and the primary R5 isolate M1 was used for infection. Drugs from each of the following groups were studied (Fig. 3): nonnucleoside reverse transcriptase inhibitors (NNRTI; efavirenz, panel a), nucleoside reverse transcriptase inhibitors (NRTI; AZT, panel b; d4T, panel c), and protease inhibitors (PI; saquinavir, panel d; nelfinavir, panel e). Virus replication was assayed by measuring p24 in the culture supernatant on day 7 after infection. Analysis of the data presented in Fig. 3 suggests that the drug combinations had at least an additive inhibitory effect on HIV-1 replication. Therefore, ITI-367 does not appear to counteract the anti-HIV activity of these approved drugs, suggesting that it can be used as an addition to a multidrug HAART cocktail.
FIG. 3.
Anti-HIV activity of combinations of ITI-367 and approved anti-HIV drugs. Cultures of tonsil-derived T lymphocytes were infected with a primary R5 HIV-1 isolate M1 and cultured in the presence of the indicated concentrations of ITI-367 in combination with various concentrations of the nonnucleoside reverse transcriptase inhibitor (NNRTI) Efavirenz (panel a), the nucleoside reverse transcriptase inhibitors (NRTIs) AZT (panel b) or d4T (panel c), or the protease inhibitors (PIs) Saquinavir (panel d) or Nelfinavir (panel e). Virus replication was analyzed by measuring p24 in the culture supernatant on day 7 after infection. The data are presented as means of the results for three independent wells.
ITI-367 inhibits HIV-1 replication in lymphoid tissue infected ex vivo.
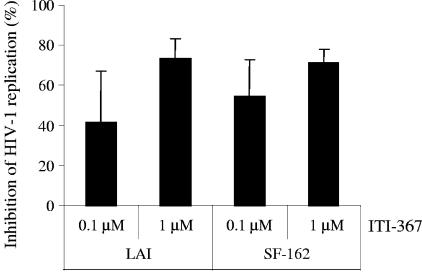
A system for culturing human tonsil tissue ex vivo (21, 22) provided an opportunity to test the anti-HIV activity of the compound in the context of intact lymphoid tissue, which is believed to be the main reservoir of replicating HIV-1 in the body of infected individuals (13, 44, 45). We infected tissue blocks with R5 (SF-162) or X4 (LAI) strains of HIV-1 and monitored viral replication by measuring p24 in the supernatant. Results presented in Fig. 4 demonstrate a potent, dose-dependent anti-HIV effect of ITI-367 in this system. Importantly, this anti-HIV activity was not associated with cytotoxicity of the compound, as demonstrated by flow cytometric analyses of cells shaken out of tissue blocks of ITI-367-treated and untreated, uninfected lymphohistocultures (data not shown). The inhibitory effect was smaller than that observed in macrophages using the same drug concentrations and was similar to the effect in T cells, consistent with predominant replication of HIV-1 in CD4+ T lymphocytes in lymphoid tissue (23).
FIG. 4.
Anti-HIV activity of ITI-367 in histocultures of human tonsils. Blocks of human tonsils were infected with X4 virus LAI or R5 strain SF-162 in the presence of the indicated concentrations of ITI-367. Virus replication was assayed on day 12 after infection by measuring p24 in culture medium. Results (mean ± standard deviation) are presented as percent inhibition of virus replication in drug-treated cultures. The results for one representative experiment out of three performed with tissue from different donors are shown.
Mutations in MA NLS-1 make HIV-1 resistant to ITI-367.
