Abstract
RNA silencing is a mechanism which higher plants and animals have evolved to defend against viral infection in addition to regulation of gene expression for growth and development. As a counterdefense, many plant and some animal viruses studied to date encode RNA silencing suppressors (RSS) that interfere with various steps of the silencing pathway. In this study, we report the first identification of an RSS from a plant double-stranded RNA (dsRNA) virus. Pns10, encoded by S10 of Rice dwarf phytoreovirus (RDV), exhibited RSS activity in coinfiltration assays with the reporter green fluorescent protein (GFP) in transgenic Nicotiana benthamiana line 16c carrying GFP. The other gene segments of the RDV genome did not have such a function. Pns10 suppressed local and systemic silencing induced by sense RNA but did not interfere with local and systemic silencing induced by dsRNA. Expression of Pns10 also increased the expression of β-glucuronidase in transient assays and enhanced Potato virus X pathogenicity in N. benthamiana. Collectively, our results establish Pns10 as an RSS encoded by a plant dsRNA virus and further suggest that Pns10 targets an upstream step of dsRNA formation in the RNA silencing pathway.
RNA silencing is a conserved defense mechanism against viruses and transposons (15, 46, 59, 60, 61, 62, 67). This mechanism, discovered in a wide variety of eukaryotic organisms, has been termed posttranscriptional gene silencing (PTGS) in plants, quelling in Neurospora, and RNA interference (RNAi) in Caenorhabditis elegans and Drosophila melanogaster (7, 8, 12, 14, 17, 22, 25, 27, 47, 59). The pathway is initially triggered by double-stranded RNAs (dsRNAs), which can be generated from exogenously introduced or endogenous transposons, transgenes, and replicating viral RNA intermediates (2, 13, 17, 20, 28). The dsRNA is recognized and cleaved into small interfering RNAs (siRNAs) of 20 to 25 nucleotides (nt) by an RNase III-like RNase called Dicer (4, 27, 56, 69, 71). The siRNAs are subsequently incorporated into an RNase complex called RNA-induced silencing complex (RISC). The siRNAs direct the RISC to target RNAs by sequence-specific base pairing. Cleavage by RISC results in elimination of the target mRNA (16, 19, 27, 40, 42, 49).
In plants, fungi, and C. elegans, RNA silencing exhibits an intriguing feature: it is non-cell autonomous, which means that RNA silencing originated at one site can transmit to remote cells or tissues to cause systemic RNA silencing. In a plant, for instance, RNA silencing signals can spread between cells through the plasmodesmata and over long distances via the vascular system to silence expression of a target gene throughout the plant (23, 43, 62, 63, 65). The exact nature of RNA silencing signals remains to be elucidated. However, RNA is likely a key component to confer sequence specificity in RNA silencing (29, 41).
Recent studies suggested that RNA silencing might play a more extended role in regulation of gene expression than expected. For instance, RNA silencing can down-regulate the expression of chalcone synthase (CHS) genes in the way of natural occurrence that results in the inhibition of seed coat pigmentation in Glycine max (soybean) (50, 57). Correspondingly, an RNA silencing suppressor can alter the phenotype of the seed coat color via suppression of RNA silencing (50, 57).
Many plant viruses have evolved a suppressor or suppressors of RNA silencing to counteract RNA silencing (37, 45, 48, 52). RNA silencing is a multistep process. Correspondingly, suppressors identified so far can interfere with this process at different steps. For instance, the helper component-proteinase (HC-Pro) of potyvirus, which was one of the first suppressors identified, interferes with RNA silencing at a step upstream of the production of siRNA (6, 36, 38). Recent studies showed that HC-Pro also affects microRNA (miRNA) biogenesis and function (10, 31, 52, 68). These results suggest that the mechanisms of HC-Pro function are complex and remain to be fully understood (3). On the other hand, the 2b protein encoded by Cucumber mosaic virus (CMV) could prevent spread of RNA silencing signals by blocking their translocation (6, 23). The p25 protein of Potato virus X (PVX) interrupts transmission of RNA silencing signals by preventing their formation (64). The p21 protein of Beet yellow virus and p19 protein of tombusvirus bind to and presumably inactivate siRNA (10, 51). Recent studies showed that p69 encoded by Turnip yellow mosaic virus suppresses RNA silencing by targeting a step upstream of dsRNA formation in the cellular RNA polymerase-dependent branch of RNA silencing (11).
Over 20 suppressors encoded by both plant and animal viruses have been identified to date (15, 33, 48, 52, 62). The diversity of currently known viral suppressors in their sequences and activities suggest that novel RNA silencing suppressors are yet to be identified, and that continuing studies on the functions of viral suppressors should contribute significantly to our understanding of the basic mechanisms of RNA silencing as well as virus-host interactions (45, 62, 67).
Viruses with dsRNA genomes are of unique importance in terms of host defense and viral counterdefense. When they are not encapsidated, these dsRNA genomes are conceivably the immediate trigger and target of host RNA silencing pathways. Whether these viruses have evolved unique or common antisilencing strategies is an outstanding question in virology. The protein σ3, encoded by a mammalian reovirus, has been shown to function as an RNA silencing suppressor (35). σ3 is the outer shell protein and can bind to dsRNA. For plant dsRNA viruses, however, RNA silencing suppressors have not been identified. We use Rice dwarf phytoreovirus (RDV) as a model system to address this question. RDV is a member of the genus Phytoreovirus, belonging to the family Reoviridae (5, 70). It replicates in rice as well as in insect vectors (Nephotettix cincticeps or Resilia dorsalis) (53). The RDV genome consists of 12 dsRNAs (S1 to S12) which encode at least seven structural proteins, P1, P2, P3, P5, P7, P8, and P9, as well as five nonstructural proteins, Pns4, Pns6, Pns10, Pns11, and Pns12 (70, 72, 73, 74). The processes and viral proteins involved in viral particle assembly have been intensively studied (24, 73). For the functions of nonstructural proteins, recent evidence demonstrated that Pns11 is a nucleic acid binding protein and Pns6 is a cell-to-cell movement protein (34).
Here we present data showing that RDV Pns10 is a suppressor of RNA silencing. It may function at a step upstream of dsRNA production and prevents the spread of systemic RNA silencing signals.
MATERIALS AND METHODS
Plasmids and agrobacterium.
