Abstract
We determined the complete genomic sequences of nine type 1 immunodeficient vaccine-derived poliovirus (iVDPV) isolates obtained over a 337-day period from a poliomyelitis patient from Taiwan with common variable immunodeficiency. The iVDPV isolates differed from the Sabin type 1 oral poliovirus vaccine (OPV) strain at 1.84% to 3.15% of total open reading frame positions and had diverged into at least five distinct lineages. Phylogenetic analysis suggested that the chronic infection was initiated by the fifth and last OPV dose, given 567 days before onset of paralysis, and that divergence of major lineages began very early in the chronic infection. Key determinants of attenuation in Sabin 1 had reverted in the iVDPV isolates, and representative isolates of each lineage showed increased neurovirulence for PVR-Tg21 transgenic mice. None of the isolates had retained the temperature-sensitive phenotype of Sabin 1. All isolates were antigenic variants of Sabin 1, having multiple amino acid substitutions within or near neutralizing antigenic sites 1, 2, and 3a. Antigenic divergence of the iVDPV variants from Sabin 1 followed two major independent evolutionary pathways. The emergence of distinct coreplicating lineages suggests that iVDPVs can replicate for many months at separate sites in the gastrointestinal tract. Some isolates had mosaic genome structures indicative of recombination across and within lineages. iVDPV excretion apparently ceased after 30 to 35 months of chronic infection. The appearance of a chronic VDPV excretor in a tropical, developing country has important implications for the strategy to stop OPV immunization after eradication of wild polioviruses.
The central strategy of the World Health Organization Global Polio Eradication Initiative is widespread use of oral poliovirus vaccine (OPV) at high rates of coverage. This strategy has reduced the global incidence of polio by over 99% since the start of the Initiative in 1988 and restricted wild poliovirus circulation to countries in western and central Africa and southern Asia (87). However, use of OPV is associated with some rare adverse events, including the appearance of cases of vaccine-associated paralytic poliomyelitis among OPV recipients and contacts (76), and the occurrence of polio outbreaks associated with circulating vaccine-derived poliovirus (cVDPV) (36). While cVDPV outbreaks can be prevented by maintenance of high rates of OPV coverage, the occurrence of vaccine-associated paralytic poliomyelitis is associated with the inherent genetic instability of the live, attenuated OPV strains (56).
In immunocompetent individuals, the risk of vaccine-associated paralytic poliomyelitis is very low, estimated in the United States at 1 case per 2.4 million OPV doses distributed (75, 76). The risk of vaccine-associated paralytic poliomyelitis is over 3,000-fold higher in patients with B-cell immunodeficiencies such as common variable immunodeficiency, X-linked agammaglobulinemia, and severe combined immunodeficiency (77). Moreover, whereas the period of poliovirus excretion is usually 2 to 6 weeks in susceptible immunocompetent individuals (2), it can be prolonged for up to 10 years or more in immunodeficient patients (35, 52, 76). Chronic poliovirus excretion (>12 months) appears to be very rare (37) and appears to be largely, but perhaps not exclusively (31, 54), associated with B-cell immunodeficiencies. Fewer than 25 immunodeficient chronic excretors have so far been identified since the introduction of OPV in 1961 (27, 76), and most of the patients have been from high-income countries (27, 76) such as the United States (35, 77), the United Kingdom (52, 53), Germany (5), Italy (11), and Japan (29, 91).
In this report, we describe a case of immunodeficient vaccine-associated paralytic poliomyelitis in a child from Taiwan diagnosed with common variable immunodeficiency (18, 69). The patient was found to have excreted type 1 immunodeficient vaccine-derived polioviruses (iVDPVs) for 10 months after onset of paralysis. From the evolution rate of the iVDPV isolate genomes, we estimated that the total period of excretion was from 30 to 35 months, and that the chronic infection most likely began with administration of the fifth and last dose of OPV. The iVDPV isolates obtained from the patient represented five main lineages derived from the common-source infection. All isolates were antigenic variants of the Sabin type 1 OPV strain (LSc 2ab; Sabin 1), but lineages differed in the pattern of amino acid substitution within or near neutralization antigenic sites. Divergence of the separate lineages appeared to have started very early in the chronic infection, with the earliest diverging lineage evolving largely independently. Some iVDPV isolates had mosaic genome structures indicative of recombination across and within lineages. Representative isolates from each lineage were tested for neurovirulence in PVR-Tg21 transgenic mice expressing the human receptor for poliovirus and were found to be either highly or moderately neurovirulent in this animal model. In addition, all isolates when grown in HeLa cells at 39.5°C had lost the temperature-sensitive phenotype of Sabin 1.
There was no evidence of spread of iVDPV to contacts of the case-patient. However, Taiwan currently maintains the high rates of OPV coverage needed to limit iVDPV spread. The appearance of a chronic VDPV excretor in a tropical, developing country underscores the challenges inherent to the development of a global strategy for cessation of OPV use after eradication of wild polioviruses.
MATERIALS AND METHODS
Patient.
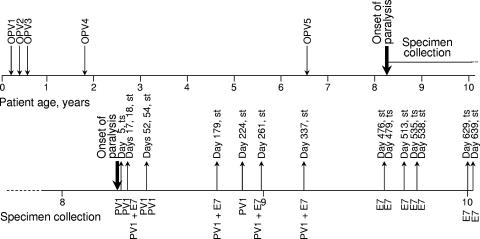
The case patient, a boy born in 1993, had received a primary series of trivalent OPV doses at ages 2, 4, 6, and 21 months and a booster dose at age 6.7 years (Fig. 1). In April 2001, at age 8 years, the patient developed acute paralysis and was diagnosed with bulbospinal poliomyelitis. Throat swabs and stool specimens were taken for virus culture, and blood specimens were taken for immunologic studies. The patient was diagnosed with common variable immunodeficiency on the basis of quantitative serum immunoglobulin readings and was placed on intravenous immunoglobulin therapy (47).
FIG. 1.
Time line summarizing immunization history, age at onset of paralysis, and time of specimen collection for the immunodeficient chronic excretor (upper line). The lower line is an expansion of the time line from the patient at age 8 years to 10.1 years showing times (in days after onset of paralysis) of collection of specimens (st, stool specimen; ts, throat swab) found to contain poliovirus type 1 (PV1) iVDPV or echovirus type 7 (E7) or both.
Virus isolation and typing.
Clinical specimens obtained from the case patient included throat swabs taken at 5, 479, 535, and 629 days after onset of paralysis, and stool specimens taken at 17, 18, 52, 54, 179, 224, 261, 337, 476, 513, 538, 639, 688, and 752 days after onset. Stool specimens were also obtained from 62 contacts, including four siblings of the case patient. Virus was isolated by culture in RD (human rhabdomyosarcoma cell line; ATCC CCL 136), L20B (mouse L cells expressing the human poliovirus receptor) (65), and HEp-2 (human cervical carcinoma cell line; ATCC CCL 23) cells (86). Echovirus type 7 (E7) isolates were initially identified by patterns of neutralization with Lim Benyesh-Melnick pools and confirmed by neutralization with E7-specific antisera and VP1 sequencing (86). Poliovirus isolates were initially characterized by immunofluorescence assay, microneutralization, diagnostic PCR (39, 40, 88), and VP1 sequencing (35, 86). Several of the poliovirus isolates had mixed-base sequences at multiple sites, and isolates from specimens taken at days 5, 17, 54, 179, 224, 261, and 337 were plaque purified before complete genomic sequencing.
Antigenic characterization.
Initial intratypic differentiation of VDPV isolates used highly specific cross-absorbed antisera in an enzyme-linked immunosorbent assay format (82). Briefly, clinical isolates were tested with two different antiserum preparations, one that reacts with Sabin 1 and another that reacts primarily with wild type 1 polioviruses. In this assay, isolates can have one of four different antigenic properties: (i) vaccine-like (reaction only with the anti-Sabin strain sera), (ii) non-vaccine-like (reaction only with anti-wild poliovirus sera), (iii) double-reactive (reaction with both anti-Sabin strain and anti-wild poliovirus sera), and (iv) nonreactive (no reaction with either anti-Sabin strain or anti-wild poliovirus sera). Only Sabin 1-related isolates have vaccine-like antigenic properties, some Sabin 1-related isolates have double reactive antigenic properties, most wild polioviruses and some Sabin 1-related antigenic variants have non-vaccine-like antigenic properties, and a small number of wild polioviruses and Sabin 1-related antigenic variants have nonreactive antigenic properties (82).
Nucleic acid sequencing.
Conditions for reverse transcription-PCR amplification and cycle sequencing were as described previously (50). Sequencing was performed in both directions, and every nucleotide position was sequenced at least once from each strand. Terminal sequences were determined by using the 5′ rapid amplification of cDNA ends (RACE) and 3′ RACE system kits (Life Technologies, Gaithersburg, Md.) according to the manufacturer's instructions.
Phylogenetic analysis.
P1/capsid, P2/noncapsid, P3/noncapsid, and complete open reading frame (ORF) sequence relationships among the nine iVDPV isolates and Sabin 1 were constructed from the corresponding regions using the maximum-likelihood method implemented in the DNAml program of the PHYLIP 3.5c package (21). The topology of the trees was obtained by majority-rule consensus among 1,000 bootstrap replicates (20, 21). The corresponding branch lengths were evaluated by likelihood ratio tests among nested models of nucleotide evolution as implemented in the program Modeltest (67). The tree with the best likelihood ratio test score was rooted to the sequences of Sabin 1 and displayed using the program TreeExplorer (K.Tamura, http://evolgen.biol.metro-u.ac.jp/TE/TE_man.html). Insertion/deletion 5′ untranslated region (5′-UTR) differences were treated as single-nucleotide substitutions.