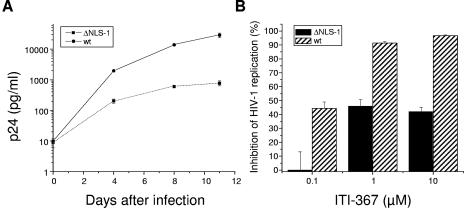
ITI-367 was designed to bind to MA within a structural groove in NLS-1 with R22, K27, Y29, and H33 predicted as contact amino acids. This binding is expected to alter the conformation of NLS-1 and thus impair its activity. Therefore, viruses carrying substitutions of the contact amino acids and/or inactivating mutations of NLS-1 are expected to acquire resistance to ITI-367 inhibitory activity. To test this inference, we used the NLHXADA strain of HIV-1, carrying substitutions of isoleucine residues for lysines in positions 26 and 27 of MA (these amino acids are critical for activity of NLS-1; see references 25 and 48). As reported previously (48), replication of this mutant virus in monocyte-derived macrophages was substantially attenuated (Fig. 5A). The sensitivity of this virus to ITI-367 was also greatly decreased compared to the virus carrying the wild-type NLS-1 (Fig. 5B). This result supports the notion that inhibitory activity of ITI-367 is targeted towards NLS-1 of MA.
FIG. 5.
HIV-1 carrying mutations in MA NLS-1 is resistant to ITI-367. (A) Monocyte-derived macrophages were infected with the wild-type NLHXADA virus (wt) or NLHXADAΔNLS-1 (ΔNLS-1) carrying substitutions of isoleucine residues for lysines in positions 26 and 27 of MA (48) and cultured for 11 days. Half of the medium was changed at days 4 and 8. Virus replication was assessed according to p24 levels in the culture supernatant and is presented as means ± the standard error of the means of the results for three independent wells. The zero time point sample was taken after the viral inoculum was washed away. (B) Triplicate cultures of macrophages were infected as for panel A, except that cells were cultivated in the presence or absence of the indicated concentrations of ITI-367. Analysis of viral replication was performed on day 8 after infection by measuring p24 in the culture medium. Inhibition of HIV-1 replication by ITI-367 was calculated for each test well by dividing the p24 value in that well by the mean p24 value of the corresponding untreated culture, expressing the result as a percent and subtracting from 100%. Results are presented as the means ± the standard errors of the means.
ITI-367 inhibits binding of the HIV-1 PIC to human importin α.
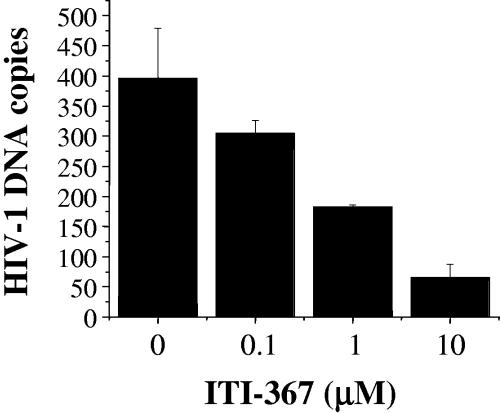
Binding to importin α is essential for nuclear import mediated by basic-type NLSs, and arylene bis(methylketone) compounds have been shown to inhibit importin α-PIC interaction (26). We therefore tested the effect of ITI-367 on binding of the HIV-1 PIC to human importin α. Jurkat cells were infected with HIV-1LAI and lysed 6 h after infection, and the postnuclear cytosolic fraction containing the PIC was incubated with increasing concentrations of ITI-367 for 2 h prior to binding to the GST-hSRP1α fusion protein. The GST-hSRP1α-PIC complexes were sedimented with glutathione-coated Sepharose beads, and the amount of viral cDNA in the sedimented fractions was evaluated by real-time PCR. As shown in Fig. 6, ITI-367 inhibited binding of HIV-1 PICs to importin α in a dose-dependent fashion. This result is consistent with the proposed mechanism of action of this group of compounds.
FIG. 6.
ITI-367 inhibits HIV-1 PIC binding to importin α. Cytoplasmic lysates of HIV-1-infected cells were incubated with importin α immobilized on Sepharose beads in the presence of the indicated concentrations of ITI-367. The amount of importin α-associated HIV-1 DNA was quantified in triplicate by real-time PCR. Results are presented as means ± the standard errors of the means of the numbers of HIV-1 DNA copies. Empty Sepharose beads retained less than 15 DNA copies. The graph shows results of one representative experiment out of two performed.
