Abstract
Coronaviruses are enveloped, positive-stranded RNA viruses considered to be promising vectors for vaccine development, as (i) genes can be deleted, resulting in attenuated viruses; (ii) their tropism can be modified by manipulation of their spike protein; and (iii) heterologous genes can be expressed by simply inserting them with appropriate coronaviral transcription signals into the genome. For any live vector, genetic stability is an essential requirement. However, little is known about the genetic stability of recombinant coronaviruses expressing foreign genes. In this study, the Renilla and the firefly luciferase genes were systematically analyzed for their stability after insertion at various genomic positions in the group 1 coronavirus feline infectious peritonitis virus and in the group 2 coronavirus mouse hepatitis virus. It appeared that the two genes exhibit intrinsic differences, the Renilla gene consistently being maintained more stably than the firefly gene. This difference was not caused by genome size restrictions, by different effects of the encoded proteins, or by different consequences of the synthesis of the additional subgenomic mRNAs. The loss of expression of the firefly luciferase was found to result from various, often large deletions of the gene, probably due to RNA recombination. The extent of this process appeared to depend strongly on the coronaviral genomic background, the luciferase gene being much more stable in the feline than in the mouse coronavirus genome. It also depended significantly on the particular genomic location at which the gene was inserted. The data indicate that foreign sequences are more stably maintained when replacing nonessential coronaviral genes.
Many different viruses have been proposed as candidate vaccine vectors and are currently being developed into live immunization vectors. Ideal vaccine vectors are characterized by low cost, safe use, easy administration, induction of long-lasting protective immunity, adaptable targeting, and high physical and genetic stability. The recent development of efficient reverse genetics systems for coronaviruses now allows us to study the potential of these viruses as vectors. Previously, we have shown that the mouse hepatitis coronavirus (MHV) accepts insertions of foreign expression cassettes at many different positions (9). In the present study the genetic stability of such coronavirus vectors is analyzed.
Coronaviruses are enveloped viruses that belong to the family Coronaviridae in the order Nidovirales. These viruses generally cause respiratory and/or intestinal infections, although some may spread systemically. They are not only pathogens of veterinary importance but also pose a threat to humans, as demonstrated by the emergence of the severe acute respiratory syndrome coronavirus. Coronaviruses contain a single-stranded, positive-sense RNA genome of about 30 kb, which is capped, polyadenylated, and infectious (13, 28). Based on antigenic and genetic criteria, they have been divided into three distinct groups. All coronaviruses contain a common set of essential genes, of which the open reading frames (ORFs) 1a and 1b, encoding the replicase functions, comprise the first two-thirds of the genome. Downstream therefrom, the essential genes encoding the structural proteins are located, invariably in the order 5′-S-E-M-N-3′. Interspersed between these genes, clusters of group-specific genes occur that are homologous within each group but differ profoundly between the groups. The group-specific genes are accessory genes, as demonstrated by the observed viability of natural and engineered deletion mutants lacking some or all of these genes (8, 16, 19, 38, 51).
ORFs 1a and 1b are translated directly from the genomic RNA (gRNA), the more downstream ORF 1b by translational read-through using a ribosomal frameshift mechanism. The other genes are translated from a 3′-coterminal nested set of subgenomic mRNAs (sgRNAs). The sgRNAs are transcribed via a discontinuous transcription process during which the body of each sgRNA is fused to a leader sequence (65 to 98 nucleotides [nt]), which is identical to the extreme 5′ end of the genome. This fusion occurs at short transcription regulatory sequences (TRSs; previously named intergenic sequences) which precede the genes and are homologous to the 3′ end of the leader sequence. The TRSs probably serve as transcription termination or pausing signals, the nascent chain either resuming transcription at the same site or being joined to the leader sequence at the 5′ end during negative-strand synthesis. The negative-strand sgRNAs subsequently function as templates for the production of the positive-strand sgRNA (44-47, 50).
The unique coronavirus transcription mechanism allows the efficient expression of foreign genes simply by inserting these genes together with a TRS into the coronaviral genome, as was demonstrated with several reporter proteins. Insertion of the green fluorescent protein (GFP) gene (5, 17, 43, 52) and of two unrelated luciferase genes (Renilla luciferase [RL] and firefly luciferase [FL]) (9) resulted in high-level expression. Little is known about the factors that determine the efficiency of foreign gene expression. It is clear, however, that the expression levels of coronavirus genes can be manipulated by changes in the TRS, in the sequences flanking it (1, 2, 6, 8, 10, 22, 25, 26, 33, 34, 36, 39, 52, 53, 56, 61), or in the position of the gene in the viral genome (10). Transcription levels of foreign genes have been demonstrated to be dependent on their genomic position as well as on the identity of the foreign gene itself (9). Expression levels generally increased when the heterologous gene was inserted closer to the 3′ end of the genome. Also, the simultaneous expression of multiple foreign genes from a single genome has been demonstrated previously (9).
Besides their efficient expression of foreign genes (5, 9, 17, 43, 52), recent studies revealed several other features that make coronaviruses attractive as vectors. Firstly, the often virulent coronaviruses can be converted into nonvirulent viruses by deleting the nonessential, group-specific genes. This was demonstrated for MHV (8), transmissible gastroenteritis virus (TGEV) (38), and feline infectious peritonitis virus (FIPV) (19). Importantly, vaccination with the FIPV deletion mutant viruses resulted in protection against an otherwise lethal FIPV challenge (19), indicating that the deletion mutant viruses are able to replicate and induce strong immune responses. Secondly, the conserved genome organization of coronaviruses can be rearranged (10). Deliberate rearrangement of the viral genes may be useful in the generation of safer vectors due to their reduced risk of generating viable viruses by recombination with circulating field viruses. Thirdly, the species and tissue tropism of coronaviruses can be manipulated by modification of their attachment and fusion protein S. Thus, MHV was retargeted to feline cells and lost the ability to infect murine cells by exchanging the ectodomain of its S protein with that of its FIPV counterpart (27). A similar, reciprocal result was obtained for FIPV (20). Manipulation of the S protein also led to changes in the tissue and cell tropism of MHV (35, 40, 54), TGEV (14), and infectious bronchitis virus (IBV) (4). Hence, the tropism of the coronavirus vector can be engineered. Fourthly, replication-competent, propagation-deficient coronaviruses have been constructed for TGEV that lack the E protein gene (37). These viruses grew to high titers in cells that complemented the TGEV E protein but failed to propagate in other cells.
The genetic stability of a viral vector is essential for its successful development as a live immunization vector. While the essential coronavirus genes are stably maintained in the virus genome, genes encoding the group-specific and/or accessory genes show a much higher variability, as is demonstrated by the occurrence of many natural viral mutants unable to express one or more of these genes (reviewed by Luytjes [31]). Furthermore, deletions in the group-specific genes were shown to occur readily in vitro, as was demonstrated for the MHV 2a gene (49) and the FIPV 7b gene (21). Apparently, these group-specific genes exert their function in vivo rather than in vitro. Little is known about the genetic stability of recombinant coronaviruses containing foreign genes, although expression of the GFP gene from different coronavirus vectors was reported to be stable for several passages (5, 17, 43, 52). In the present study, we have evaluated a number of relevant parameters for their effect on stability. By comparatively analyzing these effects on the maintenance of two different foreign genes, we established, among other features, that stability is dependent on the genomic position of the inserted gene, the genetic background (i.e., the particular virus into which the gene is inserted), and on intrinsic properties of the gene itself.
MATERIALS AND METHODS
Cells and viruses.