The RDV S10 fragment was obtained by digesting pGEM-S10 with BglII and EcoRI and cloning it into binary vector pE3 to generate pE3-S10. An S10 untranslatable mutant, in which the second Glu codon GAA was replaced with stop codon TAA, was amplified from pGEM-S10 by PCR using primers 5′-GAAGATCTCCAACATGTAAGTAGACACTGC-3′ (the premature stop codon is shown in boldfaced italics) and 5′-CGGAATTCCGTTAAGAACTGCCGCCTTTGA-3′. The PCR products were digested with BglII and EcoRI and cloned into pE3 to create pE3-S10stop. To construct pE3-GFP, the GFP fragment was obtained from pRTL2-GFP digested with HindIII and inserted into the same restriction sites of pE3. pCAMBIA1300-TAV2b and pCass3-CMV2b-GFP were kindly provided by Shouwei Ding. Each of these five binary constructs, pE3-S10, pE3-S10stop, pE3-GFP, pCAMBIA1300- TAV2b, and pCass3-CMV2b-GFP, was electroporated into Agrobacterium tumefaciens AGL-1 according to the manufacturer's instructions (Model ECM 630; BTX).
pJawohl8-gfp RNAi carrying an inverted-repeat GFP (dsRNA) insert was constructed by following the manufacturer's instructions (Invitrogen). Briefly, a “Gateway” technology of lambda-based site-specific (LR) recombination reaction was employed to obtain pENTR-GFP by mixing appropriate PCR-amplified GFP product with an entry vector, pENTR. The primers for amplifying the green fluorescent protein (GFP) were 5′-CACCATGGTGAGCAAGGGCGAGGA-3′ and 5′-GGGGTACCTTACTTGTACAGCTCGTCCA-3′. The resulting entry clone was subsequently combined with pJawohl8 RNAi to perform an LR recombination reaction, and the construct of pJawohl8-IR GFP RNAi was generated and electroporated into A. tumefaciens strain GV3101 containing pMP90RK.
To generate pGR107-S10, the PCR product of S10 amplified with primers 5′-CCATCGATATGGAAGTAGACACTGC-3′ and 5′-GCGTCGACTTAAGAACTGCCGCCTTTGA-3′ were cleaved with ClaI and SalI and inserted into pGR107 (pGR107 was kindly provided by David Baulcombe). The deletion mutant of S10 in pGR107, termed pGR107-ΔS10, was cloned by excising pGR107-S10 with Csp45I, blunted, and religated to produce pGR107-ΔS10 containing the 5′-terminal 216 nucleotides of S10. pGR107-Δ25K-S10 was generated by digesting pGR107-S10 with Bsu36I, blunted, and recircularized. The N-terminal 75 residues of 25K were included in pGR107-Δ25K-S10. pGR107 and its derivatives were electroporated into A. tumefaciens strain GV3101 containing pJIC SA_Rep.
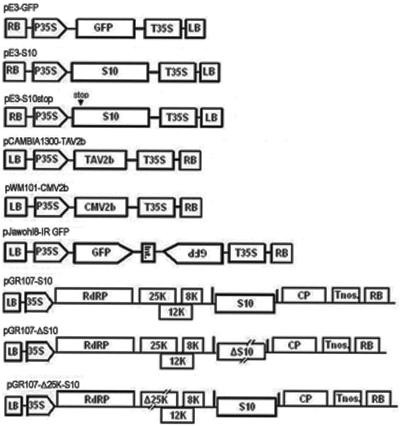
All constructs described above were verified by nucleotide sequencing. They are shown in Fig. 1.
FIG. 1.
Schematic representation of the constructs used in the present study. Shown are the common elements contained in the binary vectors' right and left borders of transfer DNA (RB and LB, respectively), the promoter of cauliflower mosaic virus (CaMV) 35S (P35S or 35S), the terminators of CaMV35S or nopaline synthase (T35S and Tnos, respectively), and genes inserted into the multicloning sites between promoters and terminators. In the diagram of pE3-S10stop, the arrow with the text “stop” above it indicates the site of the termination codon. pJawohl8-IR GFP contains inverted repeats of gfp sequences separated by an intron (Int.) from the Arabidopsis thaliana WRKY gene. In the PVX chimeras, genes are inserted between two PVX coat protein (CP) promoters shown as short vertical bars. The deletion and frameshift mutants of RDV S10 (ΔS10) and PVX 25K (Δ25K) contained in the PVX chimeras are shown as interrupted rectangles.
Agroinfiltration and GFP imaging.
The Nicotiana benthamiana plant constitutively expressing GFP transgene (line 16c; a gift from David Baulcombe) and the Agrobacterium infiltration operation have been described previously (26). The N. benthamiana line 16c plants were cultured in growth chambers at 22 to 24°C before and after infiltration. For coinfiltration, equal volumes of individual Agrobacterium cultures (optical density at 600 nm of 1) were mixed prior to infiltration. GFP fluorescence was observed under long-wavelength UV light (Black Ray model B 100A; UV Products) and photographed by using a Nikon D70 digital camera with a Y48 yellow filter.
Molecular analysis.
Quantitative and histochemical staining assays for β-glucuronidase (GUS) activity were carried out as previously described (30). Total RNAs were extracted from leaves with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. For Northern blotting analysis of GFP, PVX, S10, the 2b gene product of Tomato aspermy cucumovirus (TAV2b), and the 2b gene product of CMV (CMV2b) mRNAs, total RNA aliquots (10 μg) for each sample were separated on a 1% formaldehyde agarose gel and transferred to Hybond-N membranes (Amersham Biosciences). The membranes were hybridized with digoxigenin (DIG)-labeled probes corresponding to the full-length open reading frames (ORFs) of GFP, PVX coat protein, TAV2b, and CMV2b as well as a 300-nt fragment corresponding to the 5′ partial sequence of S10. Immunodetection was conducted by following the instructions described in the DIG system user's guide (Roche).
For Northern blot analysis of siRNAs, low-molecular-weight RNAs were enriched from total RNAs by eliminating high-molecular-weight RNA using 5% polyethylene glycol (PEG 8000) plus 0.5 M NaCl, separated on a 15% polyacrylamide-7 M urea gel, and transferred to Hybond-N membranes. The hybridization and detection of siRNA were performed as described previously (21). The probes used in the analysis of siRNA were the same as those described above for Northern blots of mRNA. For Western blot analyses of RDV Pns10 protein in the infiltrated tissues, the procedure was essentially as previously described (23, 34).
RESULTS
Identification of RDV Pns10 as an RNA silencing suppressor.
The RDV genome consists of 12 separated dsRNA segments encoding at least 12 different proteins (70). To determine whether any of these proteins might have RNA silencing suppressor (RSS) activities, we utilized an agroinfiltration bioassay (26). Each transformed agrobacterial strain carrying 1 of the 12 RDV gene segments, respectively (Fig. 1; only the S10 construct is shown; other RDV gene constructs are not shown), was mixed with a strain that carried only 35S-GFP and infiltrated into leaves of N. benthamiana line 16c. Agrobacteria harboring only the GFP gene or the 2b gene of Tomato aspermy cucumovirus (TAV2b) were used as negative and positive controls, respectively (32).