ITI-367 inhibits HIV-1 nuclear import.
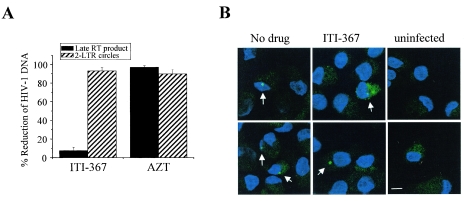
Based on the ability of ITI-367 to inhibit binding of the HIV-1 PIC to importin α, we expected it to also inhibit PIC nuclear import. To test this prediction, we analyzed the effect of the drug on the formation of late reverse transcripts (to measure the effect on reverse transcription) and 2-LTR circle forms of HIV-1 DNA. 2-LTR circular forms are commonly used as a marker for successful nuclear import of retroviral DNA (4, 8), although a recent report demonstrated that, at least in the case of murine leukemia virus, 2-LTR junctions can be detected in the cytoplasm of infected cells, albeit in rather small numbers (53). Preliminary analysis of late reverse transcripts in cells harvested 24 h and 48 h after infection did not reveal any inhibitory effect of ITI-367 (data not shown). The final analysis was performed at 72 h after infection, as 2-LTR circles could be consistently detected only at that time (56). To limit viral replication to one cycle, macrophages were cultured in the presence of anti-CD4 MAb, added after virus absorption. As shown in Fig. 7A, treatment with ITI-367 dramatically reduced the amount of 2-LTR circle forms without affecting the amount of late reverse transcripts. As expected, AZT had a similar inhibitory effect on both HIV-1 DNA forms (Fig. 7A). This result indicates that ITI-367 specifically targets HIV-1 nuclear import in macrophages, rather than an earlier step, such as virus entry or reverse transcription.
FIG. 7.
ITI-367 inhibits HIV-1 nuclear import. (A) HIV-1 DNA was analyzed 72 h after infection of macrophages with HIV-1 ADA in the presence or absence of ITI-367 (10 μM) or AZT (3 μM), using primers specific for the late-reverse transcription product or 2-LTR circle DNA. Results were normalized according to β-actin signals and are presented as means ± standard errors of the means of the reduction in DNA copy numbers from three independent experiments. (B) Monocyte-derived macrophages were infected with Alexa Fluor 488-labeled HIV-1 in the presence or absence of ITI-367 (10 μM) and were analyzed by fluorescent microscopy 72 h after infection. Nuclei were stained with TOPRO-3 iodide. Arrows point to HIV-infected cells. Two independent fields are shown for each treatment. The scale bar is 10 μm.
To visualize intracellular localization of the HIV-1 DNA in infected macrophages, we fluorescently labeled viral DNA using Alexa Fluor 488 5-dUTP added during endogenous reverse transcription (2). Analysis of untreated and drug-treated macrophages 72 h after infection with fluorescently labeled virus revealed substantially less nuclei-localized fluorescence in ITI-367-treated macrophages (Fig. 7B). This result supports the notion that ITI-367 inhibits HIV-1 nuclear translocation.