Analysis for recombination across lineages.
Discontinuities across different genomic intervals among the day 5, 18, and 52 isolates and among the day 52, 179, and 224 isolates were initially visualized by using distance plot and bootscan functions of the Simplot program (51). The program DnaSP (71) was used to localize the likely sites of recombination by alignment of polymorphic nucleotide sites and analysis of the statistical significance of discontinuities in the extent of nucleotide sequence identity. Paired estimates of corrected nucleotide substitutions and standard errors among the recombinant sequence blocks were calculated using the MEGA software package (45).
Estimation of the time of the initiating OPV dose.
The time of the initiating OPV dose was estimated from the rate of fixation of nucleotide substitutions into the nine iVDPV isolates. The maximum-likelihood estimates of the number of synonymous substitutions at synonymous sites (Ks) that accumulated from the Sabin 1 sequence were computed according to the method of Goldman and Yang (25) as implemented in the CODEML program within the PAML package (90). This method corrects for the transition/transversion rate bias, the codon frequency bias, and for multiple substitutions at a site. Corrected Ks values relative to the root sequence (Sabin 1; Ks set to zero) for each iVDPV isolate were plotted as a function of the date of sample collection. The rate of accumulation of synonymous substitutions was estimated by weighted linear regression (where the weight for each data point was proportional to the reciprocal of the error variance for the corrected Ks value) using statistical applications within the SAS system, version 9 (SAS Institute, Inc., Cary, N.C.). The date of the initiating OPV dose was estimated from the intercept on the abscissa at Ks = 0, and the 95% confidence limits around the regression line were calculated following procedures described by Sokal and Rohlf (74).
Estimation of time of divergence of iVDPV lineages.
Maximum-likelihood estimates of divergence times were obtained assuming a unique common ancestor and a single linear rate of evolution for all iVDPV lineages, as shown by the linear regression analysis. Under these assumptions, a maximum-likelihood tree was constructed using third codon positions after an exhaustive search (78), and further analyzed under the single rate dated tips model (68) implemented in the PAML package (90) with dated internodes scaled according to the time of the last OPV dose. The parental root sequence was that of the P1/capsid region of Sabin 1 and zero time was assumed to be the date of the last OPV dose (567 days before onset of paralysis).
Numbering of nucleotide and amino acid positions.
The ORF sequences of all iVDPV isolates were colinear with those of Sabin 1, but their 5′-UTR sequences were not. To facilitate comparisons, numbering of nucleotide positions of all isolates followed that described for Sabin 1 (59), with insertions assigned serial letters. Amino acid positions were indicated by the name of the viral protein and numbered consecutively from residue 1 of each protein. Amino acid substitutions were indicated by the following convention: viral protein:original residue-position-substituted residue. For example, VP1:T106A indicates a threonine-to-alanine substitution at amino acid position 106 of VP1.
Neurovirulence testing in PVR-Tg21 mice.
Neurovirulence tests on iVDPV isolates were carried out on PVR-Tg21 mice as previously described (43, 46, 73). The mice were purchased from the Central Laboratories for Experimental Animals (Kanagawa, Japan). The type 1 reference strains were Mahoney/USA41 (neurovirulent) and Sabin 1 (attenuated). Six or eight mice (equal numbers of males and females) were inoculated (30 μl/mouse) intracerebrally for each virus dilution (in 10-fold increments; range, 1.5 to 7.5 log 50% cell culture infectious dose (CCID50)/mouse). Mice were examined daily for 14 days postinoculation, and the times of paralysis or death were recorded. The virus titer that induced paralysis or death in 50% of inoculated mice was calculated by the method of Kärber (32) and expressed as CCID50/mouse.
Measurement of temperature-sensitive phenotype.
The temperature sensitivity of the iVDPV isolates was measured by the efficiency of plating at 39.5°C compared with 34.5°C. The efficiency of plating values were determined by plaque assays performed on monolayers of HeLa cells as described previously (73, 89).
Nucleotide sequence accession numbers.
Complete genomic sequences of the nine iVDPV isolates described in this study were submitted to the GenBank library under accession numbers AF538840, AF538841, AF538842, AF538843, AY928383, AY928384, AY928385, AY928386, and AY928387 (corresponding to the day 5, 17, 52, 337, 261, 224, 54, 179, and 18 isolates, respectively).
RESULTS
Clinical and epidemiologic investigations.
The last case of poliomyelitis in Taiwan associated with circulating wild poliovirus occurred in 1982, at the end of a large outbreak (1,043 reported cases with 98 deaths) associated with an imported type 1 poliovirus (13, 41). All subsequent poliovirus isolates obtained since 1982 have been derived from the oral poliovirus vaccine. Taiwan introduced immunization with OPV in 1966 and has maintained high rates of OPV coverage since 1982 and intensive surveillance for cases of acute flaccid paralysis since 1994. In 2000, all countries within the Western Pacific Region of the World Health Organization were certified as polio-free (1, 84).
The case patient developed poliomyelitis in April 2001, 19 years after the last case associated with wild poliovirus in Taiwan. Clinical records indicated that the case patient received five doses of OPV at ages 2, 4, 6, and 21 months and 6.7 years, the last dose given 567 days (∼19 months) before onset of paralysis (Fig. 1). At age 8 years, the patient developed fever and upper respiratory tract infection followed by acute left-arm paralysis. Paralysis progressed to his right arm and both legs, and further involved difficulties in swallowing, impairment of tongue and eye movement, and respiratory muscle paralysis. The clinical diagnosis was bulbospinal poliomyelitis. A month after paralysis, the patient was diagnosed with common variable immunodeficiency, a defect in antibody production (18, 69), on the basis of quantitative serum immunoglobulin readings of 270 mg/dl for immunoglobulin G (normal range: 639 to 1,349 mg/dl), 6.2 mg/dl for immunoglobulin M (normal range: 56 to 352 mg/dl), and <5.9 mg/dl for immunoglobulin A (normal range: 70 to 132 mg/dl) (15, 28). He was immediately placed on a therapeutic regimen of monthly injections of intravenous immunoglobulin. The patient continued to have atrophy and residual paralysis in both legs more than 3 years after onset of paralysis.
Clinical specimens were obtained from the patient at days 5 (throat swab), 17, 18, 52, 54, 179, 224, 261, 337, 476, 479 (throat swab), 513, 535 (throat swab), 538, 629 (throat swab), 639, 688, and 752 (all were stool specimens if not otherwise indicated) after onset (Fig. 1). Poliovirus type 1 was isolated from all specimens taken up to day 337. Echovirus type 7 and poliovirus type 1 were isolated from the day 18, 179, 261, and 337 specimens; E7 was isolated from all specimens taken from day 476 to day 639; no virus was isolated from the day 688 and day 752 specimens.
The local bureau of health investigated the polio immunization status of schoolmates of the case patient and children in his neighborhood and community (1,682 children in all). All of the children had been vaccinated with at least 3 doses of OPV, and three contact children under 5 years had received catch-up vaccinations between July and September 2000. In addition, stool specimens were taken from 62 suspected contacts of the case patient (including his four siblings) for virus isolation. None of the contacts were found to be infected with poliovirus.
Sequence properties of poliovirus isolates.
Characterization of the early poliovirus isolates (from the days 5, 17, and 18 specimens) by enzyme-linked immunosorbent assay using cross-absorbed neutralizing antisera (82) showed that all were antigenically distinct (nonreactive or non-vaccine-like) from Sabin 1, a property subsequently confirmed for all nine isolates. However, characterization by diagnostic PCR and by sequencing of the VP1 region (906 nucleotides) showed that all isolates were derived from the Sabin 1 strain. The nine isolates differed from Sabin 1 at 2.43% to 3.53% of VP1 nucleotide sequences, from each other at 0.22% (excepting the day 52 and day 54 isolates, which had identical VP1 sequences) to 5.28% of VP1 nucleotide sequences, and from contemporary wild type 1 polioviruses (including representative isolates from the 1982 Taiwan outbreak) at >18% of VP1 nucleotide sequences (34, 73) (data not shown). The Taiwan isolates were identified as iVDPVs in accordance with the World Health Organization classification scheme, where VDPVs are poliovirus clinical isolates having 1% to 15% VP1 nucleotide sequence divergence from the parental OPV strain, and iVDPVs are VDPVs isolated from patients known to have B-cell immunodeficiencies (36).
Subsequent nucleotide sequencing of the complete genomes (7,411 to 7,441 nucleotides) of all nine isolates showed that all sequences were derived from Sabin 1. Most of the genetic differences among isolates were nucleotide substitutions, 82% of which generated synonymous codons. However, three different categories of 5′-UTR deletions were also observed. The day 17 isolate had no 5′-UTR deletions compared to Sabin 1. By contrast, all other VDPV isolates had a deletion of 8 nucleotides at positions 667 to 674, and the day 5 pharyngeal isolate had an additional deletion of 22 nucleotides at positions 695 to 716. The deletions occurred within a highly variable 5′-UTR interval that appears to have little secondary structure (63). Type 1 polioviruses with deletions within this interval have been shown to be viable in cultured cells (24, 44) and in humans (49). Four 5′-UTR nucleotide substitutions (G26A, U344C, U355C, and G480A) were shared by all isolates. All of these substitutions but U344C represent reversions back to the parental Mahoney sequence (59), and all three were consistently found among type 1 cVDPV isolates from Hispaniola (34) and the Philippines (73).