Murine LR7 cells (27) and feline FCWF cells (American Type Culture Collection) were used to propagate the recombinant MHV (strain A59) and FIPV (strain 79-1146) viruses, respectively. The same cells were used for plaque purifications, one-step growth curves, and infection with the interspecies chimeric coronaviruses mFIPV (20) and fMHV (27). The generation of the recombinant viruses MHV-WT, MHV-ERLM, MHV-2aRLS, MHV-MRLN, MHV-EFLM, MHV-RLFL, MHV-FLRL, and r-wtFIPV (here designated FIPV-WT) (Fig. 1) has been described previously (9, 20).
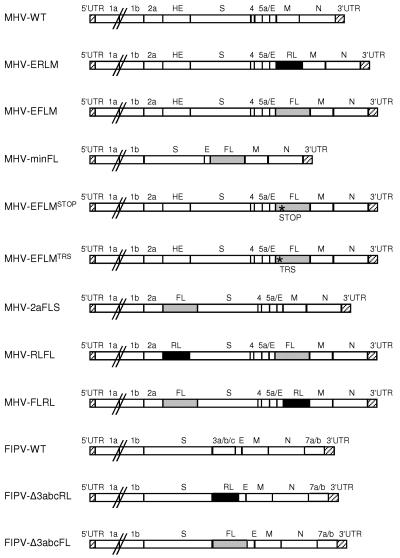
FIG. 1.
Genomic organization of the recombinant viruses. The genome structures of the recombinant wild-type MHV-A59 (MHV-WT) and FIPV (FIPV-WT) viruses and of the recombinant viruses containing either an RL or an FL expression cassette or both are depicted. The asterisks indicate either the position of a premature stop codon (MHV-EFLMSTOP) or of a mutated TRS (MHV-EFLMTRS). Numbers designate the genes encoding nonstructural proteins, while genes encoding the hemagglutinin-esterase (HE) protein, spike (S) protein, envelope (E) protein, membrane (M) protein, or nucleocapsid (N) protein are indicated by their abbreviations. The 5′- and 3′-untranslated regions (UTR) are also designated.
Plasmid constructs.
MHV transcription vectors for the production of donor RNA for targeted recombination were derived from transcription vector pMH54 (27) and derivatives thereof. pMH54, which specifies a defective MHV-A59 RNA transcript consisting of the 5′ end of the genome (467 nt) fused to codon 28 of the HE gene and running to the 3′ end of the genome (Fig. 1) was previously used for the construction of a wild-type recombinant virus (MHV-WT; Fig. 1) (8). Transcription vector pXHminFL contains the FL expression cassette between the E and the M genes while lacking ORFs 2a, HE, 4a, 4b, and 5a. To construct this vector, the FL expression cassette was cut out of pXH2711 (9) by digestion with EcoRV and XbaI, treated with a Klenow fragment of DNA polymerase I, and cloned into EcoRV-treated pXHmin (8). Transcription vector pXHEFLMSTOP was identical to pXHEFLM (9), except that a single nucleotide just downstream of the FL start codon had been deleted, resulting in a frameshift and a premature termination of the FL ORF. As a first step, a PCR was performed with primer 1664 (5′-GCTTTCCATGGAGACGCC-3′), corresponding to the sequence at the 5′ end of the FL gene but carrying a single-nucleotide deletion, and primer 509 (5′-GTCTAACATACACGGTACCTTTC-3′), which is complementary to a sequence in the 3′ end of the MHV M gene, using pXHEFLM as a template. Next the PCR fragment was cloned into pGEM-T Easy (Promega) according to the manufacturer's instructions, excised using SpeI and StyI, and cloned into XbaI- and StyI-digested pXH2711, resulting in pXH280202. In order to obtain pXHEFLMSTOP, the mutated expression cassette was removed from pXH280202 by using EcoRV and BglII, treated with Klenow polymerase, and cloned into pMH54 linearized with EcoRV. Transcription vector pXHEFLMTRS was identical to pXHEFLM, with the exception of the TRS preceding the FL gene. This plasmid was constructed as described for pXHEFLM (9), now using primer 2212 (5′-GGATATCTAATGTAAATTTTAG-3′) and primer 2213 (5′-CTAGCTAAAATTTACATTAGATATCCTGCA-3′). pXH2aFLS contains the FL gene at the position of the HE gene. To obtain this construct, first the FL gene was cut from pSPluc+ (Promega) by digestion with XbaI and AvrII and cloned into pXH2509A (9) from which the RL gene had been removed by digestion with XbaI and NheI, resulting in pXH161002. Next, the fragment excised from this latter vector by AvrII and RsrII was cloned into pMH54 treated with the same enzymes, resulting in pXH2aFLS.
FIPV transcription vectors for the production of synthetic donor RNA for targeted recombination were derived from plasmid pBRDI1 (20), which specifies an FIPV 79-1146 RNA transcript consisting of the 5′ end of the genome (681 nt) fused to the 3′ proximal end of ORF 1b (363 nt) and running to the 3′ end of the genome. This plasmid was previously used for the construction of a wild-type recombinant virus (r-wtFIPV [20]; here designated FIPV-WT; Fig. 1). The two reporter genes RL and FL were introduced into the FIPV genome under the transcriptional control of the TRS preceding gene 3a while replacing the FIPV group-specific gene cluster 3abc. To this end, the 3abc gene cluster was deleted from, and the RL and FL genes were introduced into, the plasmid pBRDI1, resulting in transcription vectors pBRDI1Δ3abc+ RL and pBRDI1Δ3abc+ FL, respectively. To construct these plasmids, a PCR product was prepared by splicing overlap extension PCR. The first PCR product was obtained using primer 1244 (5′-GCCATTCTCATTGATAAC-3′), which corresponds to part of the sequence in the 3′ end of the S gene, and primer 1514 (5′-CTGAGTCTAGAGTAGCTAGCTAATGACTAATAAGTTTAG-3′), which is complementary to the 5′ end of gene 3a and contains NheI and XbaI restriction sites. The second PCR product was prepared using primer 1245 (5′-GCTTCTGTTGAGTAATCACC-3′), which corresponds to a sequence in the 3′ end of the M gene, and primer 1513 (5′-GCTAGCTACTCTAGACTCAGGCGGTTCTAAAC-3′), which is complementary to primer 1514. The PCR products were purified, mixed, and amplified using primers 1244 and 1245. The DNA product obtained in the second round of PCR was cloned into pGEM-T Easy, resulting in pGEM-C. Both the RL and FL genes, excised from pRL-null and pSPluc+ (both from Promega) by using NheI and XbaI, were cloned into pGEM-C digested with the same enzymes, resulting in pGEM-C+ RL and pGEM-C+ FL, respectively. Finally, the fragments obtained by digestion of these latter plasmids with AflII and SnaBI were introduced into pBRDI1 treated with the same enzymes, resulting in pBRDI1Δ3abc+ RL and pBRDI1Δ3abc+ FL, respectively. All PCR products were confirmed by sequence analysis.
Generation of recombinant viruses.
Incorporation of the expression cassettes into the MHV or FIPV genome by targeted recombination was carried out as described previously (8, 20, 23). Briefly, donor RNAs transcribed from linearized MHV transcription vectors were electroporated into FCWF cells that had been infected earlier with fMHV, while donor RNAs transcribed from the linearized FIPV transcription vectors were electroporated into LR7 cells that had been infected earlier with mFIPV. The electroporated cells were then plated on a monolayer of LR7 or FCWF cells for the propagation of the recombinant MHVs or FIPVs, respectively. After 24 h of incubation at 37°C, progeny viruses released into the culture media were harvested and plaque purified twice on LR7 or FCWF cells before a passage 1 stock was grown. After confirmation of the recombinant genotypes by reverse transcriptase PCR (RT-PCR) on purified genomic RNA, a passage 2 stock was grown that was subsequently used in the experiments. In all cases, two independent recombinants were generated to verify that the observed phenotypic characteristics were the result of the intended genomic modifications.