GFP expression reached the highest level in all leaves infiltrated with GFP as well as GFP plus other genes at 2 to 3 days postinfiltration (dpi), as shown by the enhanced green fluorescence in the infiltrated patches. The green fluorescence intensity remained strong in the patches coinfiltrated with 35S-GFP plus 35S-S10 and 35S-GFP plus 35S-TAV2b, respectively, during the 6- to 9-day period of observation. Coinfiltration of GFP with S10 expressed from a PVX-derived vector also resulted in strong GFP fluorescence over a similar period of observations (Fig. 2A).
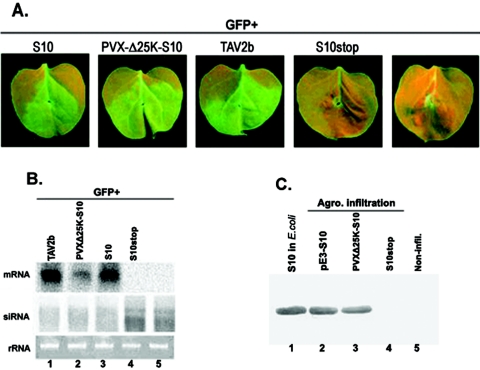
FIG. 2.
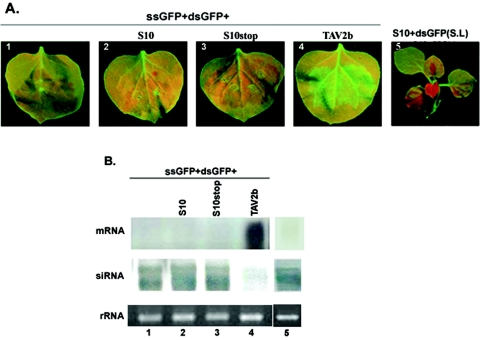
Suppression of local GFP silencing by RDV Pns10. (A) N. benthamiana line 16c plants were coinfiltrated with Agrobacterium spp. (Agro.) mixtures carrying 35S-GFP and the individual constructs indicated above each image. GFP fluorescence was viewed under long-wavelength UV light at 6 days postinfiltration (dpi). (B) Northern blot analysis of the steady-state levels of GFP mRNA and siRNA extracted from different infiltrated patches shown in panel A. The bottom gel shows rRNA with ethidium bromide staining as a loading control. (C) Western blot of total proteins from Escherichia coli and plant leaves expressing RDV Pns10, probed with polyclonal antiserum specific for Pns10. Samples from E. coli expressing S10 and those from leaf tissues expressing the S10stop mutant or from noninfiltrated (Non-infil.) leaf tissues were used as positive and negative controls, respectively.
In contrast, the green fluorescence intensity in the patches infiltrated with GFP alone or with GFP plus S10stop, in which the second codon of the S10 gene was replaced with a stop codon (see Fig. 1), declined at 3 dpi (Fig. 2A). At 6 dpi, the GFP fluorescence was hardly detectable. Similar patterns of GFP fluorescence decline were observed in patches infiltrated with GFP plus each of the other RDV ORFs (data not shown). As previously described, the disappearance of GFP fluorescence resulted from RNA silencing (6, 26, 61). In each treatment, 8 to 10 plants were infiltrated and the experiments were repeated at least three times.
Northern blot analyses revealed that the steady-state levels of GFP mRNA were much higher in tissues expressing 35S-GFP plus 35S-TAV2b or 35S-GFP plus 35S-S10 than in tissues expressing GFP alone, 35S-GFP plus 35S-S10stop (Fig. 2B), or other RDV ORFs (data not shown) at 6 dpi. Therefore, expression of S10 as well as TAV2b contributed to the stabilization of GFP mRNA that further led to elevated GFP fluorescence.
To test whether Pns10 or TAV2b increased GFP gene expression through suppression of RNA silencing, we analyzed the accumulation levels of GFP-specific siRNAs in all treatments as described in Fig. 2A. The presence of siRNAs is a hallmark of RNA silencing (25, 26). As shown in Fig. 2B, GFP siRNAs of ∼21 nt and ∼25 nt showed high levels of accumulation in leaves infiltrated with 35S-GFP alone or with 35S-GFP plus 35S-S10stop. In contrast, their levels were remarkably reduced in leaves infiltrated with 35S-GFP plus 35S-S10 or with 35S-GFP plus 35S-TAV2b (Fig. 2B).
Taken together, our data suggest that RDV S10-encoded Pns10 protein has a bona fide RSS activity. Further evidence in support of this conclusion came from Western blot analyses which showed that expression of Pns10 was directly correlated with its silencing activity (Fig. 2C).
RDV Pns10 inhibited both local and systemic RNA silencing triggered by sense GFP RNA.
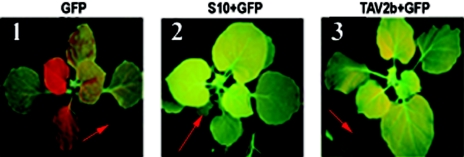
Genetic and biochemical studies indicate that RNA silencing is a complex, multistep process (3, 14, 52, 62). Furthermore, suppressors from diverse viruses interfere with different steps in the RNA silencing pathway to suppress RNA silencing (15, 45, 48, 52). The above transient bioassays showed that Pns10, as well as TAV2b, could suppress local GFP RNA silencing. Although the production of GFP siRNAs was significantly reduced, it was not completely eliminated. To determine whether Pns10 would interfere with systemic RNA silencing, we infiltrated N. benthamiana line 16c (at the four-leaf stage) as described above and monitored GFP expression or silencing in systemic leaves. In most of the plants infiltrated with 35S-GFP or with 35S-GFP plus 35S-S10stop (image not shown), the upper noninfiltrated leaves started to lose GFP fluorescence in major veins as early as 7 dpi. At 20 dpi, the whole plant lost GFP fluorescence (Fig. 3, panel 1, and Table 1, groups 1 and 2). In contrast, over 95% of noninfiltrated leaves in plants coinfiltrated with 35S-GFP plus 35S-S10s or 35S-GFP plus 35S-TAV2b retained green fluorescence over 20 dpi (Fig. 3, panels 2 and 3, and Table 1, groups 3 to 5). The steady-state levels of GFP mRNA in the noninfiltrated leaves were qualitatively correlated with GFP fluorescence (lanes 1, 2, and 4 in Fig. 4C). These results suggested that Pns10, like TAV2b, could suppress systemic RNA silencing triggered by single-stranded RNA (ssRNA) of GFP.