Emergence of multiple iVDPV lineages.
Relationships among the complete P1/capsid region sequences of the nine iVDPV isolates were summarized in a tree constructed using the maximum-likelihood algorithm (21) and rooted to the Sabin 1 sequence (Fig. 2A). The tree had a deeply branched topology, with five major branches corresponding to lineages A to E, extending from a single main lineage. The first lineage (A) to branch off from the main lineage is represented by the day 17 isolate. This isolate differed from all others by the absence of any 5′-UTR deletions. Replication of lineage A virus may have ceased soon after onset of paralysis, as no other lineage A isolates were subsequently found. All other isolates contained the 8-nucleotide 5′-UTR deletion, which probably became fixed into the virus population very early in the chronic infection. The P1/capsid sequences of the day 17 (lineage A) and day 18 (lineage B) isolates were quite distinct, differing at 3.9% of nucleotide positions. The extensive divergence of the two isolates was surprising because the viruses originated from stool specimens taken on consecutive days. However, the VP1 sequence of the original day 17 isolate was ambiguous at several positions, indicating that the virus population was a mixture of variants, and the isolate was plaque-purified before complete genomic sequencing. By contrast, the day 18 isolate sequences were unambiguous, so no plaque purification was performed. It is possible that the day 17 specimen also contained lineage B virus that was removed by the plaque-purification step.
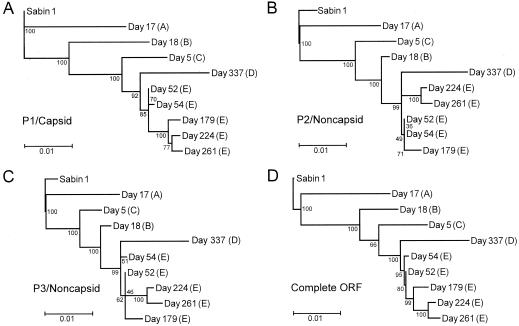
FIG. 2.
Maximum-likelihood trees summarizing sequence relationships among the Sabin 1 OPV strain (root of tree) and the nine type 1 iVDPV isolates across different genomic intervals: (A) P1/capsid region (reference interval for evolution rate calculations), (B) P2/noncapsid region, (C) P3/noncapsid region, and (D) complete ORF. Major diverging lineages are labeled A to E.
The day 18 isolate sequence appears at the tip of a long branch (Fig. 2A), suggesting that the day 18 isolate represents a second lineage that may have terminated soon after onset of paralysis. Two other distinct lineages, C and D, are represented by the day 5 and day 337 isolates, respectively. The 22-nucleotide 5′-UTR deletion apparently occurred after lineage C diverged from the evolutionary pathway leading to lineages D and E. Lineage D is defined by the sequence of the day 337 isolate, from the last poliovirus-positive stool specimen. Lineage E is defined by five closely related isolates from the day 52, day 54, day 179, day 221, and day 261 specimens. Divergence among isolates of lineage E was limited and probably occurred during the period of sampling.
All sequences of the iVDPV isolates were derived from Sabin 1. Trees of the P2/noncapsid, P3/noncapsid, and complete ORF regions had deeply branched topologies similar to the P1/capsid tree (Fig. 2). However, the four trees were not congruent, as the order of branch nodes and branch lengths differed. For example, the branch nodes of the isolate sequences were ordered day 18 (B) -day 5 (C) -day 337 (D) in the P1/capsid tree, and day 5 (C) -day 18 (B) -day 337 (D) in the P2/noncapsid and P3/noncapsid trees (Fig. 2). Bootstrap support (20, 21) for the order of these nodes was high (>90%) in all trees. Moreover, the branch length of the day 18 isolate from its node was long in the P1/capsid tree and short in the P2/noncapsid and P3/noncapsid trees.
As the window for comparison moved from the P1/capsid region to the P2/noncapsid and P3/noncapsid regions, the branch node of the day 18 isolate shifted away from the root and that of the day 5 isolate shifted toward the root. By contrast, the sequence relationships among most other isolates were similar across the ORF, apart from minor differences in tree topologies that had low bootstrap support. The exception was the day 179 isolate, within lineage E, that was most closely related to the day 224 isolate in the P1/capsid tree, but most closely related to the day 52 isolate in the P2/noncapsid and P3/noncapsid trees. Although some of the differences in tree topologies may be attributable to stochastic variability in the rates of fixation of substitutions across different lineages and genetic intervals, another mechanism probably explains the more pronounced differences.
Recombination across and within iVDPV lineages.
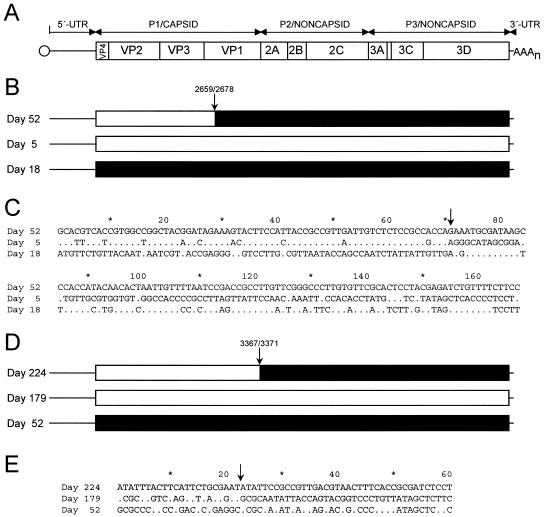
Relationships among the aligned complete ORF sequences of the day 5, day 18, and day 52 isolates were initially examined by using the distance plot and bootscan functions of the Simplot program (51), which revealed a possible recombination site between nucleotide positions 2659 and 2678, near the 5′ terminus of the VP1 region (Fig. 3B). Upstream P1/capsid region sequences (nucleotide positions 743 to 2658) of the day 52 isolate more closely matched those of the day 5 isolate (genetic distance 0.006 ± 0.002) than the day 18 isolate (genetic distance 0.035 ± 0.004), whereas the downstream ORF sequences (nucleotide positions 2679 to 7369) of the day 52 isolate more closely matched those of the day 18 isolate (genetic distance 0.007 ± 0.001) than the day 5 isolate (genetic distance 0.019 ± 0.002), a pattern suggestive of recombination.
FIG. 3.
Recombination across (panels B and C) and within (panels D and E) iVDPV lineages generating mosaic genomes. Panel A: Schematic of the poliovirus genome. The single ORF is represented by a rectangle, flanked by the 5′-UTR and 3′-UTR. Panels B and D: The most recent of the trio of isolates is represented as having the reference mosaic genome, with the sites of sequence discontinuity indicated by arrows. Sequence intervals of the reference mosaic genome most closely matching those of the preceding isolates are shaded accordingly. The 5′-UTR and 3′-UTR sequences were not included in the analyses for recombination and are represented only by lines. Panels C and E: Alignment of ORF nucleotide sites that are polymorphic among each trio of isolates. In each trio, the sequence of the most recent isolate is shown as a reference, with the sequence differences from that reference indicated below for the two preceding isolates. Polymorphic sites are numbered consecutively.
Because recombination is difficult to detect at low levels of divergence (66), recombination was further investigated using the program DnaSP (71). The sharp discontinuity in the extent of sequence identity, clearly visualized by alignment of polymorphic nucleotide sites among the ORFs of the three isolates (Fig. 3C), was very likely produced by recombination. The likelihood that the observed sequence discontinuities arose by random substitution without recombination is very low (P < 0.00001; G test with Williams's correction). Although the summary alignment (Fig. 3B) shows the day 52 isolate genome as a mosaic assembled from upstream sequences similar to those of the day 5 isolate and downstream sequences similar to those of the day 18 isolate, the actual evolutionary relationships among the three isolates is likely to be more complex. For example, runs of sequence differences in the P3/noncapsid regions of the day 18 and day 52 isolates (Fig. 3C) may signal the occurrence of additional recombination events. However, short stretches of recombinant sequences are difficult to distinguish from localized clustering of substitutions (66). More importantly, because the sequences of the parental and progeny viruses directly participating in the recombination events are unknown, and only a small number of representatives of each lineage were available for analysis, the observed mosaic structures of the genomes are explainable by several alternative pathways of recombination.
Similarly, we found that the genome of the day 224 isolate also appeared to be a mosaic, with upstream ORF sequences (nucleotide positions 743 to 3366) more closely related to those of the day 179 isolate and downstream sequences (nucleotide positions 3372 to 7369) more closely related to those of the day 52 isolate (Fig. 3D). Alignment of the polymorphic nucleotide sites (Fig. 3E) revealed discontinuities in the extent of sequence identity that are most likely attributable to recombination (P = 0.008 for random substitution without recombination; G test with Williams's correction). The statistical support for recombination among the lineage E isolates was less robust than in the previous example because the number of polymorphic sites distinguishing the ORF sequences of the three isolates was small (n = 60).
Estimation of the date of OPV exposure that initiated the chronic iVDPV infection.