One-step growth curve.
FCWF or LR7 cell monolayers (2 cm2) were infected at a multiplicity of infection (MOI) of 8. At 1 h postinfection the cells were washed three times with phosphate-buffered saline and then fed with Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Viral infectivity in culture media at different times postinfection was determined by a quantal assay on FCWF or LR7 cells, and the 50% tissue culture infectious doses (TCID50) were calculated.
Determination of FL or RL expression.
Cell monolayers (2 cm2) were infected as described above (MOI of 8). At the indicated times, the culture media were removed and cells were lysed using the appropriate buffer provided with the Firefly and Renilla Luciferase Assay Systems (Promega). Intracellular luciferase expression was measured according to the manufacturer's instructions, and relative light units (RLU) were determined in a LUMAC biocounter M2500 or a Turner Designs TD-20/20 luminometer.
Serial passaging of recombinant viruses and analysis of passage 8 virus.
Cell monolayers (25 cm2) were infected as described above at an MOI of 0.05. Eighteen hours postinfection the culture supernatants were clarified by low-speed centrifugation and used in the next low-MOI (0.05) infection as described above. The harvest obtained after eight passages (passage 8 virus) was plaque purified once, after which a stock was grown (passage 8.1 virus). Intracellular luciferase expression by the viruses obtained after the successive passages was determined as described above using an MOI of 8.
RESULTS
Stability of RL versus FL.
In our previous analysis of the expression of two unrelated luciferase genes, inserted at different positions in the MHV genome (9), the expression levels were found to be dependent on the site of insertion; higher levels were obtained when the gene was placed closer to the 3′ end of the genome. In the present study we performed a systematic analysis of the stability of foreign gene expression by coronaviruses, using the recombinant viruses described previously as well as a number of newly generated recombinant viruses. We started our analysis with MHV-ERLM and MHV-EFLM (Fig. 1), viruses containing an RL or FL gene preceded by an identical TRS and placed between the E and M genes (9). These recombinant viruses reached high titers in cell culture and expressed their luciferase genes to an extent similar to that of the structural proteins S and M (data not shown), which peaked around 8 to 9 h after infection in LR7 cells (9). Comparison with a standard curve of purified FL indicated that cells infected with MHV-EFLM contained 1 to 2 μg of luciferase per 106 cells at this time point (9). Both for MHV-ERLM and MHV-EFLM, two independently generated recombinant viruses (designated A and B) were serially passaged at a low MOI (0.05) up to passage 8. Subsequently, the intracellular luciferase expression by the viruses from the different passages was determined in a parallel infection experiment. As is clear from Fig. 2A and B, expression of the RL gene remained stable during serial passaging of both recombinant viruses; FL gene expression was stable for a few passages, but the luciferase activity produced started to decrease significantly from passage 5 onward. Comparison of the FL expression levels in cells infected with passage 2 and 8 viruses showed that expression by recombinants A and B decreased approximately 150- and 25-fold, respectively (Fig. 2A and B), while their titers increased approximately 4- to 5-fold (data not shown).
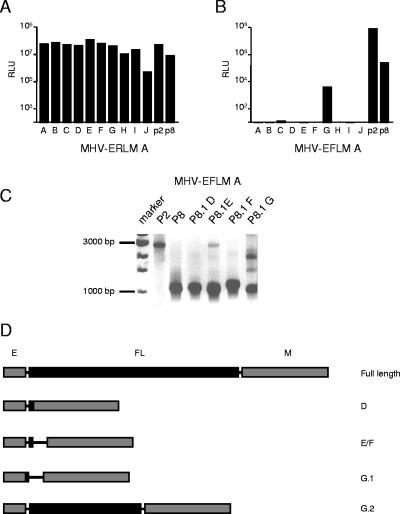
FIG. 2.
Stability of expression of RL versus FL within an otherwise identical background. Recombinant viruses MHV-ERLM (A) and MHV-EFLM (B) were passaged eight times on LR7 cells as described in Materials and Methods. Subsequently, in a parallel expression experiment, LR7 cells were infected with the recombinant viruses from the different passages at an MOI of 8. At 8.5 h postinfection, the cells were lysed and the intracellular luciferase expression was determined by using a luminometer (values are expressed in relative light units [RLU]). Independent recombinants are indicated with the letters A and B. (C) RT-PCR was used to amplify regions of genomic RNA. The RT step was performed with primer 746 (5′-CGTCTAGATTAGGTTCTCAACAATGCGG-3′), which is complementary to the 3′ end of the M gene, while the PCR was performed with primers 509 and 1091 (5′-GTTACAAACCTGAATCTCATCTTAATTCTGGTCG-3′), which are complementary to sequences in the 3′ end of the M gene and in the 5a gene, respectively. The same strategy was employed for the RL- and FL-expressing viruses. The sizes of the expected full-length PCR products are indicated. (D) The RT-PCR analysis was performed for both MHV-ERLM (recombinant A) and MHV-EFLM (recombinant A) from passages 2 (P2), 5 (P5), and 8 (P8). PCR products were analyzed by electrophoresis in 1% agarose gels stained with ethidium bromide. Sizes of the relevant DNA fragments of the marker are indicated.
To study the genetic basis of this instability of the recombinant viruses, an RT-PCR analysis was performed on genomic RNA purified from the different virus passage stocks. The strategy of this RT-PCR is depicted in Fig. 2C, while the results are shown in Fig. 2D. The RT step was performed with primer 746, which is complementary to the 3′ end of the M gene, while the PCR was performed with primers 509 and 1091, which are complementary to sequences in the M gene and in the 5a gene, respectively. The same strategy was employed for the RL- and FL-expressing viruses. For MHV-ERLM, an RT-PCR product was obtained of approximately the expected size (2,042 bp) at passage 2. A product of the same size was observed at passage 5, while after eight passages an additional product of approximately 1 kb in size was obtained. In the case of MHV-EFLM, an RT-PCR product of the expected size (2,764 bp) was observed with the passage 2 stock. However, already at passage 5 only a smaller RT-PCR product was obtained, while a similar result was observed after eight passages. Analysis of the independent recombinants gave similar results. The outcome of the experiments indicates the occurrence of large deletions in both the RL and FL genes, which were detected earlier in the FL than in the RL gene.
Analysis of passage 8 virus.
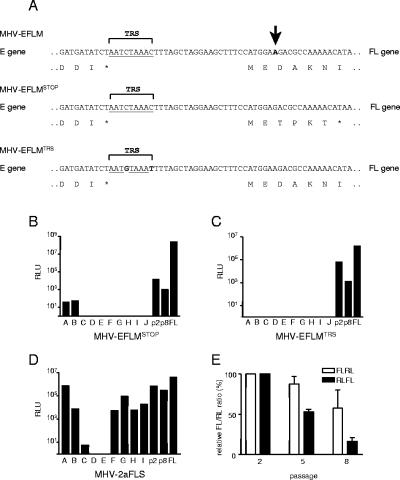
The results obtained with the RT-PCR analysis (Fig. 2D) confirm the corresponding luciferase expression profiles (Fig. 2A and B), showing that the expression of the RL gene was much more stable than that of the FL gene. However, while expression of the RL gene appeared to be stable for eight passages, the RT-PCR analysis revealed the appearance of deletions. Similarly, while full-length RT-PCR products could not be obtained for MHV-EFLM at passages 5 and 8, luciferase activity could still be easily detected after infection of cells with these viruses. This apparent discrepancy is probably the result of smaller PCR products being preferentially amplified over larger PCR products. In order to get more insight into the identity of these RL or FL gene-containing viruses, the passage 8 recombinant viruses were analyzed in more detail. To this end, these viruses were plaque purified once, after which stocks of a number of cloned viruses were grown. Subsequently, the intracellular luciferase expression by these viruses was determined. Passage 2 and passage 8 viruses were used as references (Fig. 3A and B). Nine out of 10 plaque-purified viruses expressed RL to the same extent as MHV-ERLM from passage 2 (Fig. 3A). Only one virus exhibited a lower luciferase expression level. However, for MHV-EFLM, 9 out 10 plaque-purified viruses were negative in their luciferase expression. Only one virus expressed a substantial FL activity. The data indicate that upon serial passaging, viruses that have lost their FL expression accumulate. The residual FL activity observed after passage 8 can be attributed to a minority of viruses still being able to express their foreign gene to some extent. The results confirm that the FL expression is less stable upon serial passaging than the RL expression.