FIG. 3.
Effects of RDV Pns10 on systemic GFP silencing. The lower leaves (indicated with red arrows in each panel) at the four-leaf stage were coinfiltrated with agrobacteria containing 35S-ssGFP plus 35S-S10/TAV2b. The images were taken under UV light at 15 dpi.
TABLE 1.
Statistics of systemic GFP silencing in the infiltration assay
| Group | Construct | Infiltrated plant (n) | Systemic silencing (n) | % Silencing |
|---|---|---|---|---|
| 1 | ssGFP | 40 | 38 | 95 |
| 2 | S10stop+ssGFP | 36 | 33 | 91.68 |
| 3 | S10+ssGFP | 40 | 2 | 5 |
| 4 | PVXΔ25K-S10+ssGFP | 21 | 1 | 4.76 |
| 5 | TAV2b+ssGFP | 30 | 1 | 3.3 |
| 6 | S10(B)+ssGFP(T)a | 15 | 1 | 6.67 |
| 7 | S10(U)+ssGFP(L)b | 17 | 1 | 5.88 |
| 8 | S10(T)+ssGFP(B) | 15 | 15 | 100 |
| 9 | S10(L)+ssGFP(U) | 13 | 13 | 100 |
| 10 | ssGFP+dsGFP | 21 | 21 | 100 |
| 11 | S10+ssGFP+dsGFP | 23 | 23 | 100 |
| 12 | TAV2b+ssGFP+dsGFP | 21 | 19 | 90.48 |
(B) and (T) indicate the base and tip portions of infiltrated leaves, respectively.
(U) and (L) indicate the upper and lower infiltrated leaves, respectively.
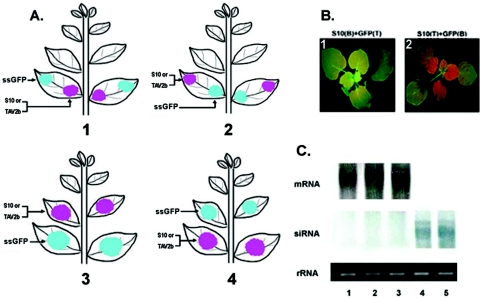
FIG. 4.
Distantly expressed RDV Pns10 blocks or inactivates systemic silencing signals. (A) Schematics showing infiltration of the indicated DNAs into different parts of a leaf or different leaves on a plant of N. benthamiana line 16c. (B) Images 1 and 2 show the different effects of Pns10 on systemic GFP silencing when the plants are infiltrated as depicted in images 1 and 2 of panel A. Letters B and T in parentheses denote the base and tip portions of the leaf infiltrated with the indicated DNA, respectively. (C) Northern blot analysis of both GFP mRNA and siRNA from samples shown in Fig. 3 and panel B. Lanes 1, 2, and 3 correspond to samples shown in panels 2 and 3 of Fig. 3 and image 1 of panel B, respectively. Lanes 4 and 5 correspond to samples shown in panel 1 of Fig. 3 and image 2 of panel B, respectively. All RNAs were extracted from plants at 15 dpi.
Pns10 blocked or inactivated the spread of mobile RNA silencing signals triggered by sense GFP RNA.
We used an assay described by Guo and Ding (23) to dissect whether Pns10 suppressed systemic RNA silencing by inactivating or blocking the spread of mobile RNA silencing signals. As illustrated in Fig. 4A, the agrobacterial suspension cells expressing the suppressor gene (35S-S10 or 35S-TAV2b) and 35S-GFP (ssRNA) were simultaneously but separately infiltrated into different parts of a leaf or into leaves at different positions of the same plant. Specifically, the cells were infiltrated into the base or tip portion of the same leaf (Fig. 4A, images 1 and 2) or the upper or lower leaves of the same plant (Fig. 4A, images 3 and 4). We reasoned that if a suppressor blocks or inactivates (Fig. 4A, images 1 and 3) the spread of RNA silencing signals produced in the regions infiltrated with 35S-ssGFP, systemic RNA silencing would not occur in the upper noninfiltrated leaves. The reverse configuration of infiltration depicted in Fig. 4A (images 2 and 4) served as a control.
The results are summarized in Table 1, and representative images are shown in Fig. 4B. The infiltration arranged as those shown in groups 6 and 7 (Table 1; illustrated in Fig. 4A, images 1 and 3), in which Pns10 or TAV2b was expressed in the zone between source and recipient tissues of the RNA silencing signals, resulted in the suppression of systemic GFP RNA silencing induced by ssGFP (Fig. 4B, image 1; the image depicted as image 3 in panel A is not shown). In contrast, systemic GFP silencing occurred in plants infiltrated in manners as depicted in the rest of the group (Table 1, groups 8 and 9, Fig. 4A, images 2 and 4, and B, image 2). The data revealed that Pns10 inhibited systemic RNA silencing by blocking or inactivating the spread of silencing signals triggered by sense GFP RNA. TAV2b exhibited the same function as Pns10 in these assays (data not shown). Northern blot analyses confirmed the observation of GFP fluorescence (Fig. 4C, images 3 and 5).
Pns10 did not suppress local or systemic RNA silencing triggered by GFP dsRNA.
It has been well established that generation of RNA silencing signals precedes siRNA formation (26, 39, 41, 61, 64). Our results suggest that Pns10 suppresses silencing by inhibiting the generation or spread of RNA silencing signals or both. To determine whether Pns10 would interfere with generation of RNA silencing signals, we tested the effect of Pns10 on GFP dsRNA-triggered silencing. We infiltrated leaves of transgenic N. benthamiana plant line 16c with Agrobacterium spp. strains harboring 35S-ssGFP (sense GFP RNA), 35S-dsGFP (IR-GFP), and a binary vector containing S10, S10stop, or TAV2b under the control of the 35S promoter. As shown in Fig. 5A (images 1 to 3), leaves infiltrated with 35S-ssGFP plus 35S-dsGFP or with 35S-ssGFP plus 35S-dsGFP plus 35S-S10 (or 35S-S10stop) lost GFP fluorescence at 5 dpi, indicating strong local GFP RNA silencing. Furthermore, systemic GFP RNA silencing developed in these plants at 14 dpi (Fig. 5A, image 5, Table 1, groups 10 and 11; S10stop+ssGFP+dsGFP is not shown). Therefore, Pns10 did not suppress local and systemic silencing induced by dsRNA. In contrast, enhanced GFP fluorescence was sustained in leaves infiltrated with 35S-ssGFP plus 35S-dsGFP plus 35S-TAV2b as long as the leaves were alive, indicating that TAV2b suppressed local GFP RNA silencing triggered by dsRNA (Fig. 5A, image 4). However, TAV2b did not suppress systemic RNA silencing induced by dsRNA (Table 1, group 12; image not shown).