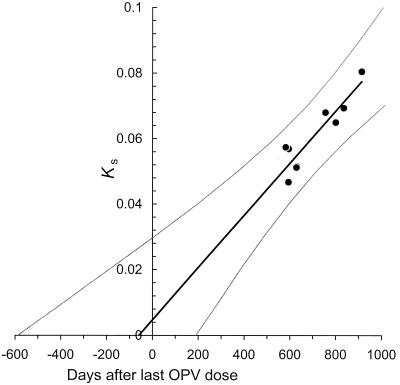
The topologies of the trees indicated that genetic divergence from Sabin 1 increased with the time of sampling (Fig. 2). An approximately linear relationship (R2 = 0.84) was obtained when the number of synonymous substitutions (Ks) in the P1/capsid region was plotted as a function of the times of collection (zero time: date of last OPV dose) for the nine specimens (Fig. 4). The linear relationship suggests that all lineages were derived from a single OPV dose. The date of the initiating OPV dose, estimated by extrapolation to the time when Ks = 0, was 59 days before the fifth and last OPV dose (95% confidence interval: 588 days before to 190 days after the last OPV dose). The rate of P1/capsid evolution estimated from the slope of the regression line was (2.92 ± 1.25) × 10-2 synonymous substitutions per synonymous site per year, a value very similar to those previously obtained for poliovirus P1/capsid region or VP1 sequences (34, 35, 50, 53). When we performed similar analyses based upon total nucleotide substitutions (Kt) in the P1/capsid region, the intercept was 172 days before the last OPV dose and the evolution rate at all sites was estimated to be (1.14 ± 0.45) × 10-2 substitutions per site per year (data not shown).
FIG. 4.
Estimate of time of the initiating OPV dose from rate of accumulation of synonymous substitutions into the P1/capsid region of the nine type 1 iVDPV isolates. Ks, number of synonymous substitutions at synonymous sites in the P1/capsid region (Sabin 1 sequence set to zero substitutions). The evolution rate was estimated by weighted linear regression (R2 = 0.84) as described in Materials and Methods. The curves flanking the regression line trace the 95% confidence limits for the time estimates calculated from Ks values.
Although the two analyses yielded comparable estimates for the time of start of the chronic infection, we generally prefer to base our estimates upon the rate of fixation of synonymous substitutions, which we assume to accumulate at a nearly constant rate by random genetic drift. We also prefer to use the P1/capsid region for our calculations because it is the largest interval within the ORF that can generally be assumed to share a recent common ancestry, as recombination of noncapsid region sequences with heterologous species C enteroviruses frequently occurs in circulating wild polioviruses (10, 49) and cVDPVs (34, 70, 73, 89). However, because sequences in all genomic intervals were derived from a recent common Sabin 1 ancestor, we were also able to estimate the date of the initiating dose using the evolutionary rate data for the complete ORF (estimated date: 95 days after the last OPV dose; evolution rate: [3.70 ± 0.67] × 10-2 synonymous substitutions per synonymous site per year; R2 = 0.95) as well as for VP1 (estimated date: 146 days before the last OPV dose; evolution rate: [2.41 ± 1.58] × 10-2 synonymous substitutions per synonymous site per year; R2 = 0.64) (data not shown). Recombination across iVDPV lineages would not substantially alter the overall rate estimates provided that all iVDPV lineages evolved at nearly equivalent rates within the same genomic intervals. These evolution rate estimates strongly implicate the fifth and last OPV dose, given 567 days before onset of paralysis, as the OPV exposure event that initiated the chronic iVDPV infection.
Estimated times of divergence of different iVDPV lineages.
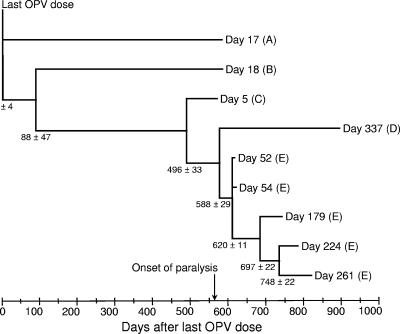
The topologies of the trees (Fig. 2) suggested that the five major coreplicating iVDPV lineages diverged from each other before or soon after the onset of paralysis. For example, lineage A, whose branch node is near the root in all four trees, apparently diverged from the main lineage very early in the chronic infection. Other lineages diverged subsequently. To estimate the dates of divergence of the major iVDPV lineages, we assumed that all lineages diverged at a single rate of evolution and we used the single rate dated tips model (68) to convert genetic distances into units of time. We further assumed that the chronic infection began with the last OPV dose given 567 days before onset of paralysis. We based our estimates of divergence time upon the sequence relationships within the interval of nucleotides 743 to 2658, the upstream 72% of the P1/capsid region that we assume to have evolved principally by progressive fixation of nucleotide substitutions (Fig. 3B).
We obtained a global evolution rate for third codon positions (representing ∼91% of all synonymous substitutions) of (3.05 ± 0.34) × 10-2 substitutions per site per year for the partial (1916-nucleotide) P1/capsid interval compared. The internal nodes on the phylogenetic tree (Fig. 5) represent point estimates of the times of divergence of lineages A to E during the chronic infection. The topology of the partial P1/capsid tree (Fig. 5) was very similar to that of the complete P1/capsid tree (Fig. 2A) except for differences in the relative branch lengths of lineages B and C. Lineage A was estimated to have diverged from lineages B to E immediately after administration of the last OPV dose. Lineage B diverged from the remaining lineages around day 88, and lineage C around day 496, of the chronic infection. Additional divergences were estimated to have occurred after the onset of paralysis (at day 567 of the chronic infection), with lineages D and E diverging around day 588, and further divergences within lineage E around days 620, 697, and 748 (Fig. 5). Selection of other genomic intervals would have yielded different dates of divergence because of the effects of recombination. However, comparisons across the capsid interval we selected yielded the most deeply branched tree (compare Fig. 2 and 5) and the earliest dates for divergence of lineages A and B. Moreover, most of our phylogenetic comparisons are restricted to P1/capsid region sequences, which encode the defining biological properties of poliovirus (10, 34, 49, 89).
FIG. 5.
Estimated times of divergence of major iVDPV lineages based upon the rate of substitution at third-codon positions within the P1/capsid region interval of nucleotides 743 to 2658. Genetic distances and standard errors were scaled under the single rate dated tips model (68) to the time of specimen collection (time zero: date of last OPV dose) as described in Materials and Methods. Note that isolates are identified by the day of specimen collection, not by the day after the last OPV dose.
Antigenic divergence of iVDPV lineages.
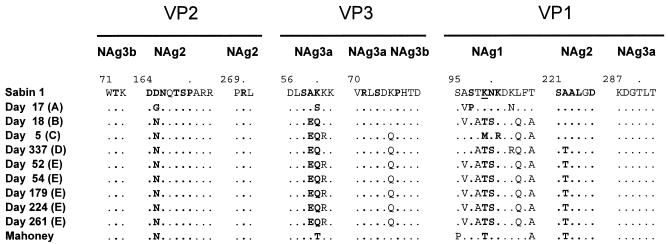
All iVDPV isolates were antigenic variants of the Sabin 1 strain. The day 17 isolate showed non-vaccine-like antigenic properties in the enzyme-linked immunosorbent assay using type-specific cross-absorbed antisera (82), whereas all of the other isolates were nonreactive. The antigenic differences are probably attributable to the numerous amino acid substitutions clustered within or near surface loops forming neutralizing antigenic (NAg) sites 1, 2, and 3a (7, 55, 62) (Fig. 6). Multiple (three to six) amino acid substitutions mapped to the interval forming most of NAg-1 (residues VP1:95 to 106, B-C loop), 1 to 2 substitutions in NAg-2 (at residue VP2:65, E-F loop; and residue VP1:222, G-H loop), and 1 to 4 substitutions within or near NAg-3 (at residues VP3:58 to 60, and VP3:75; loop preceding the B strand). All isolates had amino acid substitutions at site VP3:K060. Substitution at that site is associated with the antigenic reversion (from vaccine-like to non-vaccine-like) of Sabin 1-derived isolates tested with neutralizing monoclonal antibodies (7).
FIG. 6.
Sequences of amino acid residues within or bounding neutralizing antigenic sites 1 (VP1:95 to 106), 2 (VP2:164 to 173; VP2:269 to 271; VP1:221 to 226), 3a (VP3:56 to 62; VP3:70 to 74; VP1:287 to 292), and 3b (VP2:71 to 73; VP3:75 to 79). Residues defining the type 1 poliovirus neutralizing antigenic sites by mutations conferring resistance to neutralization by monoclonal antibodies (7, 55, 62, 83) are indicated by boldface type. The trypsin cleavage site in NAg-1 (VP1:K99), characteristic of Sabin 1 (22), is underlined.
No substitutions were found in surface residues of NAg-3b (VP2:T72 and VP3:P76) or in parts of NAg-2 (VP2:270) and NAg-3a (VP1:287 to 292) (55, 62) (Fig. 6). Four of the substitutions (VP2:D165N, VP1:K99T, VP1:T106A, and VP1:A222T) restored the amino acid of the parental Mahoney strain, and four other substitutions (VP2:D165G, VP3:A59E, VP3:K60Q, and VP1:N100S) were identical to the monoclonal antibody escape mutations that were originally used to define the NAg sites of Sabin 1 (62, 83). The trypsin cleavage site in NAg-1 (VP1:K99), characteristic of Sabin 1 (22), was consistently eliminated by amino acid substitution (day 5 isolate, VP1:K99M; days 18 to 337 isolates, VP1:K99T) in all but the day 17 isolate. Amino acid substitutions outside of the surface loops were generally conservative and mapped to β-sheet structures.