FIG. 3.
Analysis of cloned MHV-ERLM and MHV-EFLM after serial passaging. Passage 8 viruses from MHV-ERLM (A) and MHV-EFLM (B) were plaque purified once, after which stocks were grown on LR7 cells (passage 8.1, A to J). Intracellular luciferase expressed in LR7 cells after infection with these plaque-purified viruses was determined. Passage 2 (p2) and passage 8 (p8) viruses were used as references. (C) RT-PCR was used to amplify regions of genomic RNA as described in the legend to Fig. 2. The analysis was performed for MHV-EFLM (recombinant A) from passage 2 (P2), from passage 8 (P8), and from the plaque-purified viruses (P8.1, E to G). Sizes of the relevant DNA fragments of the marker are indicated. (D) The PCR products were cloned and sequenced. A schematic representation of the sequencing results is shown. Genes E and M are indicated by gray boxes, while the inserted expression cassette is shown in black. The open reading frame that will be translated theoretically from the inserted sequences (or what is left of it) is indicated with a black box. The plaques from which these sequences were derived are indicated on the right.
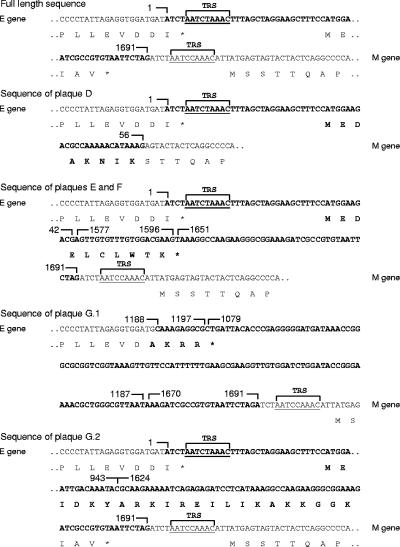
Next, the sequence of the FL expression cassette of four MHV-EFLM plaque-purified viruses (P8.1E to P8.1G) was analyzed. Thus, RT-PCRs were performed (Fig. 3C) as described above. The RT-PCR analyses of passage 2 and 8 viruses, taken as controls, gave the same results as shown in Fig. 2D. As expected, the predominant RT-PCR products obtained for the plaque-purified viruses were all approximately 1 kb in size, although in some cases (P8.1E and P8.1G) some larger products could also be observed. The RT-PCR products were purified, cloned, and sequenced. A schematic representation of the sequencing results is shown in Fig. 3D, while the sequences themselves are shown in Fig. 4. For comparison, the sequence of the beginning and the end of the full-length FL expression cassette is also shown in Fig. 4. The sequence analysis of the plaque D virus revealed the occurrence of a large deletion, comprising almost the entire FL gene insert, except for the first 56 nt, and also comprising the 5′ end of the M gene, including its TRS. As a result, the first two codons of the M gene of this virus are replaced by the first eight codons of the FL gene. In addition, the resulting M gene is under the transcriptional control of the TRS originally preceding the FL gene. Sequences obtained from the plaque E and F viruses were identical. Two deletions of nt 43 to 1576 and 1597 to 1650 of the FL expression cassette had removed almost the entire FL gene, leaving a small ORF encoding a putative polypeptide of 10 amino acids under the control of the inserted TRS. Two sequences were retrieved from the plaque G virus (G.1 and G.2). The sequence of G.1 most likely corresponds with the approximately 1-kb predominant RT-PCR product seen in Fig. 3C. Again, almost the entire FL gene has been deleted; however, the remaining part of the FL sequence appeared to be drastically shuffled. In addition, the extreme 3′ end of the E gene had been replaced with a short FL sequence derived from the middle of the FL gene. As a result, the last two amino acids of the E gene (DI) have been replaced by the amino acid sequence AKRR. The sequence of G.2 probably corresponds to the RT-PCR product of approximately 1.5 kb in size. Here a single deletion of nt 944 to 1623 was found, removing most of the 3′ half of the FL gene. Notably, in several plaque-purified viruses (E, F, and G2) the TRS of the FL expression cassette has been maintained, which supposedly gives rise to an additional sgRNA during infection. Furthermore, the heterogeneity in the deleted sequences indicates that the removal is random rather than involving specific sequence motifs.
FIG. 4.
Sequence analysis of the viruses plaque purified from MHV-EFLM passage 8. The detailed sequencing results from the MHV-EFLM viruses derived from plaques E, D, F, and G are shown. The 5′ end of the E gene and the 3′ end of the M gene are indicated as well as the encoded amino acid sequence. The sequence of the inserted expression cassette is shown in boldface (nt 1 to 1691). The TRSs are indicated. Viruses derived from plaque D only contain the first 56 nt from the expression cassette fused in frame to the 3′ end of the M gene. Viruses derived from plaques E and F had lost nt 43 to 1576 and 1597 to 1650 from the expression cassette. Two sequences were obtained from plaque G (G1 and G2). The G.1 sequence shows various deletions as well as rearrangements in the expression cassette. As a result, the E gene has been extended with a few residues. The G.2 sequence shows one large deletion of nt 944 through 1624.
Loss of the FL gene is not due to genome size limitations.
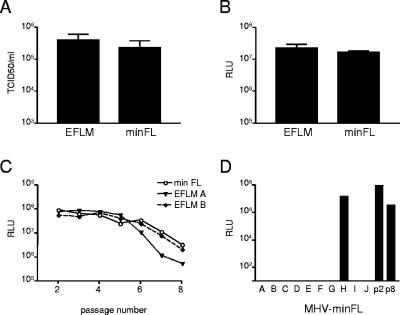
Since the FL gene is approximately 700 nt larger than the RL gene, we hypothesized that the observed difference in their genetic stability might be the result of the genome size being limiting, possibly because of packaging constraints. Alternatively, the foreign sequence itself, its genomic position, the transcription of an extra sgRNA, or the resulting protein product may influence the stable maintenance of the gene. To study these different possibilities, several new recombinant viruses were generated. First, a recombinant virus was generated (MHV-minFL; Fig. 1) in which the 2a, HE, 4a, 4b, and 5a genes were deleted, while maintaining the position of the FL gene insert in the virus genome between the E and the M genes. So while approximately 1.7 kb of genetic material was inserted, a total of 2.8 kb was deleted, resulting in a recombinant virus with a genome that is still approximately 1.1 kb smaller than the parental virus MHV-A59. Previously, we demonstrated that viruses lacking all of the nonessential genes were able to replicate efficiently in vitro (8). Indeed, MHV-minFL reached very similar titers to those of MHV-EFLM after a high-MOI infection (Fig. 5A) and expressed very similar levels of FL (Fig. 5B). However, upon serial passaging, MHV-minFL displayed an instability of its foreign gene expression similar to that of MHV-EFLM (Fig. 5C). Analysis of the FL expression of viruses plaque-purified from the passage 8 stock showed a result similar to that for MHV-EFLM (compare Fig. 5D to 3B). Nine out of 10 plaque-purified viruses were completely negative in their luciferase expression. Only one virus showed expression of FL. As the reduction in genome size did not result in a more stable phenotype, the total length of the genome is probably not the factor limiting the genetic stability of the FL-expressing viruses.