FIG. 5.
Pns10 does not suppress local and systemic GFP silencing triggered by GFP dsRNA. (A) Images 1 to 4 indicate that the local leaves of N. benthamiana line 16c plants were triple infiltrated with agrobacteria carrying the constructs indicated above the images. Image 1 presents coinfiltration with ssGFP+dsGFP as a negative control. The images were taken at 5 dpi. Image 5 indicates systemic GFP silencing in the systemic leaves (shown as S.L in parentheses) induced by coinfiltration with S10 plus dsGFP. The image was photographed at 15 dpi. (B) Northern blot analyses of mRNA and siRNA in the leaves shown in images 1 to 5 of panel A and using probes specific for GFP. Ethidium bromide staining of rRNA shows loading controls.
Northern blot analyses showed negligible accumulation of GFP mRNA and high accumulation of GFP-specific siRNAs in leaves infiltrated with 35S-ssGFP plus 35S-dsGFP and 35S-ssGFP plus 35S-dsGFP plus 35S-S10/35S-S10stop (Fig. 5B, lanes 1 to 3) and in systemic leaves infiltrated with 35S-S10 plus 35S-dsGFP (Fig. 5B, lane 5), providing further evidence that Pns10 did not suppress either local or systemic RNA silencing triggered by dsRNA of GFP. On the other hand, leaves infiltrated with 35S-ssGFP plus 35S-dsGFP plus 35S-TAV2b showed high accumulation of GFP mRNA and much reduced accumulation of siRNA (Fig. 5B, lane 4).
Pns10 enhanced both transient and stable gene expression.
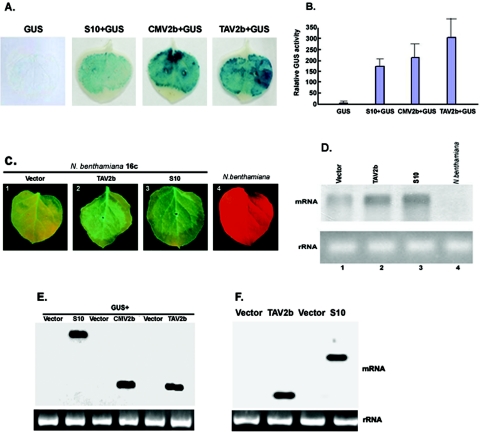
To further compare the RNA silencing suppression capacity of Pns10 with that of other known suppressors, the following experiments were carried out by using transiently expressed β-glucuronidase (GUS) as a reporter. N. benthamiana plants were infiltrated with Agrobacterium spp. containing 35S-GUS alone or 35S-GUS mixed with 35S-S10, 35S-TAV2b, or 35S-CMV2b. As shown in Fig. 6A, GUS activity was barely visible in tissues expressing GUS alone at 6 dpi. In contrast, strong GUS activities were observed in tissues infiltrated with 35S-GUS plus 35S-S10, 35S-GUS plus 35S-TAV2b, or 35S-GUS plus 35S-CMV2b. These results indicated that Pns10, like TAV2b and CMV2b, could inhibit RNA silencing triggered by an exogenously introduced gene in nontransgenic plants.
FIG. 6.
Pns10 boosts levels of transient and transgenic gene expression. (A) Enhanced activity of β-glucuronidase (GUS). Wild-type N. benthamiana plants were coinfiltrated with mixtures of agrobacteria containing constructs indicated above the images. Single infiltration of 35S-GUS served as a control. The leaves were analyzed for GUS activities at 5 dpi. (B) Quantitative comparisons of relative GUS activities in leaves infiltrated with different DNA constructs. Each bar represents the means of five measurements for each treatment plus standard deviations. (C) Enhanced GFP expression in transgenic N. benthamiana line 16c. Leaves infiltrated with the indicated DNA constructs were photographed under UV light at 3 dpi. (D) Northern blot analysis of GFP mRNA from the samples displayed in panel C. (E) Northern blot analysis of S10, TAV2b, and CMV2b mRNAs from leaves shown in panel A. (F) Northern blot analysis of S10 and TAV2b mRNAs from leaves shown in panel C.
The RNA silencing suppression effects of Pns10, CMV2b, and TAV2b were quantitatively compared. Total proteins were extracted from all samples, and GUS activities were measured via a spectrometric method (30). As shown in Fig. 6B, GUS activity in the presence of Pns10, CMV2b, and TAV2b was approximately 38-, 46-, and 66-fold higher, respectively, than that in the absence of these suppressors.
Recent evidence suggests that RNA silencing has a more general role in the regulation of gene expression in addition to its role in host defense against viral infection (50, 57). The above results from GUS assays prompted us to carry out the following experiments to determine whether Pns10 could also enhance expression of a stably integrated transgene. Leaves of N. benthamiana line 16c were infiltrated with 35S-S10 and 35S-TAV2b, respectively. In each experiment, enhanced GFP fluorescence was evident at 2 dpi and sustained for up to 9 dpi, in comparison with GFP fluorescence in leaves infiltrated with an empty vector (Fig. 6C). RNA gel blot analyses showed increased accumulation of GFP mRNA in leaves expressing Pns10 or TAV2b compared to leaves expressing the empty vector (Fig. 6D). GFP-specific siRNAs were not detected in these tissues (data not shown).
To verify that the above enhanced gene expression effects were genuinely caused by the expression of Pns10, TAV2b, and CMV2b, Northern blots were used to examine the tissues utilizing probes specific for S10, TAV2b, and CMV2b, respectively. As shown in Fig. 6E and F, S10, TAV2b, and CMV2b were expressed and their mRNAs levels are similar, suggesting that the expression of S10, TAV2b, and CMV2b was responsible for the enhanced reporter gene expression. Taken together, these results indicated that Pns10 as well as TAV2b and CMV2b enhanced both transient and stable gene expression.
Pns10 is a pathogenicity determinant in the PVX heterologous system.