The NAg sites of the day 17 isolate (lineage A; non-vaccine-like antigenicity) were less highly substituted (5 amino acid substitutions) than those of all other iVDPV isolates (9 to 11 amino acid substitutions; nonreactive antigenicities), and only one substitution (VP1:A96V; NAg-1) was shared with any of the other isolates (lineages B and E) (Fig. 6). The non-vaccine-like antigenicity of the day 17 isolate indicates retention of the capacity to bind the anti-wild poliovirus cross-absorbed sera used in the standard enzyme-linked immunosorbent assay intratypic differentiation test (81, 86), a capacity lost by the more highly substituted nonreactive isolates. Lineage A appears to have followed an independent pathway of antigenic divergence from Sabin 1, consistent with its early divergence from the other lineages. The NAg sites of lineages B to E appear to have shared an early common pathway of antigenic divergence from Sabin 1. However, the NAg-1 sequences of the day 5 isolate (lineage C) differed from the lineage B, D, and E isolates at several positions, and may represent another pathway for antigenic evolution within NAg-1. The NAg sites of all five lineage E isolates were identical (Fig. 6), consistent with the view that the antigenic evolution had stabilized in the late stages of the chronic infection.
Neurovirulence of iVDPV isolates in PVR-Tg21 transgenic mice.
Sabin 1 is an attenuated derivative of the neurovirulent Mahoney strain (59, 72). Important determinants of the attenuation phenotype in Sabin 1 include nucleotide G480 in the 5′-UTR and amino acid residues VP4:S65 (encoded by U935), VP3:M325 (encoded by A2438), VP1:T106 (encoded by A2795), and VP1:F134 (encoded by U2879) (9, 33). Reversion of the 5′-UTR determinant, G480A, occurred in all iVDPV isolates (Table 1). Reversion of the capsid determinant, VP1:T106A, occurred in eight of the nine isolates, and reversion of another capsid determinant, VP3:M325L, occurred in seven of the nine isolates. Two determinants, VP4:S65 and VP1:F134, did not revert.
TABLE 1.
Nucleotide and amino acid substitutions at critical determinants of the attenuation and temperature-sensitive phenotypes of Sabin 1 and the corresponding phenotypes of representative Taiwan iVDPV isolates
| Virus | Lineage | 5′-UTR 480c | Viral protein:amino acid positiona,b
|
3′-UTR 7441d | Log PD50 | EOPe | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VP4:65c,d | VP3:325c,d | VP1:88d | VP1:106c | VP1:134c | 3D:73d | ||||||
| Sabin 1 | G | S | M | A | T | F | H | G | >8.0f | 0.009 | |
| Day 17 | A | A | S | M | A | T | F | Y | A | 3.7 | 1.0 |
| Day 18 | B | A | S | M | A | A | F | Y | A | 3.5 | 1.0 |
| Day 5 | C | A | S | L | A | A | F | Y | A | 2.7 | 1.3 |
| Day 337 | D | A | S | L | A | A | F | Y | A | 4.8 | 0.7 |
| Day 52 | E | A | S | L | A | A | F | Y | A | 5.6 | 0.8 |
| Day 261 | E | A | S | L | A | A | F | Y | G | 3.3 | 1.1 |
| Mahoney | A | A | L | T | A | L | Y | A | 2.8 | 1.0 | |
Amino acid substitutions correspond to the following nucleotide substitutions: VP4:S65A, U935G; VP3:M325L, A2438U; VP1:A88T, G2741A; VP1:T106A, A2795G; VP1:F134L, U2879C; and 3D:H73Y, C6203U.
Amino acid residues at all six critical sites were identical in all five lineage E isolates.
Site of major determinant of attenuation phenotype in Sabin 1.
Site of major determinant of temperature-sensitive phenotype in Sabin 1.
EOP, efficiency of plating, 39.5°C/34.5°C.
Isolates representing each lineage were tested for neurovirulence in PVR-Tg21 transgenic mice expressing the human receptor for poliovirus (Table 1). All six iVDPV isolates tested were more neurovirulent than Sabin 1, but only the day 5 pharyngeal isolate (lineage C) appeared to be as highly neurovirulent as the reference Mahoney strain. The day 17 (lineage A), day 18 (lineage B), and day 261 (lineage E) isolates were also highly neurovirulent, but the day 337 (lineage D) and day 52 (lineage E) isolates required substantially higher virus titers to induce paralysis or death in the transgenic mice. Surprisingly, the highly neurovirulent day 5 and day 261 isolates had identical alleles at the five critical sites as the less neurovirulent day 52 and day 337 isolates. Moreover, the day 52 isolate appears to be a recombinant between viruses closely related to the day 5 and day 18 isolates, but was markedly less neurovirulent than either (Table 1). These observations suggest that different genetic backgrounds may modify the effects of reversion at critical determinants of the attenuation phenotype, consistent with the composite nature of the determinants of the attenuation phenotype in Sabin 1 (26). As has recently been noted for type 1 cVDPV isolates from the Philippines (73), evolution toward increased neurovirulence is not necessarily irreversible for derivatives of Sabin 1.
Temperature sensitivity of iVDPV isolates.
All three Sabin strains have a temperature-sensitive phenotype, producing lower virus yields at supraoptimal temperatures than wild polioviruses (58, 61). Major determinants of the temperature-sensitive phenotype of Sabin 1 include two amino acid residues associated with the attenuation phenotype (VP4:S65 and VP3:M325; see above), and minor determinants include two amino acids not associated with the attenuation phenotype, VP1:A88 (encoded by G2741) and 3D:H73 (encoded by C6203), along with nucleotide G7441 in the 3′-UTR (9, 64). As described above, the determinant of the temperature-sensitive (and attenuation) phenotypes, VP4:S65, did not revert, whereas the determinant VP3:M325 reverted in all but two iVDPV isolates (Table 1).
Another capsid determinant of the temperature-sensitive phenotype, VP1:A88, also did not revert. By contrast, reversions 3D:H73Y and G7441A occurred in all nine isolates, but an A7441G backmutation restored the Sabin 1 allele in the day 261 isolate. Despite the incomplete reversion at all sites conferring the temperature-sensitive phenotype, plaque yields in HeLa cells for the nine iVDPV isolates were similar at 39.5°C and 34.5°C (Table 1). Under our experimental conditions, the iVDPV isolates were like the Mahoney strain and unlike the temperature-sensitive Sabin 1 strain, which had a ∼100-fold decrease in plaque counts at the elevated temperature (Table 1). Thus, the type 1 iVDPV isolates from Taiwan resembled the type 1 cVDPV isolates from Hispaniola (34) and the Philippines (73), which had also lost both the attenuation and temperature-sensitive phenotypes.
DISCUSSION
The genetic relationships among the type 1 iVDPV isolates described in this report highlight the dynamics of poliovirus evolution during chronic infection of an immunodeficient person. At least five distinct type 1 iVDPV lineages, derived from a single initiating OPV dose, emerged over the estimated 30 months of the chronic infection. Divergence of separate lineages began at the start of the infection and continued for at least 18 months thereafter. The five main lineages observed represent the minimum that may have emerged during the chronic infection, as some lineages may have terminated before the collection of the first clinical specimen, estimated to have been taken ∼19 months after the start of the chronic infection. Moreover, only nine isolates were obtained over 337 days after onset of paralysis, and all but two of the isolates had been plaque purified before sequencing, so that any potentially wider diversity of the viruses in the original clinical specimens may have escaped detection.
Genetic divergence was associated with changes in the antigenic surface of the viruses. Antigenic evolution of the first diverging lineage (lineage A) appeared to be less extensive than, and independent of, the evolution of the other lineages. Antigenic evolution of lineages B to E followed a common early pathway, with an initial burst of substitutions in or near NAg sites 1, 2, and 3a that probably occurred within the first 3 months of the chronic infection. Antigenic evolution of lineages B, D, and E apparently stabilized afterwards, as only four polymorphic amino acid residues were found among the NAg sites of these lineages, and the antigenic surface of all lineage E isolates apparently remained unchanged during the 209 days of replication from day 52 to day 261. By contrast, the NAg-1 sequences of the day 5 isolate (lineage C) differed from the lineage B and E isolates at five sites, possibly signaling the existence of a diverging pathway for antigenic evolution.
Extensive genetic and antigenic diversity may be a characteristic feature of iVDPV isolates from long-term chronic poliovirus excretors. Stool specimens from other immunodeficient patients with prolonged infections have also been shown to contain mixtures of divergent iVDPV variants (29, 35, 53; R. Park, unpublished data). By contrast, the genetic diversity of vaccine-related or wild poliovirus isolates from immunocompetent individuals is typically low, presumably because the duration of each infection is short. cVDPV isolates from outbreaks similarly have low genetic diversity, as the duration of their infections and modes of transmission are probably the same as for wild polioviruses (34, 70, 73, 89). Although some diversification of wild type 3 poliovirus in a single immunocompetent person has been reported, the extent of divergence was far lower than what has been observed with immunodeficient long-term chronic excretors (42).
The emergence of divergent lineages suggests that the iVDPV had established separate sites of replication within the gastrointestinal tract. Poliovirus replicates in the lymphoid tissue of the oropharynx (tonsils), the small intestine (the Peyer's patches in the ileum), and in the mesenteric lymph nodes (8). The isolation of virus from both throat swabs and stool specimens is indicative of prolonged iVDPV replication in both the oropharynx and in the intestinal tract. It is interesting that the day 5 throat isolate (lineage C) was more closely related to the stool isolates of lineages B, D, and E than was the day 17 stool isolate (lineage A). The deep branch structure of the tree suggests that the separate lineages could replicate independently for extended periods of time (for at least 19 months in the case of lineage A virus). However, the detection of recombination across lineages indicates that the tissue compartmentalization of different lineages was incomplete. It is possible that recombination within and across lineages occurs frequently, but it is difficult to distinguish recombinants from multiple substitution mutants if the parental viruses are very closely related. Such recombinants become even more difficult to recognize if the intervals exchanged are short or if multiple rounds of genetic exchange had occurred.