FIG. 5.
Stability of foreign gene expression of MHV-minFL. (A) LR7 cells were infected at an MOI of 8. At 8.5 h postinfection, the viral infectivity in the culture medium was determined by a quantal assay on LR7 cells, and TCID50 values were calculated. (B) In the same experiment the intracellular expression of the FL gene (in RLU) was determined by using a luminometer. Standard deviations are indicated. (C) The stability of foreign gene expression upon serial passaging was determined as described in the legend to Fig. 2 and compared to that of MHV-EFLM (D). The analysis of individual clones of MHV-minFL (passage 8) was performed as described in the legend to Fig. 3.
Instability is not linked to the FL protein or its sgRNA.
The insertion of an expression cassette into the coronavirus genome leads to the synthesis of additional sgRNA transcripts and of an additional protein in infected cells. Either one or both of these processes might have negative consequences for the virus life cycle. Though we know of no toxic effects, the FL protein might adversely interfere with infection. Similarly, the active transcription of an extra sgRNA might affect viral replication. Indeed, MHV-EFLM appeared to reach slightly lower titers in a one-step growth curve compared to MHV-WT and MHV-ERLM (9). To study these possibilities, we constructed two recombinant viruses: one in which a single nucleotide just downstream of the FL start codon was deleted (MHV-EFLMSTOP) and one in which the TRS immediately upstream of the luciferase gene was mutated (MHV-EFLMTRS) (Fig. 1 and 6). The single-nucleotide deletion in MHV-EFLMSTOP causes a frame shift, after which the reading frame runs into a stop codon resulting in a very short, 6-amino-acid predicted translation product (Fig. 6A). Theoretically a firefly luciferase protein lacking the first 29 amino acids could be synthesized, only by leaky scanning, when translation would start at the third methionine the ribosome encounters. Indeed, MHV-EFLMSTOP displayed a much lower luciferase expression level (1,000-fold), while it reached titers similar to those of MHV-EFLM (data not shown). Figure 6B shows that serial passaging of MHV-EFLMSTOP resulted in a decrease in the FL activity (compare passages 2 and 8). Analysis of viruses plaque purified from passage 8 demonstrated that 8 out of 10 viruses were completely negative in their luciferase expression. The remaining two viruses exhibited luciferase expression slightly above background levels. These results indicate that the dramatic reduction in FL protein synthesis did not result in a more stable maintenance of the FL sequences during serial passaging. The 2-nucleotide mutation in MHV-EFLMTRS was predicted to abolish FL sgRNA transcription (56). Luciferase expression was indeed strongly reduced to about 4% of MHV-EFLM, while the ability of the virus to replicate was not affected (data not shown). Upon passaging of MHV-EFLMTRS, the expression of its FL gene decreased (Fig. 6C; compare passages 2 and 8), which was confirmed again by the analysis of the plaque-purified viruses from passage 8. None of the viruses was able to express detectable levels of FL, indicating that the stability of FL expression of MHV-EFLMTRS was comparable to or lower than that of MHV-EFLM. These results indicate that manipulation of the foreign gene transcription and/or protein expression levels did not improve the preservation of the foreign sequences.
FIG. 6.
Stability of foreign gene expression of other recombinant viruses. (A) The sequences of the 3′ end of the FL expression cassettes of MHV-EFLM, MHV-EFLMSTOP, and MHV-EFLMTRS are shown. The asterisk represents a stop codon. The TRSs are indicated. The single nucleotide deleted in MHV-EFLMSTOP is shown in boldface and is indicated with an arrow in the sequence of MHV-EFLM. The nucleotides mutated in the TRS in MHV-EFLMTRS are shown in boldface and are not underlined. MHV-EFLMSTOP (B), MHV-EFLMTRS (C), and MHV-2aFLS (D) were passaged eight times as described in Materials and Methods. The viruses from passage 8 were analyzed as described in the legend to Fig. 3. As a reference, the intracellular FL expression of MHV-EFLM (passage 2) is also shown (FL). (E) MHV-FLRL and MHV-RLFL were passaged eight times as described, and viruses from passages 2, 5, and 8 were analyzed. LR7 cells were infected at an MOI of 8. At 8.5 h postinfection, the intracellular expression of RL and FL was determined, and the FL/RL ratio was calculated. Standard deviations are indicated.
Stability of FL expression depends on the site of insertion.
In all the viruses analyzed so far, the foreign gene was expressed from a cassette inserted between the E and the M genes. Although expression of the RL gene appeared to be relatively stable from this position, the observed instability of the FL gene might somehow be related to this site of insertion. Therefore, a recombinant virus was constructed in which an identical FL expression cassette was placed at a different position, upstream of the S gene (MHV-2aFLS; Fig. 1), replacing the sequences of the HE pseudogene (32). Expression of the FL gene from this position was approximately 5- to 10-fold lower than from its more downstream position between the E and the M genes. Very similar results have previously been obtained for the RL gene (9). Upon serial passaging of MHV-2aFLS, only a very limited decrease in the expression of the FL gene was observed when passage 2 and 8 viruses were compared (Fig. 6D). Analysis of viruses plaque-purified after eight passages showed that the majority of the viruses were able to express appreciable levels of luciferase activity. Apparently, the stability of FL expression is dependent on the genomic position of the FL gene. We also analyzed the stability of RL expression from different genomic positions. It appeared that a similar stability was observed for MHV-ERLM, when the RL expression cassette was placed between the M and the N gene or upstream of the S gene (data not shown).
To confirm the position-dependent stability of FL expression, viruses containing both the RL and the FL genes (9) were also passaged. Subsequently, the ratio of the FL and RL expression levels was determined at passages 2, 5, and 8. MHV-FLRL contains the FL gene between the 2a and the S genes, while the RL gene is inserted between the E and the M genes. For MHV-RLFL, the reciprocal holds true. The ratio between the FL and RL expression levels, which was high for MHV-RLFL and much lower for MHV-FLRL, was set at 100% for the passage 2 viruses. Upon serial passaging, the ratio decreased for both recombinant viruses, indicating that, regardless of the genomic position of the foreign genes, the expression of the RL gene was more stable than that of the FL gene (Fig. 6E). However, the FL/RL ratio decreased more rapidly for MHV-RLFL than for MHV-FLRL, indicating that expression of the FL gene was much more unstable when placed at the more downstream position. These results confirm that the RL gene is more stably maintained than the FL gene and that the stability of the FL gene is position dependent.
Stability of FIPV luciferase expression.