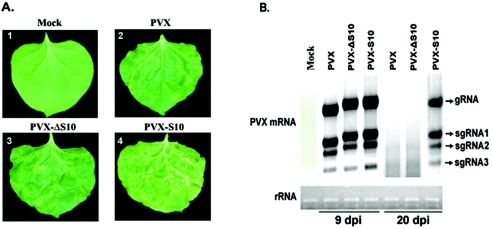
Results from the above experiments indicated that RDV Pns10 was a silencing suppressor but did not reveal whether this function would have biological significance for viral infection. The biological role of Pns10 in RDV infection cannot be investigated at this stage, because infectious RDV cDNA clones are not yet available to allow reverse genetics studies of gene functions. Therefore, we tested the biological function of Pns10 in a heterologous viral system, based on the finding that coinfection of the same host plant by two different viruses could synergistically enhance viral symptoms (44). This phenomenon is usually attributed to suppression of RNA silencing (14, 44, 58). To investigate the impact of Pns10 on the replication and infection of a heterologous virus, we utilized a PVX vector (pGR107) to express S10 and an ORF frame-shift mutant (ΔS10). PVX infection of N. benthamiana caused mild mosaic symptoms (6, 44, 54, 58). Seedlings of N. benthamiana plants (four-leaf stage) were infiltrated with PVX (pGR107), PVX-S10, and PVX-ΔS10, respectively. All inoculated leaves were asymptomatic. However, symptoms were visible in systemically infected leaves as early as 5 dpi. Variations in symptom severity were observed in individual infections. Generally, PVX-S10 caused more severe symptoms than PVX or PVX-ΔS10 did.
At 9 dpi, chlorotic and necrotic mottling was observed in most (90% of infected plants from a total of 31 tested plants) of systemically infected leaves inoculated with PVX-S10 and sustained throughout the life of the plants. However, the symptoms caused by PVX or PVX-ΔS10 developed initially as veinal chlorosis between 6 and 9 dpi and subsequently as mild chlorotic spots in some leaves. Interestingly, some leaves (82% of inoculated plants) became asymptomatic as a result of recovery from viral infection (Fig. 7A).
FIG. 7.
RDV Pns10 enhances pathogenicity of chimeric PXV. (A) Image 1 shows a leaf from a mock-infected plant. Leaves infected with PVX (image 2) or PVXΔS10 (image 3) show mild disease symptoms as a few scattered chlorotic speckles, whereas leaves infected with PSV-S10 show extensive necrotic mottles (image 4). (B) RNA gel blot analysis of accumulation of PVX genomic (gRNA) and subgenomic mRNAs (sgRNA1 to sgRNA3) at 9 and 20 dpi. Equal loading of total RNA was verified by spectrometric measurement and ethidium bromide staining of 28S RNA as indicated at the bottom of the panel.
To verify that the more severe symptoms in the presence of Pns10 indeed resulted from suppression of virus-induced gene silencing, RNA gel blot was employed to examine the steady-state levels of PVX coat protein mRNA. To correlate with degrees of symptom severity, the levels of mRNA were examined at successive time intervals between 9 and 20 dpi. At 9 dpi, the concentration of PVX mRNA was approximately the same in plants infected with PVX-S10, PVX-ΔS10, or PVX. At 20 dpi, however, the PVX mRNA accumulated to a higher level in plants infected with PVX-S10 than that in plants infected with PVX or with PVX-ΔS10 (Fig. 7B).
Taken together, these results indicate that Pns10 had a synergetic effect on PVX infection in N. benthamiana. Furthermore, this effect was much more prominent in systemically infected leaves, presumably as a result of Pns10 inhibition of RNA silencing or virus-induced gene silencing at a step in the spread of silencing signals. Therefore, the silencing suppressor function of Pns10 is important for pathogenicity in the heterologous system tested, but the role of Pns10 in reovirus infection has yet to be established.
DISCUSSION
The RNA silencing-based defense and counterdefense interplay has important ramifications for the replication of dsRNA viruses, because the dsRNA genome itself could conceivably be the immediate trigger and target of host RNA silencing when it is not encapsidated. How such viruses deal with host RNA silencing is poorly understood. In this study, a coinfiltration assay in GFP transgenic N. benthamiana line 16c was used to identify the gene from the RDV genome, a plant dsRNA virus, that would encode a function to suppress RNA silencing. We tested the functions of all 12 genes of RDV and determined that S10 encodes a suppressor (Pns10) of RNA silencing. The suppression activity is derived from Pns10 protein but not from its mRNA, because an early termination mutant or a truncated mutant of S10 could not suppress RNA silencing in coinfiltration assays. The results from the coinfiltration assay were further confirmed when Pns10 was expressed from a PVX vector with disabled silencing suppressor functions. Finally, Pns10 functions as a pathogenicity determinant in the PVX heterologous system, but its role in reovirus infection has yet to be determined. This is the first silencing suppressor identified to date from a plant dsRNA virus.
Pns10 suppresses local and systemic GFP RNA silencing induced by GFP sense RNA. This suppressor could reduce, but not eliminate, siRNA in the local and systemic RNA silencing suppression assays, suggesting that Pns10 functions similarly to turnip yellow mosaic virus p69 (11) by interfering with initial stages of RNA silencing. Supporting this hypothesis, Pns10 suppresses ssRNA-induced RNA silencing but fails to suppress dsRNA-induced RNA silencing. Presumably, presence of Pns10 prevents formation of GFP dsRNA from GFP sense RNA. Therefore, silencing cannot be initiated and silencing signals cannot be produced. Whether Pns10 interferes with the function of cellular RNA-dependent RNA polymerase or coeffectors (1, 9, 12, 18, 41, 64) remains an outstanding issue. Our results show that Pns10 as an RSS does not suppress dsRNA-induced silencing. If RDV genomic dsRNAs can trigger RNA silencing, the apparent question is how Pns10 prevents the viral dsRNAs from being degraded by RNA silencing machinery. There is evidence demonstrating that RDV genomic dsRNAs synthesize inside the core particles and bind to minor core protein P7 even when the core particles are collapsed (75 and Y. Li, unpublished data). Consequently, these encapsidated dsRNAs could not trigger RNA silencing.
Besides suppressing an early step of silencing initiation, Pns10 has the capacity to block spread of silencing signals when it is expressed between the source and recipient tissues for the signals generated by ssRNA triggers. Significantly, Pns10 does not suppress systemic spread of silencing induced by dsRNA triggers. Interestingly, TAV2b also inhibited systemic silencing induced by ssRNA but not by dsRNA. Although the underlying mechanisms remain to be understood, these observations imply that the nature of systemic silencing signals may be different depending on the silencing triggers (41, 55). Furthermore, the ability of Pns10 to block systemic silencing signals, like what CMV2b does (23), when it is expressed in a distant location from where silencing signals are generated raises the important question of whether Pns10 exerts its effects by trafficking from the expressing cells into neighboring cells to physically interact with the signals or by generating yet-to-be-identified plant responses to inhibit propagation of the signals.
Our results also indicate that Pns10 can enhance the levels of transient and stable gene expression as much as several other well-established suppressors, such as CMV2b and TAV2b. The data expand the list of silencing suppressors that may be utilized for high-throughput expression of foreign genes in higher plants (66).