Although the patient was concurrently infected with E7 for many months, there was no evidence of genetic exchange between the two coinfecting enteroviruses. The lack of genetic exchange may be attributable to barriers to recombination between human enteroviruses of species B (E7) and species C (including polioviruses) (10, 60). Like the iVDPVs, the E7 isolates also showed evidence of extensive evolution and establishment of separate lineages (J.-Y. Yang, unpublished results).
The occurrence of chronic iVDPV infections may permit observation of processes that normally occur during acute poliovirus infections, but are otherwise undetectable because of the short duration of those infections. For example, intertypic recombination among the three Sabin strains is well documented (12, 23, 48) and frequently occurs in immunocompetent individuals fed trivalent OPV (17, 38), but natural intratypic recombination among OPV variants has been more difficult to demonstrate. Recently, Cherkasova et al. presented evidence consistent with intratypic recombination between coevolving lineages of virus derived from Sabin 2 (14). It is likely that intratypic recombination occurs constantly during natural poliovirus infections, as it does under experimental conditions in cell culture (16). Consistent with experimental findings (80), it is also likely that intratypic recombination in humans occurs at much higher frequencies than intertypic recombination. Similarly, it has long been known that polioviruses can colonize different sites in the gastrointestinal tract, as evidenced by the frequent isolation of virus both from stool specimens and throat swabs taken from the same immunocompetent patient. However, most poliovirus infections are cleared before the emergence of multiple variant lineages becomes evident.
Chronic poliovirus infections of immunodeficient persons may differ from acute infections involving person-to-person transmission of polioviruses in other respects. For example, the NAg sites of circulating wild polioviruses and cVDPVs appear to be more stable than those of iVDPVs from chronic excretors. The well-documented antigenic lability of the Sabin OPV strains (55, 58, 82) appears to only partially account for these differences (J. Jorba et al., unpublished results). Fluctuating antibody levels during immunoglobulin therapy may be an important contributing factor for the rapid antigenic evolution of iVDPVs.
The dynamics of genetic and phenotypic variation described here for iVDPVs are analogous to processes that typically occur during chronic infections established by other rapidly evolving viruses, among which human immunodeficiency virus type 1 has been the most extensively studied. Underlying mechanisms for the rapid evolution of human immunodeficiency virus type 1 in chronically infected patients include the emergence of multiple virus lineages, the proliferation of antigenic variants, the localization of different lineages to separate tissue compartments, and recombination across lineages (57, 92).
The two categories of VDPVs present different risks for the World Health Organization strategy to cease OPV use after global eradication of wild polioviruses (4, 19). The emergence and spread of cVDPVs can be prevented by maintenance of high rates of poliovirus vaccine coverage (36). By contrast, the emergence of iVDPVs in immunodeficient patients is rare and sporadic (27, 37) and cannot be prevented by high levels of population immunity to polioviruses. The potential for person-to-person spread of iVDPVs is unknown, because most iVDPV infections detected so far have occurred in well-immunized communities, and secondary infections have rarely been detected. It is possible that the Taiwan iVDPV had spread to close contacts but that all infections had cleared by the time the immunodeficient patient showed signs of paralysis and the case was investigated. It appears likely that iVDPVs can spread to contacts at least as efficiently as virus excreted by many healthy OPV recipients (6, 75). However, it is unknown whether iVDPVs have the same transmissibility as cVDPVs, which appear to be biologically indistinguishable from wild polioviruses (34, 70, 73, 89).
The risks of emergence of long-term chronic iVDPV excretors vary in different settings (4). Many high-income countries in Europe and North America have shifted from OPV to IPV, thereby effectively preventing the emergence of new vaccine-associated paralytic poliomyelitis cases and iVDPV infections (3). The survival rates of patients with B-cell immunodeficiencies are highest in high-income countries because of the availability of supportive immunoglobulin therapy. In such settings, the occurrence of chronic iVDPV infections is rare even among patients with B-cell immunodeficiencies, and most chronically infected patients either spontaneously cease iVDPV excretion or die from complications of their immunodeficiency (37). Nonetheless, some patients in high-income countries appear to have been infected with iVDPVs for up to 10 years or more and either experienced the late onset of paralytic poliomyelitis (5, 35) or remained asymptomatic carriers of iVDPVs (52).
Although most reports of chronic iVDPV excretors have come from high-income countries (5, 11, 29, 35, 37, 52, 53), recent reports of iVDPV infections have come from other middle-income countries such as Argentina (30), Thailand (79), and Peru (85). Middle-income countries with improving levels of sanitation and rising access to medical care may be at increasing (although very low) risk for the occurrence of immunodeficient chronic iVDPV excretors. This risk will continue as long as OPV is used. By contrast, lower-income countries appear to be at the lowest risk for the occurrence of immunodeficient chronic iVDPV excretors because conditions do not favor the survival of persons with B-cell immunodeficiencies (27). It thus appears especially important for middle- and high-income countries that improved methods for detecting iVDPV infections be developed and that effective therapies for clearing chronic poliovirus infections be found.
Acknowledgments
We thank Tsan-Chang Tseng and Tsuey-Li Lin (Taiwan CDC) and Ray Campagnoli, Naomi Dybdahl-Sissoko, Deborah Moore, Annelet Vincent, and A. J. Williams (CDC, Atlanta) for excellent technical assistance.
This work was supported, in part, by grants from the National Research Program for Genomic Medicine (93-0324-19-F-00-00-00-35), National Science Council, Taiwan; Center for Disease Control Research Project (DOH91-DC-2022), Department of Health, Taiwan; and by grants-in-aid for Promotion of Polio Eradication from the Ministry of Health, Labor and Welfare, Japan. H.S. was supported, in part, by grants-in-aid for Development of Expanded Programme on Immunization and Accelerating Measles Control in the Polio-free Era from the Ministry of Health, Labor and Welfare, Japan. A.U. was supported by grants-in-aid from the Japan Society for the Promotion of Sciences.
REFERENCES
- 1.Adams, T. 2000. Farewell to polio in the Western Pacific. Bull. W.H.O. 78:1375. [PMC free article] [PubMed]
- 2.Alexander, J. P., Jr., H. E. Gary, Jr., and M. A. Pallansch. 1997. Duration of poliovirus excretion and its implications for acute flaccid paralysis surveillance: a review of the literature. J. Infect. Dis. 175(Suppl. 1):S176-S182. [DOI] [PubMed] [Google Scholar]
- 3.Alexander, L. N., J. F. Seward, T. A. Santibanez, M. A. Pallansch, O. M. Kew, D. R. Prevots, P. M. Strebel, J. Cono, M. Wharton, W. A. Orenstein, and R. W. Sutter. 2004. Vaccine policy changes and epidemiology of poliomyelitis in the United States. JAMA 292:1696-1701. [DOI] [PubMed] [Google Scholar]
- 4.Aylward, R. B., and S. L. Cochi. 2004. Framework for evaluating the risks of paralytic poliomyelitis after global interruption of wild poliovirus transmission. Bull. W.H.O. 82:40-46. [PMC free article] [PubMed] [Google Scholar]
- 5.Bellmunt, A., G. May, R. Zell, P. Pring-Akerblom, W. Verhagen, and A. Heim. 1999. Evolution of poliovirus type I during 5.5 years of prolonged enteral replication in an immunodeficient patient. Virology 265:178-184. [DOI] [PubMed] [Google Scholar]
- 6.Benyesh-Melnick, M., J. L. Melnick, W. E. Rawls, I. Wimberley, J. Barrera-Oro, E. Ben-Porath, and V. Rennick. 1967. Studies on the immunogenicity, communicability, and genetic stability of oral poliovaccine administered during the winter. Am. J. Epidemiol. 86:112-136. [DOI] [PubMed] [Google Scholar]
- 7.Blondel, B., R. Crainic, O. Fichot, G. Dufraisse, A. Candréa, D. Diamond, M. Girard, and F. Horaud. 1986. Mutations conferring resistance to neutralization with monoclonal antibodies in type 1 poliovirus can be located outside or inside the antibody-binding site. J. Virol. 57:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodian, D., and D. M. Horstmann. 1965. Polioviruses, p. 430-473. In F. L. Horsfall and I. Tamm (ed.), Viral and rickettsial infections of man, 4th ed. J. B. Lippincott, Philadelphia, Pa.