DISCUSSION
In this study we explored the anti-HIV activity of ITI-367, a drug selected by CADD to target the first NLS of the HIV-1 MA. Our results demonstrate that the drug inhibits the preintegration step of HIV-1 replication, blocks HIV-1 PIC binding to importin α (Fig. 6), and reduces nuclear import of HIV-1 DNA (Fig. 7). This activity is specific for HIV-1, as replication of another human retrovirus, HTLV-I, was not affected by the compound (Fig. 2). The drug was identified from a final list of 87 compounds predicted by CADD to bind within a structural groove generated by tyrosine 29 present in the first (N terminal) NLS of the matrix protein (25). The notion that ITI-367 targets NLS-1 of the MA is supported by our finding that the virus carrying inactivating mutations in NLS-1 is resistant to the drug (Fig. 5B). Some inhibition by high ITI-367 concentrations (1 μM and higher) may be due either to residual activity of the NLS-1 in the NLHXADAΔNLS-1 virus or to an additional activity of the drug that has not been revealed in other tests. The final proof for the target site of ITI-367 will require X-ray crystallography. Potential contact amino acids (R22, K27, Y29, and H33) are very well conserved in HIV-1 strains, suggesting that they are critical for viral fitness. The conserved nature of ITI-367 contact amino acids makes it harder for the virus to obtain resistance to this drug through mutation of the drug-binding site. Consistent with this notion, our efforts to generate ITI-367-resistant virus in vitro produced only a partially resistant virus after prolonged cultivation in the presence of suboptimal concentrations of the drug (data not shown).
We show that ITI-367 effectively inhibited virus replication in macrophages and T lymphocytes (Fig. 2) and in ex vivo cultured lymphoid tissue (Fig. 4). The T lymphocytes were approximately 10-fold-less sensitive to the effects of the drug than primary macrophage cultures. This difference may be due in part to the proliferative capacity of tonsil-derived lymphocytes, which limits anti-HIV activity of nuclear import inhibitors (26), as well as poor uptake of the compound by T cells compared to that by macrophages (unpublished observation). Importantly, ITI-367 did not antagonize the activity of anti-HIV drugs currently used for treatment of HIV-infected patients (Fig. 3), suggesting that this class of compounds can be used as a combination therapy in HAART.
The expected mode of action of ITI-367 is inhibition of MA NLS-dependent nuclear import of the HIV-1 PIC. The same activity has been described for ITI-002, an arylene bis(methylketone) compound (12). However, these two drugs inhibit nuclear import by different mechanisms: ITI-002 inactivates basic-type NLSs by forming Schiff bases with adjacent lysine residues (1), whereas ITI-367 is expected to disrupt the conformation of NLS-1.
The fact that such a potent inhibition of HIV-1 infection can be achieved by a drug targeting MA NLS is not surprising, since mutations within MA NLSs are sufficient to inhibit HIV replication (25). However, given that several other factors (i.e., DNA flap, IN, Vpr) likely contribute to HIV-1 nuclear import (43), the data we present may point to a paradox in the regulation of this necessary step in the virus life cycle. One possible explanation that would reconcile this paradox is that these factors (i.e., DNA flap, IN, MA, Vpr) work either sequentially or synergistically to regulate PIC nuclear import. Alternatively, the contribution of some of these factors might be in the amplification of the nuclear import process, as proposed for Vpr (47). The possibility that MA may be shed upon maturation of the PICs (16) suggests that MA might be required during the earliest phase of the PIC nuclear import, possibly even at the step of translocation towards the nucleus. This model is supported by a demonstrated role of MA in PIC interaction with actin (5), an important step in formation of the HIV-1 reverse transcription complex and its subsequent transport to the nuclear pore (43).
In conclusion, our study introduces a novel class of compounds that inhibit nuclear targeting of the HIV-1 preintegration complexes. These inhibitors may supplement our inventory of anti-HIV therapeutic agents and further improve the potency of HAART. The step of nuclear import follows reverse transcription and precedes integration, and the product of each reaction is the immediate substrate for the subsequent one. Therefore, one can expect a cooperative effect of RT, IN, and nuclear import inhibitors. Addition of the latter two classes of compounds to currently used anti-HIV drug cocktails may help achieve a complete long-term suppression of viral replication. As a class, nuclear import-blocking compounds may be useful not only against HIV, but also against other viruses that require nuclear import for replication, such as hepatitis B virus or herpesviruses. As the development of such compounds progresses, they are likely to provide a valuable enrichment of our arsenal of antiviral drugs.