Pns10 does not share significant sequence or structural similarities with the other characterized RNA silencing suppressors. The σ3 protein, which is a structural protein (outer shell protein) of a mammalian reovirus, is the only other suppressor identified to date in a dsRNA virus (35). Thus, dsRNA viruses from both plants and animals encode silencing suppressors, although they may differ in forms (i.e., structural or nonstructural) and other aspects of biological functions. Identification of silencing suppressors for dsRNA viruses establishes a new avenue of research to investigate how these important and unique viruses interact with their hosts to achieve infection.
Acknowledgments
We thank David Baulcombe for providing plasmid pGR107 and A. tumefaciens strain GV3101 containing pJIC SA_Rep and Shou-Wei Ding for providing plasmids pCass3-CMV2b-GFP and pCAMBIA1300-TAV2b. We also thank Bekir Ülker for providing plasmid pJawohl8-RNAi and Stephen Chisholm for critical reading of the manuscript.
This work was supported by a National Outstanding Youth grant (no. 30125004), a National Science Foundation of China grant (30428021), and the National High Technology Program, China (863; no. 2001AA212131), and 211 Programs from Peking University.
REFERENCES
- 1.Ahlquist, P. 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296:1270-1273. [DOI] [PubMed] [Google Scholar]
- 2.Bass, B. L. 2000. Double-stranded RNA as a template for gene silencing. Cell 101:235-238. [DOI] [PubMed] [Google Scholar]
- 3.Baulcombe, D. C. 2004. RNA silencing in plants. Nature 431:356-363. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 5.Boccardo, G., R. G. Milne. 1984. Plant reovirus group. Descriptions of plant viruses, AAB 294. [Online.] http://www.dpvweb.net/dpv/showdpv.php?dpvno=294.
- 6.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Caplen, N. J., J. Fleenor, A. Fire, and R. A. Morgan. 2000. dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene 252:95-105. [DOI] [PubMed] [Google Scholar]
- 8.Caplen, N. J., S. Parrish, F. Imani, A. Fire, and R. A. Morgan. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98:9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrington, J. C. 2000. Moving targets. Nature 408:150-151. [DOI] [PubMed] [Google Scholar]
- 10.Chapman, E. J., A. I. Prokhnevsky, K. Gopinath, V. V. Dolja, and J. C. Carrington. 2004. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18:1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J., X. L. Wan, D. X. Xie, J. R. Peng, and Shou Wei Ding. 2004. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell 16:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cogoni, C., and G. Macino. 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399:166-169. [DOI] [PubMed] [Google Scholar]
- 13.Dalmay, T., A. J. Hamilton, E. Mueller, and D. C. Baulcombe. 2000. Potato virus X amplicons in Arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell 12:369-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding, S. W. 2000. RNA silencing. Curr. Opin. Biotechnol. 11:152-156. [DOI] [PubMed] [Google Scholar]
- 15.Ding, S. W., H. W. Li, R. Lu, F. Li, and W. X. Li. 2004. RNA silencing: a conserved antiviral immunity of plants and animals. Virus Res. 102:109-115. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001b. 2001b. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 18.Forrest, E. C., C. Carlo, and G. Macino. 2004. The RNA-dependent RNA polymerase, QDE-1, is a rate-limiting factor in post-transcriptional gene silencing in Neurospora crassa. Nucleic Acids Res. 32:2123-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frederick, M., Jr. 2000. RNA degradation and models for post-transcriptional gene silencing. Plant Mol. Biol. 43:261-273. [DOI] [PubMed] [Google Scholar]
- 20.Geley, S., and C. Muller. 2004. RNAi: ancient mechanism with a promising future. Exp. Gerontol. 39:985-998. [DOI] [PubMed] [Google Scholar]
- 21.Goto, K., A. Kanazawa, M. Kusaba, and C. Masuta. 2003. A simple and rapid method to detect plant siRNAs using nonradiactive probes. Plant Mol. Biol. Rep. 21:51-58. [Google Scholar]
- 22.Grishok, A., H. Tabara, and C. C. Mello. 2000. Genetic requirements for inheritance of RNAi in C. elegans. Science 287:2494-2497. [DOI] [PubMed] [Google Scholar]
- 23.Guo, H. S., and S. W. Ding. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagiwara, K., T. Higashi, K. Namba, T. Uehara-Ichiki, and T. Omura. 2003. Assembly of single-shelled cores and double-shelled virus-like particles after baculovirus expression of major structural proteins P3, P7 and P8 of Rice dwarf virus. J. Gen. Virol. 84:981-984. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 26.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 28.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 29.Himber, C., P. Dunoyer, G. Moissiard, C. Ritzenthaler, and O. Voinnet. 2003. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jefferson, R. A., T. A. Kavanagh, and M. W. Bevan. 1987. GUS fusion: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasschau, K. D., Z. X. Xie, E. Allen, C. Llave, E. J. Chapman, K. A. Krizan, and J. C. Carrington. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4:205-217. [DOI] [PubMed] [Google Scholar]
- 32.Li, H. W., A. P. Lucy, H. S. Guo, W. X. Li, L. H. Ji, S. M. Wong, S. W. Ding. 1999. Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 18:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, H. W., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 34.Li, Y., Y. M. Bao, C. H. Wei, Z. S. Kang, Y. W. Zhong, P. Mao, G. Wu, Z. L. Chen, J. Schiemann, and R. S. Nelson. 2004. Rice dwarf phytoreovirus segment S6-encoded nonstructural protein has a cell-to-cell movement function. J. Virol. 78:5382-5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lichner, Z., D. Silhavy, and J. Burgya′n. 2003. Double-stranded RNA-binding proteins could suppress RNA interference-mediated antiviral defences. J. Gen. Virol. 84:975-980. [DOI] [PubMed] [Google Scholar]
- 36.Llave, C., K. D. Kasschau, and J. C. Carrington. 2000. Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97:13401-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu, R., A. Folimonov, M. Shintaku, W. X. Li, B. W. Falk, W. O. Dawson, and S. W. Ding. 2004. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA 101:15742-15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mallory, A. C., L. Ely, T. H. Smith, R. Marathe, R. Anandalakshmi, M. Fagard, H. Vaucheret, G. Pruss, L. Bowman, and V. B. Vance. 2001. HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13:571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mallory, A. C., S. Mlotshwa, L. H. Bowman, and V. B. Vance. 2003. The capacity of transgenic tobacco to send a systemic RNA silencing signal depends on the nature of the inducing transgene locus. Plant J. 35:82-92. [DOI] [PubMed] [Google Scholar]