- 9.Bouchard, M. J., D. H. Lam, and V. R. Racaniello. 1995. Determinants of attenuation and temperature sensitivity in the type 1 poliovirus Sabin strain. J. Virol. 69:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, B. A., M. S. Oberste, K. Maher, and M. Pallansch. 2003. Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the non-capsid coding region. J. Virol. 77:8973-8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buttinelli, G., V. Donati, S. Fiore, J. Marturano, A. Plebani, P. Balestri, A. R. Soresina, R. Vivarelli, F. Delpeyroux, J. Martín, and L. Fiore. 2003. Nucleotide variation in Sabin type 2 poliovirus from an immunodeficient patient with poliomyelitis. J. Gen. Virol. 84:1215-1221. [DOI] [PubMed] [Google Scholar]
- 12.Cammack, N., A. Phillips, G. Dunn, V. Patel, and P. D. Minor. 1988. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology 167:507-514. [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 1983. Update: poliomyelitis outbreak —Taiwan. Morb. Mortal. Wkly. Rep. 32:384-386. [PubMed] [Google Scholar]
- 14.Cherkasova, E. A., M. L. Yakovenko, G. V. Rezapkin, E. A. Korotkova, O. E. Ivanova, T. P. Eremeeva, L. I. Krasnoproshina, N. I. Romanenkova, N. R. Rozaeva, L. Sirota, V. I. Agol, and K. M. Chumakov. 2005. Spread of vaccine-derived poliovirus from a paralytic case in an immunodeficient child: an insight into the natural evolution of oral polio vaccine. J. Virol. 79:1062-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conley, M. E., L. D. Notarangelo, and A. Etzioni. 1999. Diagnostic criteria for primary immunodeficiencies. Clin. Immunol. 93:190-197. [DOI] [PubMed] [Google Scholar]
- 16.Cooper, P. D. 1968. A genetic map of poliovirus temperature-sensitive mutants. Virology 35:584-596. [DOI] [PubMed] [Google Scholar]
- 17.Cuervo, N. S., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, R. Crainic, and F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunningham-Rundles, C., and C. Bodian. 1999. Common variable immunodeficiency: clinical and immunological features of 248 patients. Clin. Immunol. 92:34-48. [DOI] [PubMed] [Google Scholar]
- 19.Dowdle, W. R., E. de Gourville, O. M. Kew, M. A. Pallansch, and D. J. Wood. 2003. Polio eradication: the OPV paradox. Rev. Med. Virol. 13:277-291. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein, J. 2004. Inferring phylogenies. Sinauer Associates, Sunderland, Mass.
- 21.Felsenstein, J. 1993. PHYLIP (phylogeny inference package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 22.Fricks, C. E., J. P. Icenogle, and J. M. Hogle. 1985. Trypsin sensitivity of the Sabin strain type 1 poliovirus: cleavage sites in virions and related particles. J. Virol. 54:856-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furione, M., S. Guillot, D. Otelea, J. Balanant, A. Candrea, and R. Crainic. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated poliomyelitis. Virology 196:199-208. [DOI] [PubMed] [Google Scholar]
- 24.Gmyl, A. P., E. V. Pilipenko, S. V. Maslova, G. A. Belov, and V. I. Agol. 1993. Functional and genetic plasticities of the poliovirus genome: quasi-infectious RNAs modified in the 5′-untranslated region yield a variety of pseudorevertants. J. Virol. 67:6309-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldman, N., and Z. Yang. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11:725-736. [DOI] [PubMed] [Google Scholar]
- 26.Gromeier, M., and A. Nomoto. 2002. Determinants of poliovirus pathogenesis, p. 367-379. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 27.Halsey, N. A., J. Pinto, F. Espinosa-Rosales, M. A. Faure-Fontenia, E. da Silva, A. J. Khan, A. D. B. Webster, P. D. Minor, G. Dunn, E. Asturias, H. Hussain, M. A. Pallansch, O. M. Kew, J. Winkelstein, R. Sutter, and the Polio Project Team. 2004. Search for poliovirus carriers in persons with primary immune deficiency diseases in the United States, Mexico, Brazil, and the United Kingdom. Bull. W.H.O. 82:3-8. [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton, R. G. 1997. Human immunoglobulins, p. 65-109. In M. S. Leffell, A. D. Donnenberg, and N. R. Rose (ed.), Handbook of human immunology. CRC Press, Boca Raton, Fla.
- 29.Hara, M., Y. Saito, T. Komatsu, H. Kodama, W. Abo, S. Chiba, and T. Nakao. 1981. Antigenic analysis of polioviruses isolated from a child with agammaglobulinemia and paralytic poliomyelitis after Sabin vaccine administration. Microbiol. Immunol. 25:905-913. [DOI] [PubMed] [Google Scholar]
- 30.Hidalgo, S., M. Garcia Erro, D. Cisterna, and M. C. Freire. 2003. Paralytic poliomyelitis caused by a vaccine-derived polio virus in an antibody-deficient Argentinean child. Pediatr. Infect. Dis. J. 22:570-572. [PubMed] [Google Scholar]
- 31.Hovi, T., N. Lindholm, C. Savolainen, M. Stenvik, and C. Burns. 2004. Evolution of wild-type 1 poliovirus in two healthy siblings excreting the virus over a period of 6 months. J. Gen. Virol. 85:369-377. [DOI] [PubMed] [Google Scholar]
- 32.Kärber, G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch. Exp. Pathol. Pharmakol. 162:480-483. [Google Scholar]
- 33.Kawamura, N., M. Kohara, S. Abe, T. Komatsu, K. Tago, M. Arita, and A. Nomoto. 1989. Determinants in the 5′ noncoding region of poliovirus Sabin 1 RNA that influence the attenuation phenotype. J. Virol. 63:1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kew, O. M., V. Morris-Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. André, E. Blackman, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, H. Shimizu, T. Yoneyama, T. Miyamura, H. van der Avoort, M. S. Oberste, D. Kilpatrick, S. Cochi, M. Pallansch, and C. de Quadros. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 35.Kew, O. M., R. W. Sutter, B. Nottay, M. McDonough, D. R. Prevots, L. Quick, and M. Pallansch. 1998. Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J. Clin. Microbiol. 36:2893-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kew, O. M., P. F. Wright, V. I. Agol, F. Delpeyroux, H. Shimizu, N. Nathanson, and M. A. Pallansch. 2004. Circulating vaccine-derived polioviruses: current state of knowledge. Bull. W.H.O. 82:16-23. [PMC free article] [PubMed] [Google Scholar]
- 37.Khetsuriani, N., D. R. Prevots, L. Quick, M. E. Elder, M. Pallansch, O. Kew, and R. W. Sutter. 2003. Persistence of vaccine-derived polioviruses among immunodeficient persons with vaccine-associated paralytic poliomyelitis. J. Infect. Dis. 188:1845-1852. [DOI] [PubMed] [Google Scholar]
- 38.Kilpatrick, D. R., K. Ching, J. Iber, R. Campagnoli, C. J. Freeman, N. Mishrik, M. A. Pallansch, and O. M. Kew. 2004. Multiplex PCR method for identifying recombinant vaccine-related polioviruses. J. Clin. Microbiol. 42:4313-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kilpatrick, D. R., B. Nottay, C.-F. Yang, S.-J. Yang, E. da Silva, S. Peñaranda, M. Pallansch, and O. Kew. 1998. Serotype-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy. J. Clin. Microbiol. 36:352-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kilpatrick, D. R., B. Nottay, C.-F. Yang, S.-J. Yang, M. N. Mulders, B. P. Holloway, M. A. Pallansch, and O. M. Kew. 1996. Group-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy. J. Clin. Microbiol. 34:2990-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim-Farley, R. J., G. Rutherford, P. Lichfield, S. T. Hsu, W. A. Orenstein, L. B. Schonberger, K. J. Bart, K. J. Lui, and C. C. Lin. 1984. Outbreak of paralytic poliomyelitis, Taiwan. Lancet ii:1322-1324. [DOI] [PubMed] [Google Scholar]
- 42.Kinnunen, L., T. Pöyry, and T. Hovi. 1991. Generation of virus genetic lineages during an outbreak of poliomyelitis. J. Gen. Virol. 72:2483-2489. [DOI] [PubMed] [Google Scholar]
- 43.Koike, S., C. Taya, T. Kurata, S. Abe, I. Ise, H. Yonekawa, and A. Nomoto. 1991. Transgenic mice susceptible to poliovirus. Proc. Natl. Acad. Sci. USA 88:951-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuge, S., and A. Nomoto. 1987. Construction of viable deletion and insertion mutants of the Sabin strain of type 1 poliovirus: function of the 5′ noncoding sequence in viral replication. J. Virol. 61:1478-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetic analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 46.Li, J., L.-B. Zhang, T. Yoneyama, H. Yoshida, H. Shimizu, K. Yoshii, M. Hara, T. Nomura, H. Yoshikura, T. Miyamura, and A. Hagiwara. 1996. Genetic basis of the neurovirulence of type 1 polioviruses isolated from vaccine-associated paralytic patients. Arch. Virol. 141:1047-1054. [DOI] [PubMed] [Google Scholar]
- 47.Lin, S.-J., H.-C. Chao, K.-W. Chang, and D.-C. Yan. 2003. Effect of interleukin 15 and interleukin 2 on anti-CD3-induced T-cell activation and apoptosis in children with common variable immunodeficiency. Ann. Allergy Asthma Immunol. 91:65-70. [DOI] [PubMed] [Google Scholar]
- 48.Lipskaya, G. Y., A. R. Muzychenko, O. K. Kutitova, S. V. Maslova, M. Equestre, S. G. Drozdov, R. Perez Bercoff, and V. I. Agol. 1991. Frequent isolation of intertypic poliovirus recombinants with serotype 2 specificity from vaccine-associated polio cases. J. Med. Virol. 35:290-296. [DOI] [PubMed] [Google Scholar]