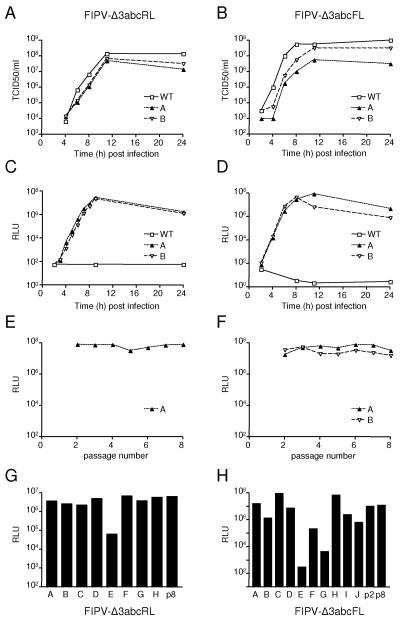
FIPV causes a progressive, usually lethal disease in cats. Recently, we demonstrated that deletion of the nonessential genes located between the S and the E genes or downstream of the N gene resulted in viable viruses that were strongly attenuated in cats. Furthermore, vaccination with the FIPV deletion mutant viruses resulted in protection against (otherwise) lethal FIPV challenge (19). The deletion mutant viruses were demonstrated to induce strong immune responses, comparable to wild-type viruses. In our view, these viruses might well be of use as multivalent vectors for protection against other feline infectious diseases. Therefore, we decided to explore the expression of foreign genes by FIPV in more detail. Furthermore, it gave us the opportunity to compare the stability of foreign gene expression between two different coronaviral vectors. In view of the observed position-dependent stability of the expression of the FL gene, with the highest stability being exhibited when the gene was introduced at the position of a nonessential gene, we decided to introduce the luciferase genes at the position of the nonessential gene cluster 3abc, combining the introduction of the foreign sequences with the deletion of this cluster. The RL and FL genes were each placed under control of the natural TRS that precedes cluster 3abc, resulting in FIPV-Δ3abcRL and FIPV-Δ3abcFL, respectively (Fig. 1). The viral recombinants were analyzed for their in vitro growth characteristics in FCWF cells. Pairs of independently generated recombinants were compared with a recombinant wild-type virus FIPV-WT. FIPV-Δ3abcRL reached high titers comparable to those of FIPV-WT, while the titers of FIPV-Δ3abcFL were somewhat lower at all time points (Fig. 7A and B). In a parallel experiment, the intracellular expression of the luciferases was analyzed. All recombinant viruses, but not FIPV-WT, expressed high levels of either RL or FL, which peaked at 8 to 10 h postinfection (Fig. 7C and D). The expression levels were similar to those found for the MHV recombinants. Luciferase expression could be demonstrated as early as 1.5 h postinfection (data not shown).
FIG. 7.
Stability of FIPV foreign gene expression. (A and B) Single-step growth kinetics of FIPV recombinants. FCWF cells were infected with each recombinant FIPV at an MOI of 8. Virus infectivity in the culture medium was determined at different times postinfection by a quantal assay on LR7 cells, and TCID50 values were calculated. Independently generated recombinants are indicated with the letters A and B. (C and D) In the same experiment, the intracellular expression of RL or FL (in relative light units [RLU]) was determined by using a luminometer. (E and F) The stability of foreign gene expression upon serial passaging was determined as described in the legend to Fig. 2, except that FCWF cells were used. (G and H) The analysis of individual clones of passage 8 viruses was performed as described in the legend to Fig. 3, except that FCWF cells were used. WT, wild type.
Both types of viruses were passaged at low MOI (0.05) up to passage 8. Subsequently, the luciferase expression by the viruses of the different passages was determined in a parallel infection experiment in FCWF cells. The expression of both RL and of FL appeared stable upon serial passaging (Fig. 7E and F). To study the stability of the RL- or FL-gene-containing viruses further, the recombinant viruses, obtained after eight passages, were analyzed in more detail. They were plaque purified once, after which stocks were grown. Subsequently, the intracellular luciferase expression of the plaque-purified viruses was determined. Passage 2 and/or passage 8 viruses were used as references (Fig. 7G and H). Seven out of eight plaque-purified viruses expressed RL to the same extent as passage 8 FIPV-Δ3abcRL. Only one plaque-purified virus was somewhat decreased in its luciferase expression. Also, most FIPV-Δ3abcFL plaque-purified viruses had maintained their high-FL expression level. Only in 3 out of 10 viruses was the FL expression decreased (viruses E, F, and G). Expression of the FL gene appeared to be more stable by FIPV-Δ3abc than by MHV-EFLM and was similar to that of MHV-2aFLS.
DISCUSSION
Over the past years coronaviruses have been shown to accept and express foreign genes (5, 9, 17, 43, 52), making them attractive as candidate vectors for vaccination and therapy. Interest in viruses for medical purposes has increased considerably during the last decades due to their amenability to genetic manipulation. While this seemed to apply particularly to DNA viruses and retroviruses, it has become clear that there may be a role as well for genuine RNA viruses. These viruses, however, are inherently more prone to genetic instability, and this might particularly be the case for coronaviruses in view of their documented high frequency of recombination (29). In the present work we have therefore initiated a study of the maintenance of foreign genes inserted into MHV and FIPV. Our results show that there are intrinsic differences in the genetic stability of different genes but also that the loss of expression depends significantly on the genomic background and on the particular genomic location of the inserted gene.
A key observation in our study was the consistently greater stability of RL compared to FL. Though the FL instability varied depending on conditions, the RL gene was maintained much better under all circumstances. Several possible reasons for this difference were already excluded. Firstly, although the FL gene is larger than the RL gene, its stability did not improve by additional deletion of the nonessential genes, suggesting that the genome size is not limiting nor that packaging constraints play a role. Secondly, the difference in RL and FL transcription (9) is not likely to play a role in the observed difference in stability. Although the FL gene was transcribed to a greater extent than the RL gene when the expression cassettes, which contain identical TRSs, were inserted between the E and the M gene (9), lowering FL transcription 25-fold by mutating its TRS did not result in a more stable phenotype. Thirdly, the FL and RL genes obviously encode proteins with different properties. Lowering the amount of expressed FL protein by more than 1,000-fold, however, did not result in a more stable phenotype. Therefore, the FL instability is not likely to be the result of the FL protein interfering in some way with the virus life cycle.
The maintenance of inserted genes obviously depends on the viral mutation and recombination rate. The observation that the larger FL gene is less stable than the RL gene might be the result of statistics, as bigger genes will be more vulnerable to deletions. Alternatively, the foreign genes may contain sequences that make them more susceptible to mutation or recombination during coronavirus replication, as recombination events within a foreign gene are dependent on the degree of base pairing between the foreign gene and the (recombinant) virus genome sequence (29). The greater instability of the FL gene might also be associated with its higher GC content (46.7%) compared to that of the RL gene (36.5%) and of the coronavirus genomes (MHV, 41.8%; FIPV, 38.1%). Indeed, for poliovirus vectors it was shown that manipulation of the GC content increased the genetic stability of foreign inserts (30). However, the GFP gene, reported to be stably expressed after insertion into TGEV and MHV (5, 17, 43, 52), has a GC content of 61 to 64%. Foreign inserts may also contain RNA secondary structures, which are prone to undergo RNA recombination. Recombination has been demonstrated to occur more frequently at regions of predicted RNA secondary structure in the MHV S gene (42). RNA recombination hot spots could, however, not be readily observed for the FL gene.
Besides the viral mutation and recombination rates, the selective advantage of newly generated virus variants is also an important determinant for the stability of foreign gene expression. Foreign inserts are likely to contain hidden “signals” like secondary RNA structures or protein binding sites that may affect viral replication or transcription. Indeed, while aberrant sgRNAs were produced after insertion of the RL and the FL genes, they were much more prominent in the latter case (9). In addition, the molar ratio of the genuine coronavirus sgRNAs and their encoded proteins was more disturbed after insertion of the FL than of the RL gene (9). Correspondingly, insertion of the FL gene appeared to result in a slightly larger replicative disadvantage than insertion of the RL gene both for MHV (9) and for FIPV (this study). During passaging, MHV carrying the FL gene between the E and M genes lost its foreign sequence and reached higher titers. However, for the recombinant FIPVs differences in the RL and FL stability were less pronounced.