Acknowledgments
We thank Robyn Rufner and the GWU imaging facility for help with fluorescent microscopy. We appreciate the help of Leonid Margolis (NIH/NICHD) with experiments involving ex vivo cultured lymphoid tissue. The HIV-1 p24 Gag monoclonal reagent (no. 24-2) was obtained from Michael H. Malim through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This work was supported in part by NIH grants R01 AI040386 and R01 AI033776 (M.B.) and R44 AI047782 (O.H. and R.L.).
REFERENCES
- 1.Al Abed, Y., L. Dubrovsky, B. Ruzsicska, M. Seepersaud, and M. Bukrinsky. 2002. Inhibition of HIV-1 nuclear import via schiff base formation with arylene bis(methylketone) compounds. Bioorg. Med. Chem. Lett. 12:3117-3119. [DOI] [PubMed] [Google Scholar]
- 2.Borroto-Esoda, K., and L. R. Boone. 1991. Equine infectious anemia virus and human immunodeficiency virus DNA synthesis in vitro: characterization of the endogenous reverse transcriptase reaction. J. Virol. 65:1952-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. Hiv-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 4.Bowerman, B., P. O. Brown, J. M. Bishop, and H. E. Varmus. 1989. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 3:469-478. [DOI] [PubMed] [Google Scholar]
- 5.Bukrinskaya, A., B. Brichacek, A. Mann, and M. Stevenson. 1998. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J. Exp. Med. 188:2113-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukrinsky, M. 2004. A hard way to the nucleus. Mol. Med. 10:1-5. [PMC free article] [PubMed] [Google Scholar]
- 7.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinsky, M. I., N. Sharova, M. P. Dempsey, T. L. Stanwick, A. G. Bukrinskaya, S. Haggerty, and M. Stevenson. 1992. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. USA 89:6580-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butera, S. T., V. L. Perez, B. Y. Wu, G. J. Nabel, and T. M. Folks. 1991. Oscillation of the human immunodeficiency virus surface receptor is regulated by the state of viral activation in a CD4+ cell model of chronic infection. J. Virol. 65:4645-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 12.Dubrovsky, L., P. Ulrich, G. J. Nuovo, K. R. Manogue, A. Cerami, and M. Bukrinsky. 1995. Nuclear localization signal of HIV-1 as a novel target for therapeutic intervention. Mol. Med. 1:217-230. [PMC free article] [PubMed] [Google Scholar]
- 13.Embretson, J., M. Zupancic, J. L. Ribas, A. Burke, P. Racz, K. Tenner-Racz, and A. T. Haase. 1993. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature 362:359-362. [DOI] [PubMed] [Google Scholar]
- 14.Emerman, M., M. Bukrinsky, and M. Stevenson. 1994. HIV-1 infection of non-dividing cells. Nature 369:107-108. [DOI] [PubMed] [Google Scholar]
- 15.Ewing, T. J., S. Makino, A. G. Skillman, and I. D. Kuntz. 2001. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J. Comput.-Aided Mol. Des. 15:411-428. [DOI] [PubMed] [Google Scholar]
- 16.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folks, T. M., K. A. Clouse, J. Justement, A. Rabson, E. Duh, J. H. Kehrl, and A. S. Fauci. 1989. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc. Natl. Acad. Sci. USA 86:2365-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freed, E. O., G. Englund, and M. A. Martin. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 69:3949-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedler, A., N. Zakai, O. Karni, Y. C. Broder, L. Baraz, M. Kotler, A. Loyter, and C. Gilon. 1998. Backbone cyclic peptide, which mimics the nuclear localization signal of human immunodeficiency virus type 1 matrix protein, inhibits nuclear import and virus production in nondividing cells. Biochemistry 37:5616-5622. [DOI] [PubMed] [Google Scholar]
- 21.Glushakova, S., B. Baibakov, L. B. Margolis, and J. Zimmerberg. 1995. Infection of human tonsil histocultures: a model for HIV pathogenesis. Nat. Med. 1:1320-1322. [DOI] [PubMed] [Google Scholar]