- 49.Liu, H.-M., D.-P. Zheng, L.-B. Zhang, M. S. Oberste, O. M. Kew, and M. A. Pallansch. 2003. Serial recombination during circulation of type 1 wild-vaccine recombinant polioviruses in China. J. Virol. 77:10994-11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu, H.-M., D.-P. Zheng, L.-B. Zhang, M. S. Oberste, M. A. Pallansch, and O. M. Kew. 2000. Molecular evolution of a type 1 wild-vaccine poliovirus recombinant during widespread circulation in China. J. Virol. 74:11153-11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacLennan, C., G. Dunn, A. P. Huissoon, D. S. Kumararatne, J. Martín, P. O'Leary, R. A. Thompson, H. Osman, P. Wood, P. Minor, D. J. Wood, and D. Pillay. 2004. Failure to clear persistent vaccine-derived neurovirulent poliovirus infection in an immunodeficient man. Lancet 363:1509-1513. [DOI] [PubMed] [Google Scholar]
- 53.Martín, J., G. Dunn, R. Hull, V. Patel, and P. D. Minor. 2000. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J. Virol. 74:3001-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martín, J., K. Odoom, G. Tuite, G. Dunn, N. Hopewell, G. Cooper, C. Fitzharris, K. Butler, W. W. Hall, and P. D. Minor. 2004. Long-term excretion of vaccine-derived poliovirus by a healthy child. J. Virol. 78:13839-13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Minor, P. D. 1990. Antigenic structure of picornaviruses. Curr. Top. Microbiol. Immunol. 161:121-154. [DOI] [PubMed] [Google Scholar]
- 56.Minor, P. D., and J. W. Almond. 2002. Poliovirus vaccines: molecular biology and immune response, p. 381-390. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 57.Morris, A., M. Marsden, K. Halcrow, E. S. Hughes, R. P. Brettle, J. E. Bell, and P. Simmonds. 1999. Mosaic structure of the human immunodeficiency virus type 1 genome infecting lymphoid cells and the brain: evidence for frequent in vivo recombination events in the evolution of regional populations. J. Virol. 73:8720-8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakano, J. H., M. H. Hatch, M. L. Thieme, and B. Nottay. 1978. Parameters for differentiating vaccine-derived and wild poliovirus strains. Prog. Med. Virol. 24:78-206. [PubMed] [Google Scholar]
- 59.Nomoto, A., T. Omata, H. Toyoda, S. Kuge, H. Horie, Y. Kataoka, Y. Genba, Y. Nakano, and N. Imura. 1982. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc. Natl. Acad. Sci. USA 79:5793-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oberste, M. S., K. Maher, and M. A. Pallansch. 2004. Evidence for frequent recombination within species human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J. Virol. 78:855-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Omata, T., M. Kohara, S. Kuge, T. Komatsu, S. Abe, B. L. Semler, A. Kameda, H. Itoh, M. Arita, E. Wimmer, and A. Nomoto. 1986. Genetic analysis of the attenuation phenotype of poliovirus type 1. J. Virol. 58:348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Page, G. S., A. G. Mosser, J. M. Hogle, D. J. Filman, R. R. Rueckert, and M. Chow. 1988. Three-dimensional structure of poliovirus serotype 1 neutralizing determinants. J. Virol. 62:1781-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palmenberg, A. C., and J.-Y. Sgro. 1997. Topological organization of picornaviral genomes: statistical prediction of RNA structural signals. Semin. Virol. 8:231-241. [Google Scholar]
- 64.Paul, A. V., J. Mugavero, J. Yin, S. Hobson, S. Schultz, J. H. van Boom, and E. Wimmer. 2000. Studies on the attenuation phenotype of polio vaccines: poliovirus RNA polymerase derived from Sabin type 1 sequence is temperature sensitive in the uridylylation of VPg. Virology 272:72-84. [DOI] [PubMed] [Google Scholar]
- 65.Pipkin, P. A., D. J. Wood, V. R. Racaniello, and P. D. Minor. 1993. Characterisation of L cells expressing the human poliovirus receptor for the specific detection of polioviruses in vitro. J. Virol. Methods 41:333-340. [DOI] [PubMed] [Google Scholar]
- 66.Posada, D. 2002. Evaluation of methods for detecting recombination from DNA sequences: empirical data. Mol. Biol. Evol. 19:708-717. [DOI] [PubMed] [Google Scholar]
- 67.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 68.Rambaut, A. 2000. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics 16:395-399. [DOI] [PubMed] [Google Scholar]
- 69.Rosen, F. S., M. D. Cooper, and R. J. P. Wedgwood. 1995. The primary immunodeficiencies. N. Engl. J. Med. 333:431-440. [DOI] [PubMed] [Google Scholar]
- 70.Rousset, D., M. Rakoto-Andrianarivelo, R. Razafindratsimandresy, B. Randriamanalina, S. Guillot, J. Balanant, P. Mauclère, and F. Delpeyroux. 2003. Recombinant vaccine-derived poliovirus in Madagascar. Emerg. Infect. Dis. 9:885-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rozas, J., J. C. Sánchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 72.Sabin, A. B., and L. R. Boulger. 1973. History of Sabin attenuated poliovirus oral live vaccine strains. J. Biol. Stand. 1:115-118. [Google Scholar]
- 73.Shimizu, H., B. Thorley, F. J. Paladin, K. A. Brussen, V. Stambos, L. Yuen, A. Utama, Y. Tano, M. Arita, H. Yoshida, T. Yoneyama, A. Benegas, S. Roesel, M. Pallansch, O. Kew, and T. Miyamura. 2004. Circulation of type 1 vaccine-derived poliovirus in the Philippines in 2001. J. Virol. 78:13512-13521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sokal, R. R., and F. J. Rohlf. 1995. Biometry, 3 ed. W. H. Freeman and Co., New York, NY.
- 75.Strebel, P. M., R. W. Sutter, S. L. Cochi, R. J. Biellik, E. W. Brink, O. M. Kew, M. A. Pallansch, W. A. Orenstein, and A. R. Hinman. 1992. Epidemiology of poliomyelitis in the United States one decade after the last reported case of indigenous wild virus-associated disease. Clin. Infect. Dis. 14:568-579. [DOI] [PubMed] [Google Scholar]
- 76.Sutter, R. W., O. M. Kew, and S. L. Cochi. 2004. Poliovirus vaccine -live, p. 651-705. In S. A. Plotkin and W. A. Orenstein (ed.), Vaccines, 4th ed. W.B. Saunders Company, Philadelphia, Pa.
- 77.Sutter, R. W., and R. Prevots. 1994. Vaccine-associated paralytic poliomyelitis among immunodeficient persons. Infect. Med. 11:426-438. [DOI] [PubMed] [Google Scholar]
- 78.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinaeur Associates, Sunderland, Mass.
- 79.Tharmaphornpilas, P. 2005. Vaccine-derived poliovirus, Thailand, 2003. Emerg. Infect. Dis. 11:777-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tolskaya, E. A., L. I. Romanova, M. S. Kolesnikova, and V. I. Agol. 1983. Intertypic recombination in poliovirus: genetic and biochemical studies. Virology 124:121-132. [DOI] [PubMed] [Google Scholar]
- 81.van der Avoort, H. G., J. H. Reimerink, A. Ras, M. N. Mulders, and A. M. van Loon. 1995. Isolation of epidemic poliovirus from sewage during the 1992-3 type 3 outbreak in The Netherlands. Epidemiol. Infect. 114:481-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van der Avoort, H. G. A. M., B. P. Hull, T. Hovi, M. A. Pallansch, O. M. Kew, R. Crainic, D. J. Wood, M. N. Mulders, and A. M. van Loon. 1995. A comparative study of five methods of intratypic differentiation of polioviruses. J. Clin. Microbiol. 33:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wiegers, K., H. Uhlig, and R. Dernick. 1988. Evidence for a complex structure of neutralization antigenic site I of poliovirus type 1 Mahoney. J. Virol. 62:1845-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.World Health Organization. 2000. Certification of poliomyelitis eradication: Western Pacific Region. Wkly. Epidemiol. Rec. 75:399-400. [PubMed] [Google Scholar]
- 85.World Health Organization. 2004. Laboratory surveillance for wild and vaccine-derived polioviruses, January 2003—June 2004. Wkly. Epidemiol. Rec. 79:393-398. [PubMed] [Google Scholar]
- 86.World Health Organization. 2004. Polio laboratory manual, 4th ed. World Health Organization, Geneva, Switzerland.
- 87.World Health Organization. 2005. Progress towards interruption of wild poliovirus transmission, January 2004 to March 2005. Wkly. Epidemiol. Rec. 80:149-155. [PubMed] [Google Scholar]
- 88.Yang, C.-F., L. De, B. P. Holloway, M. A. Pallansch, and O. M. Kew. 1991. Detection and identification of vaccine-related polioviruses by the polymerase chain reaction. Virus Res. 20:159-179. [DOI] [PubMed] [Google Scholar]
- 89.Yang, C.-F., T. Naguib, S.-J. Yang, E. Nasr, J. Jorba, N. Ahmed, R. Campagnoli, H. van der Avoort, H. Shimizu, T. Yoneyama, T. Miyamura, M. A. Pallansch, and O. Kew. 2003. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt, 1983 to 1993. J. Virol. 77:8366-8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 91.Yoneyama, T., A. Hagiwara, M. Hara, and H. Shimojo. 1982. Alteration in oligonucleotide fingerprint patterns of the viral genome in poliovirus type 2 isolated from paralytic patients. Infect. Immun. 37:46-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang, L., L. Rowe, T. He, C. Chung, J. Yu, W. Yu, A. Talal, M. Markowitz, and D. D. Ho. 2002. Compartmentalization of surface envelope glycoprotein of human immunodeficiency virus type 1 during acute and chronic infection. J. Virol. 76:9465-9473. [DOI] [PMC free article] [PubMed] [Google Scholar]