The maintenance of a foreign gene is determined by its intrinsic properties (see above) as well as by its genomic position. This was clearly demonstrated by the recombinant MHVs containing the FL gene at different positions in the virus genome. A recombinant MHV in which the FL gene was inserted between the E and the M gene displayed a much less stable phenotype than a virus in which the FL gene was inserted upstream at the position of a nonessential gene. Also, in the case of FIPV the expression of the FL gene appeared to be relatively stable when the foreign gene was replacing a nonessential gene cluster. In another approach, a foreign gene (encoding GFP) was inserted into the MHV genome as a 3′ extension of the S gene (3). The foreign genetic material was, however, rapidly lost because the hybrid S/GFP protein appeared to be incorporated less efficiently into coronavirus particles than the wild-type S protein. Again, a more stable expression of the GFP gene from coronavirus genomes was observed when it replaced nonessential genes (5, 17, 43, 52). When the GFP gene was inserted into the TGEV genome downstream of the N gene but not replacing nonessential genes, a highly unstable virus was obtained (52). Consistently, attempts to insert the GFP gene downstream of the N gene into the MHV genome did not result in viable viruses expressing GFP (24). These results clearly demonstrate that the maintenance of foreign genes is determined in part by the insertion location in the coronavirus genome. They also suggest that foreign sequences are probably maintained more stably when replacing nonessential genes.
The potential of arteriviruses, with coronaviruses belonging to the order Nidovirales, to function as expression vectors has also been studied to some extent. Short foreign epitopes were genetically fused to the M or the N protein (11, 18). While three serial passages did not result in the loss of the foreign genetic material fused to the M gene, analysis of the recombinant viruses with the extended N protein demonstrated that these viruses had already lost the epitope at passage 4. The epitope could only be stably maintained for four passages when it was expressed as a precursor protein fused to the N protein, from which it was autoproteolitically cleaved. Introduction of the complete GFP gene into the arterivirus genome resulted in a highly unstable recombinant virus (12); deletion mutant viruses were already detected by RT-PCR after two passages. The poor ability of arteriviruses to accommodate foreign genetic material may be due to packaging constraints imposed by their icosahedral nucleocapsid structures. Other features that make these viruses unattractive as vectors are the absence of nonessential genes, the overlap of all adjacent ORFs, and the consequent embedding of each TRS within the preceding gene. In contrast, coronaviruses have helical nucleocapsid structures, which may be less restrictive with respect to genome size. They have a genome twice as large as that of arteriviruses containing several nonessential genes. While some coronaviruses also show an overlap of some of their genes, for others, like MHV, this is not the case.
While the stability of foreign gene expression is high for DNA viruses, that of RNA viruses is low, as RNA-dependent RNA polymerases lack a proofreading mechanism and have a high error rate (15, 57, 58). Furthermore, the genomic stability of positive-strand RNA viruses appears to be lower than that of negative-strand RNA viruses, probably because of the almost complete lack of recombination in the latter (15). RNA recombination may dramatically alter or delete portions of the RNA genome (41, 48, 55, 60). Coronaviruses display a very high recombination rate, which probably functions as a natural evolutionary strategy (29). Consistently, our sequencing results hardly demonstrated any point mutations but rather the occurrence of large deletions, while the remaining sequences were sometimes even scrambled. Deletions were also observed after fusion of GFP to the carboxy-terminal tail of the MHV S protein (3). The observed deletions are hypothesized to be the result of intra- or intermolecular recombination events. In contrast, in negative-strand RNA viruses, point mutations rather than large deletions have been observed after insertion of foreign sequences. In a recombinant vesicular stomatitis virus containing a G protein carboxy-terminally extended with GFP, expression of the GFP sequence was rapidly lost through point mutations that introduced a stop codon (7). Similarly, vesicular stomatitis virus-driven expression of a foreign gene inserted between the N and P genes was lost by point mutations that prevented transcription of the inserted gene (59).
In spite of the relatively high genomic instability of positive-strand RNA viruses, the maintenance of foreign sequences can be significantly improved, as was demonstrated for polioviruses by manipulation of the GC content (30) and for coronaviruses by selection of an appropriate foreign insert and a particular genomic location. Clearly, however, more insight into the factors affecting the genetic stability of foreign inserts will be required for the further development of coronaviruses as vectors.
Acknowledgments
We thank Linda van Genne, Jeroen Stoop, and Haukeline Volders for technical assistance during part of the work and Berend Jan Bosch and Raoul de Groot for stimulating discussions. Furthermore, we gratefully acknowledge the continuing support of Jeske Leeters.
This work was supported by grants from The Netherlands Organization for Scientific Research (NWO-VIDI-700.54.421) to C.A.M.d.H. and The Netherlands Foundation for Applied Sciences to B.J.H.
REFERENCES
- 1.Alonso, S., A. Izeta, I. Sola, and L. Enjuanes. 2002. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 76:1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, S., and S. Makino. 1998. Characterizations of coronavirus cis-acting RNA elements and the transcription step affecting its transcription efficiency. Virology 243:198-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch, B. J., C. A. M. de Haan, and P. J. M. Rottier. 2004. The coronavirus spike glycoprotein, extended carboxy terminally with the green fluorescent protein, is assembly competent. J. Virol. 78:7369-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casais, R., B. Dove, D. Cavanagh, and P. Britton. 2003. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J. Virol. 77:9084-9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis, K. M., B. Yount, and R. S. Baric. 2002. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 76:1422-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtis, K. M., B. Yount, A. C. Sims, and R. S. Baric. 2004. Reverse genetic analysis of the transcription regulatory sequence of the coronavirus transmissible gastroenteritis virus. J. Virol. 78:6061-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalton, K. P., and J. K. Rose. 2001. Vesicular stomatitis virus glycoprotein containing the entire green fluorescent protein on its cytoplasmic domain is incorporated efficiently into virus particles. Virology 279:414-421. [DOI] [PubMed] [Google Scholar]
- 8.de Haan, C. A., P. S. Masters, X. Shen, S. Weiss, and P. J. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Haan, C. A., L. van Genne, J. N. Stoop, H. Volders, and P. J. Rottier. 2003. Coronaviruses as vectors: position dependence of foreign gene expression. J. Virol. 77:11312-11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haan, C. A., H. Volders, C. A. Koetzner, P. S. Masters, and P. J. Rottier. 2002. Coronaviruses maintain viability despite dramatic rearrangements of the strictly conserved genome organization. J. Virol. 76:12491-12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries, A. A., A. L. Glaser, M. J. Raamsman, C. A. de Haan, S. Sarnataro, G. J. Godeke, and P. J. Rottier. 2000. Genetic manipulation of equine arteritis virus using full-length cDNA clones: separation of overlapping genes and expression of a foreign epitope. Virology 270:84-97. [DOI] [PubMed] [Google Scholar]
- 12.de Vries, A. A., A. L. Glaser, M. J. Raamsman, and P. J. Rottier. 2001. Recombinant equine arteritis virus as an expression vector. Virology 284:259-276. [DOI] [PubMed] [Google Scholar]
- 13.de Vries, A. A., M. C. Horzinek, P. J. Rottier, and R. J. de Groot. 1997. The genome organization of the nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin. Virol. 8:33-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enjuanes, L., I. Sola, F. Almazan, J. Ortego, A. Izeta, J. M. Gonzalez, S. Alonso, J. M. Sanchez, D. Escors, E. Calvo, C. Riquelme, and C. Sanchez. 2001. Coronavirus derived expression systems. J. Biotechnol. 88:183-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figlerowicz, M., M. Alejska, and A. Kurzynska-Kokorniak. 2003. Genetic variability: the key problem in the prevention and therapy of RNA-based virus infections. Med. Res. Rev. 23:488-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer, F., D. Peng, S. T. Hingley, S. R. Weiss, and P. S. Masters. 1997. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J. Virol. 71:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1998. Construction of a mouse hepatitis virus recombinant expressing a foreign gene. Adv. Exp. Med. Biol. 440:291-295. [DOI] [PubMed] [Google Scholar]