- 22.Glushakova, S., B. Baibakov, J. Zimmerberg, and L. B. Margolis. 1997. Experimental HIV infection of human lymphoid tissue: correlation of CD4+ T cell depletion and virus syncytium-inducing/non-syncytium- inducing phenotype in histocultures inoculated with laboratory strains and patient isolates of HIV type 1. AIDS Res. Hum. Retrovir. 13:461-471. [DOI] [PubMed] [Google Scholar]
- 23.Glushakova, S., J. C. Grivel, K. Suryanarayana, P. Meylan, J. D. Lifson, R. Desrosiers, and L. Margolis. 1999. Nef enhances human immunodeficiency virus replication and responsiveness to interleukin-2 in human lymphoid tissue ex vivo. J. Virol. 73:3968-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gulizia, J., M. P. Dempsey, N. Sharova, M. I. Bukrinsky, L. Spitz, D. Goldfarb, and M. Stevenson. 1994. Reduced nuclear import of human immunodeficiency virus type 1 preintegration complexes in the presence of a prototypic nuclear targeting signal. J. Virol. 68:2021-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haffar, O. K., S. Popov, L. Dubrovsky, I. Agostini, H. Tang, T. Pushkarsky, S. G. Nadler, and M. Bukrinsky. 2000. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. J. Mol. Biol. 299:359-368. [DOI] [PubMed] [Google Scholar]
- 26.Haffar, O. K., M. D. Smithgall, S. Popov, P. Ulrich, A. G. Bruce, S. G. Nadler, A. Cerami, and M. I. Bukrinsky. 1998. CNI-H0294, a nuclear importation inhibitor of the human immunodeficiency virus type 1 genome, abrogates virus replication in infected activated peripheral blood mononuclear cells. Antimicrob. Agents Chemother. 42:1133-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamid, R., Y. Rotshteyn, L. Rabadi, R. Parikh, and P. Bullock. 2004. Comparison of alamar blue and MTT assays for high through-put screening. Toxicol. In Vitro 18:703-710. [DOI] [PubMed] [Google Scholar]
- 28.Harada, S., Y. Koyanagi, and N. Yamamoto. 1985. Infection of HTLV-III /LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science 229:563-566. [DOI] [PubMed] [Google Scholar]
- 29.Heinzinger, N. K., M. I. Bukrinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 32.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanner, S. B., and O. K. Haffar. 1995. HIV-1 down-regulates CD4 costimulation of TCR/CD3-directed tyrosine phosphorylation through CD4/p56lck dissociation. J. Immunol. 154:2996-3005. [PubMed] [Google Scholar]
- 34.Katz, R. A., J. G. Greger, P. Boimel, and A. M. Skalka. 2003. Human immunodeficiency virus type 1 DNA nuclear import and integration are mitosis independent in cycling cells. J. Virol. 77:13412-13417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis, P. F., and M. Emerman. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Massiah, M. A., M. R. Starich, C. Paschall, M. F. Summers, A. M. Christensen, and W. I. Sundquist. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 244:198-223. [DOI] [PubMed] [Google Scholar]
- 37.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 38.Matthews, S., P. Barlow, J. Boyd, G. Barton, R. Russell, H. Mills, M. Cunningham, N. Meyers, N. Burns, and N. Clark. 1994. Structural similarity between the p17 matrix protein of HIV-1 and interferon-gamma. Nature 370:666-668. [DOI] [PubMed] [Google Scholar]
- 39.Matthews, S., P. Barlow, N. Clark, S. Kingsman, A. Kingsman, and I. Campbell. 1995. Refined solution structure of p17, the HIV matrix protein. Biochem. Soc. Trans. 23:725-729. [DOI] [PubMed] [Google Scholar]
- 40.Merl, S., B. Kloster, J. Moore, C. Hubbell, R. Tomar, F. Davey, D. Kalinowski, A. Planas, G. Ehrlich, and D. Clark. 1984. Efficient transformation of previously activated and dividing T lymphocytes by human T cell leukemia-lymphoma virus. Blood 64:967-974. [PubMed] [Google Scholar]