- 40.Martinez, J., A. Patkaniowska, H. Urlaub, R. Luhrmann, and T. Tuschl. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110:563-574. [DOI] [PubMed] [Google Scholar]
- 41.Mlotshwa, S., O. Voinnet, M. F. Mette, M. Matzke, H. Vaucheret, S. W. Ding, G. Pruss, and V. B. Vance. 2002. RNA silencing and the mobile silencing signal. Plant Cell 14(Suppl.):S289-S301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nykanen, A., B. Haley, and P. D. Zamore. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107:309-321. [DOI] [PubMed] [Google Scholar]
- 43.Palauqui, J. C., T. Elmayan, J. M. Pollien, and H. Vaucheret. 1997. Systemic acquired silencing: transgene-specific posttranscriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16:4738-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pruss, G., X. Ge, X. M. Shi, J. C. Carrington, and V. B. Vance. 1997. Synergism: the potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell. 9:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qi, Y. J., X. H. Zhong, A. Itaya, and B. Ding. 2004. Dissecting RNA silencing in protoplasts uncovers novel effects of viral suppressors on the silencing pathway at the cellular level. Nucleic Acids Res. 32:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ratcliff, F., B. D. Harrison, and D. C. Baulcombe. 1997. A similarity between viral defense and gene silencing in plants. Science 276:1558-1560. [DOI] [PubMed] [Google Scholar]
- 47.Romano, N., and G. Macino. 1992. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6:3343-3353. [DOI] [PubMed] [Google Scholar]
- 48.Roth, B. M., G. J. Pruss, and V. B. Vance. 2004. Plant viral suppressors of RNA silencing. Virus. Res. 102:97-108. [DOI] [PubMed] [Google Scholar]
- 49.Schwarz, D. S., G. Hutvagner, B. Haley, and P. D. Zamore. 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10:537-548. [DOI] [PubMed] [Google Scholar]
- 50.Senda, M., C. Masuta, S. Ohnishi, K. Goto, A. Kasai, T. Sano, Jin-Sung Hong, and S. MacFarlaned. 2004. Patterning of virus-infected glycine max seed coat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell 16:807-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silhavy, D., A. Molnar, A. Lucioli, G. Szittya, C. Hornyik, M. Tavazza, and J. Burgyan. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silhavy, D., and J. Burgya′n. 2004. Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 9:1360-1385. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki, N., M. Sugamara, T. Kusano, H. Mori, and Y. Matsuura. 1994. Immunodetection of rice dwarf phytoreoviral proteins in both insect and plant hosts. Virology 202:41-48. [DOI] [PubMed] [Google Scholar]
- 54.Teresa, R. M., O. Voinnet, and D. C. Baulcombe. 1998. Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Timmons, L., H. Tabara, C. C. Mello, and A. Z. Fire. 2003. Inducible systemic RNA silencing in Caenorhabditis elegans. Mol. Biol. Cell 14:2972-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuschl, T., P. D. Zamore, R. Lehmann, D. P. Bartel, and P. A. Sharp. 1999. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 13:3191-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuteja, J. H., S. J. Clough, W. C. Chan, and L. O. Vodkin. 2004. Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in glycine max. Plant Cell 16:819-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vance, V. B., P. H. Berger, J. C. Carrington, A. G. Hunt, and X. M. Shi. 1995. 5′ Proximal potyviral sequences mediate potato virus X/potyviral synergistic disease in transgenic tobacco. Virology 206:583-590. [DOI] [PubMed] [Google Scholar]
- 59.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants-defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 60.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 61.Voinnet, O. 2002. RNA silencing: small RNAs as ubiquitous regulators of gene expression. Curr. Opin. Plant Biol. 5:444-451. [DOI] [PubMed] [Google Scholar]
- 62.Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nature 6:206-220. [DOI] [PubMed] [Google Scholar]
- 63.Voinnet, O., and D. C. Baulcombe. 1997. Systemic signalling in gene silencing. Nature 389:553. [DOI] [PubMed] [Google Scholar]
- 64.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157-167. [DOI] [PubMed] [Google Scholar]
- 65.Voinnet, O., P. Vain, S. Angell, and D. C. Baulcombe. 1998. Systemic spread of sequence-specific transgene RNA degradation is initiated by localized introduction of ectopic promoterless DNA. Cell 95:177-187. [DOI] [PubMed] [Google Scholar]
- 66.Voinnet, O., S. Rivas, P. Mestre, and D. C. Baulcombe. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33:949-956. [DOI] [PubMed] [Google Scholar]
- 67.Wang, M. B., and M. Metzlaff. 2005. RNA silencing and antiviral defense in plants. Curr. Opin. Plant Biol. 8:216-222. [DOI] [PubMed] [Google Scholar]
- 68.Waterhouse, P. M., and C. A. Helliwell. 2003. Exploring plant genomes by RNA-induced gene silencing. Nature 4:29-38. [DOI] [PubMed] [Google Scholar]
- 69.Xie, Z. X., K. D. Kasschau, and J. C. Carrington. 2003. Negative feedback regulation of Dicer-like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13:784-789. [DOI] [PubMed] [Google Scholar]
- 70.Xu, H., Y. Li, Z. J. Mao, Z. Wu, L. Qu, C. An, X. Ming, J. Schiemann, R. Casper, and Z. L. Chen. 1998. Rice dwarf phytovirus segment s11 encodes a nucleic acid binding protein. Virology 240:267-272. [DOI] [PubMed] [Google Scholar]
- 71.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, F., Y. Li, Y. F. Liu, C. C. An, and Z. L. Chen. 1997. Molecular cloning, sequencing, and functional analysis and expression in E. coli of major core protein gene (S3) of rice dwarf virus Chinese isolate. Acta Virol. 141:161-168. [PubMed] [Google Scholar]
- 73.Zheng, H. H., L. Yu, C. H. Wei, D. W. Hu, Y. P. Shen, Z. L. Chen, and Y. Li. 2000. Assembly of double-shelled, virus-like particles in transgenic rice plants expressing two major structural proteins of Rice dwarf virus. J. Virol. 74:9808-9810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhong, B. X., A. Kikuchi, Y. Moriyasu, T. Higashi, K. Hagiwara, and T. Omura. 2003. A minor outer capsid protein, P9, of Rice dwarf virus. Arch. Virol. 148:2275-2280. [DOI] [PubMed] [Google Scholar]
- 75.Zhong, B. X., D. A. Roth, Y. F. Zhu, T. Omura. 2004. An assembly model of Rice dwarf virus particle. Sci. China Ser. C Life Sci. 47:92-100. [DOI] [PubMed] [Google Scholar]