- 18.Groot Bramel-Verheije, M. H., P. J. Rottier, and J. J. Meulenberg. 2000. Expression of a foreign epitope by porcine reproductive and respiratory syndrome virus. Virology 278:380-389. [DOI] [PubMed] [Google Scholar]
- 19.Haijema, B. J., H. Volders, and P. J. Rottier. 2004. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 78:3863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haijema, B. J., H. Volders, and P. J. Rottier. 2003. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 77:4528-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrewegh, A. A., H. Vennema, M. C. Horzinek, P. J. Rottier, and R. J. de Groot. 1995. The molecular genetics of feline coronaviruses: comparative sequence analysis of the ORF7a/7b transcription unit of different biotypes. Virology 212:622-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiscox, J. A., K. L. Mawditt, D. Cavanagh, and P. Britton. 1995. Investigation of the control of coronavirus subgenomic mRNA transcription by using T7-generated negative-sense RNA transcripts. J. Virol. 69:6219-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsue, B., T. Hartshorne, and P. S. Masters. 2000. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 74:6911-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsue, B., and P. S. Masters. 1999. Insertion of a new transcriptional unit into the genome of mouse hepatitis virus. J. Virol. 73:6128-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeong, Y. S., J. F. Repass, Y. N. Kim, S. M. Hwang, and S. Makino. 1996. Coronavirus transcription mediated by sequences flanking the transcription consensus sequence. Virology 217:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joo, M., and S. Makino. 1992. Mutagenic analysis of the coronavirus intergenic consensus sequence. J. Virol. 66:6330-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai, M. M. C. 1996. Recombination in large RNA viruses: coronaviruses. Semin. Virol. 7:381-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, S. G., D. Y. Kim, B. H. Hyun, and Y. S. Bae. 2002. Novel design architecture for genetic stability of recombinant poliovirus: the manipulation of G/C contents and their distribution patterns increases the genetic stability of inserts in a poliovirus-based RPS-Vax vector system. J. Virol. 76:1649-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luytjes, W. 1995. Coronavirus gene expression, p. 33-54. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 32.Luytjes, W., P. J. Bredenbeek, A. F. Noten, M. C. Horzinek, and W. J. Spaan. 1988. Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology 166:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makino, S., and M. Joo. 1993. Effect of intergenic consensus sequence flanking sequences on coronavirus transcription. J. Virol. 67:3304-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makino, S., M. Joo, and J. K. Makino. 1991. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J. Virol. 65:6031-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navas, S., S. H. Seo, M. M. Chua, J. D. Sarma, E. Lavi, S. T. Hingley, and S. R. Weiss. 2001. Murine coronavirus spike protein determines the ability of the virus to replicate in the liver and cause hepatitis. J. Virol. 75:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ontiveros, E., L. Kuo, P. S. Masters, and S. Perlman. 2001. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology 289:230-238. [DOI] [PubMed] [Google Scholar]
- 37.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 76:11518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ortego, J., I. Sola, F. Almazan, J. E. Ceriani, C. Riquelme, M. Balasch, J. Plana, and L. Enjuanes. 2003. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology 308:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozdarendeli, A., S. Ku, S. Rochat, G. D. Williams, S. D. Senanayake, and D. A. Brian. 2001. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 75:7362-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips, J. J., M. M. Chua, E. Lavi, and S. R. Weiss. 1999. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J. Virol. 73:7752-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romanova, L. I., V. M. Blinov, E. A. Tolskaya, E. G. Viktorova, M. S. Kolesnikova, E. A. Guseva, and V. I. Agol. 1986. The primary structure of crossover regions of intertypic poliovirus recombinants: a model of recombination between RNA genomes. Virology 155:202-213. [DOI] [PubMed] [Google Scholar]
- 42.Rowe, C. L., J. O. Fleming, M. J. Nathan, J. Y. Sgro, A. C. Palmenberg, and S. C. Baker. 1997. Generation of coronavirus spike deletion variants by high-frequency recombination at regions of predicted RNA secondary structure. J. Virol. 71:6183-6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarma, J. D., E. Scheen, S. H. Seo, M. Koval, and S. R. Weiss. 2002. Enhanced green fluorescent protein expression may be used to monitor murine coronavirus spread in vitro and in the mouse central nervous system. J. Neurovirol. 8:381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sawicki, D., T. Wang, and S. Sawicki. 2001. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 82:385-396. [DOI] [PubMed] [Google Scholar]
- 45.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-219. [DOI] [PubMed] [Google Scholar]
- 47.Schaad, M. C., and R. S. Baric. 1993. Evidence for new transcriptional units encoded at the 3′ end of the mouse hepatitis virus genome. Virology 196:190-198. [DOI] [PubMed] [Google Scholar]
- 48.Schlesinger, S., and B. G. Weiss. 1994. Recombination between Sindbis virus RNAs. Arch. Virol. Suppl. 9:213-220. [DOI] [PubMed] [Google Scholar]
- 49.Schwarz, B., E. Routledge, and S. G. Siddell. 1990. Murine coronavirus nonstructural protein ns2 is not essential for virus replication in transformed cells. J. Virol. 64:4784-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sethna, P. B., S. L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sola, I., S. Alonso, C. Sanchez, J. M. Sanchez-Morgado, and L. Enjuanes. 2001. Expression of transcriptional units using transmissible gastroenteritis coronavirus derived minigenomes and full-length cDNA clones. Adv. Exp. Med. Biol. 494:447-451. [DOI] [PubMed] [Google Scholar]
- 52.Sola, I., S. Alonso, S. Zuniga, M. Balasch, J. Plana-Duran, and L. Enjuanes. 2003. Engineering the transmissible gastroenteritis virus genome as an expression vector inducing lactogenic immunity. J. Virol. 77:4357-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sola, I., J. L. Moreno, S. Zuniga, S. Alonso, and L. Enjuanes. 2005. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J. Virol. 79:2506-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thackray, L. B., and K. V. Holmes. 2004. Amino acid substitutions and an insertion in the spike glycoprotein extend the host range of the murine coronavirus MHV-A59. Virology 324:510-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tolskaya, E. A., L. I. Romanova, V. M. Blinov, E. G. Viktorova, A. N. Sinyakov, M. S. Kolesnikova, and V. I. Agol. 1987. Studies on the recombination between RNA genomes of poliovirus: the primary structure and nonrandom distribution of crossover regions in the genomes of intertypic poliovirus recombinants. Virology 161:54-61. [DOI] [PubMed] [Google Scholar]
- 56.van der Most, R. G., R. J. de Groot, and W. J. Spaan. 1994. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J. Virol. 68:3656-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ward, C. D., and J. B. Flanegan. 1992. Determination of the poliovirus RNA polymerase error frequency at eight sites in the viral genome. J. Virol. 66:3784-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ward, C. D., M. A. Stokes, and J. B. Flanegan. 1988. Direct measurement of the poliovirus RNA polymerase error frequency in vitro. J. Virol. 62:558-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wertz, G. W., R. Moudy, and L. A. Ball. 2002. Adding genes to the RNA genome of vesicular stomatitis virus: positional effects on stability of expression. J. Virol. 76:7642-7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White, K. A., and P. D. Nagy. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acids Res. Mol. Biol. 78:187-226. [DOI] [PubMed] [Google Scholar]
- 61.Zuniga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78:980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]