- 41.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nadler, S. G., D. Tritschler, O. K. Haffar, J. Blake, A. G. Bruce, and J. S. Cleaveland. 1997. Differential expression and sequence-specific interaction of karyopherin alpha with nuclear localization sequences. J. Biol. Chem. 272:4310-4315. [DOI] [PubMed] [Google Scholar]
- 43.Nisole, S., and A. Saib. 2004. Early steps of retrovirus replicative cycle. Retrovirology 1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pantaleo, G., C. Graziosi, J. F. Demarest, L. Butini, M. Montroni, C. H. Fox, J. M. Orenstein, D. P. Kotler, and A. S. Fauci. 1993. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature 362:355-358. [DOI] [PubMed] [Google Scholar]
- 45.Pantaleo, G., C. Graziosi, and A. S. Fauci. 1993. The role of lymphoid organs in the pathogenesis of HIV infection. Semin. Immunol. 5:157-163. [DOI] [PubMed] [Google Scholar]
- 46.Piller, S. C., L. Caly, and D. A. Jans. 2003. Nuclear import of the pre-integration complex (PIC): the Achilles heel of HIV? Curr. Drug Targets 4:409-429. [DOI] [PubMed] [Google Scholar]
- 47.Popov, S., M. Rexach, L. Ratner, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates docking of the HIV-1 preintegration complex to the nuclear pore complex. J. Biol. Chem. 273:13347-13352. [DOI] [PubMed] [Google Scholar]
- 48.Popov, S., M. Rexach, G. Zybarth, N. Reiling, M. A. Lee, L. Ratner, C. M. Lane, M. S. Moore, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 17:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rexach, M., and G. Blobel. 1995. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83:683-692. [DOI] [PubMed] [Google Scholar]
- 51.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidtmayerova, H., G. J. Nuovo, and M. Bukrinsky. 1997. Cell proliferation is not required for productive HIV-1 infection of macrophages. Virology 232:379-384. [DOI] [PubMed] [Google Scholar]
- 53.Serhan, F., M. Penaud, C. Petit, T. Leste-Lasserre, S. Trajcevski, D. Klatzmann, G. Duisit, P. Sonigo, and P. Moullier. 2004. Early detection of a two-long-terminal-repeat junction molecule in the cytoplasm of recombinant murine leukemia virus-infected cells. J. Virol. 78:6190-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simon, J. H., R. A. Fouchier, T. E. Southerling, C. B. Guerra, C. K. Grant, and M. H. Malim. 1997. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J. Virol. 71:5259-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stanley, S. K., T. M. Folks, and A. S. Fauci. 1989. Induction of expression of human immunodeficiency virus in a chronically infected promonocytic cell line by ultraviolet irradiation. AIDS Res. Hum. Retrovir. 5:375-384. [DOI] [PubMed] [Google Scholar]
- 56.Triques, K., and M. Stevenson. 2004. Characterization of restrictions to human immunodeficiency virus type 1 infection of monocytes. J. Virol. 78:5523-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Maele, B., J. De Rijck, E. De Clercq, and Z. Debyser. 2003. Impact of the central polypurine tract on the kinetics of human immunodeficiency virus type 1 vector transduction. J. Virol. 77:4685-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von Schwedler, U., R. S. Kornbluth, and D. Trono. 1994. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc. Natl. Acad. Sci. USA 91:6992-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, and B. H. Hahn. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 60.Weinberg, J. B., T. J. Matthews, B. R. Cullen, and M. H. Malim. 1991. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J. Exp. Med. 174:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westervelt, P., D. B. Trowbridge, L. G. Epstein, B. M. Blumberg, Y. Li, B. H. Hahn, G. M. Shaw, R. W. Price, and L. Ratner. 1992. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J. Virol. 66:2577-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]