Abstract
In order to identify cellular proteins required for early stages of retroviral replication, a high volume screening with mammalian somatic cells was performed. Ten pools of chemically mutagenized Chinese hamster ovary (CHO-K1) cells were challenged with a murine leukemia virus (MLV) vector pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G), and cells that failed to be transduced were enriched by cell sorting. Each pool yielded a clonally derived cell line with a 5-fold or greater resistance to virus infection, and five cell lines exhibited a >50-fold resistance. These five cell lines were efficiently infected by a human immunodeficiency virus vector pseudotyped with VSV-G. When engineered to express the TVA receptor for subgroup A avian sarcoma and leukosis virus (ASLV-A), the five cell lines were resistant to infection with a MLV vector pseudotyped with the ASLV-A envelope protein but were fully susceptible to infection with an ASLV-A vector. Thus, the defect in these cells resides after virus-cell membrane fusion and, unlike those in other mutant cell lines that have been described, is specific for the MLV core. To identify the specific stages of MLV infection that are impaired in the resistant cell lines, real-time quantitative PCR analyses were employed and two phenotypic groups were identified. Viral infection of three cell lines was restricted before reverse transcription; in the other two cell lines, it was blocked after reverse transcription, nuclear localization, and two-long terminal repeat circle formation but before integration. These data provide genetic evidence that at least two distinct intracellular gene products are required specifically for MLV infection. These cell lines are important tools for the biochemical and genetic analysis of early stages in retrovirus infection.
The retroviral life cycle is a multistep process, divided into early and late stages (reviewed in reference 8). The early stages consist of virus binding to a cellular receptor, fusion of viral and cellular membranes, delivery of the viral core into the cytoplasm, reverse transcription of the positive-strand RNA genome to generate a doubled-stranded DNA product, translocation of viral nucleoprotein complexes to the nucleus, and integration of the viral DNA into the host cell genome to generate a provirus. Interaction of the viral envelope protein (Env) with cellular receptors is the primary determinant for retroviral entry (reviewed in reference 8). However, the events that occur following virus-cell membrane fusion and that lead to proviral DNA establishment, especially those involving cellular factors, are only partially understood (18). Cellular factors other than receptors that have been implicated as playing important roles at distinct early steps of retroviral replication include actin, microtubules, importin-7, HMGa1, LAP-2α and the barrier-to-autointegration factor (9, 11, 14, 28, 40). A putative serine kinase that phosphorylates murine leukemia virus (MLV) p12 has also been implicated in cytoplasm-to-nucleus transport of that virus (44, 45). Yet other cellular factors, including Fv-1, Trim-5α, and APOBEC3G, act to restrict the early steps of retroviral replication (3, 4, 36, 38).
It is likely that other, as-yet-unidentified cellular factors contribute to other steps of retroviral replication. Indeed, a recently described cell-free uncoating assay has implicated cellular factor involvement in the uncoating step of retroviral infection which occurs immediately after membrane fusion and leads to the formation of an active reverse transcription complex (31). Moreover, the involvement of multiple cellular factors is consistent with the dynamic nature of the reverse transcription complex during the progression of infection (12, 13). Genetic studies have also implicated cellular factors as playing a positive role in early stages of retroviral infection. Two lines of chemically mutagenized Rat-2 fibroblast cell lines (R3-2 and R4-7) were identified under negative selection conditions following multiple rounds of challenge with a mixture of amphotropic and ecotropic MLV vectors (17). Cell line R3-2 was approximately 1,000-fold resistant to infection by these vectors, a defect that mapped to a stage following reverse transcription but before nuclear localization (17). Cell line R4-4 was approximately 100-fold resistant to infection by these vectors, and the block in this cell line occurred before the earliest step of reverse transcription (17). The defects in cell lines R3-2 and R4-4 were judged to be common to all retroviruses, since these cell lines restricted the early steps of entry by MLV and human immunodeficiency virus type 1 (HIV-1) vectors (17). Two cDNAs that complement the defect in cell line R4-4 have been isolated. One cDNA represents an antisense transcript to the transcriptional coactivator CAPER, and the other is a sense transcript of the central portion of the VL30 endogenous retrovirus-like element. Since neither of these cDNAs is predicted to give rise to a protein product, it is not yet clear how they exert their complementing effects (16).
In an attempt to identify other cellular factors that participate in the early steps of retroviral replication, we have employed a high-throughput somatic cell mutagenesis-based approach. This approach involved mutagenizing Chinese hamster ovary (CHO-K1) cells with the frameshift mutagen ICR-191 and subjecting the cells to multiple rounds of challenge with an MLV vector that was pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G). After each round of infection, the cells were subjected to either fluorescent or magnetic sorting, leading to single-cell cloning. This screening led to the isolation of two classes of mutant cell lines that have a defect that lies either upstream of reverse transcription or appears to lie downstream of nuclear localization but before proviral DNA establishment. These data suggest a role for novel cellular proteins in facilitating MLV-specific early steps of replication.
MATERIALS AND METHODS
Cell culture and virus production.
CHO-K1 cells (ATCC CCL-61) were cultured in F-12 medium supplemented with 10% bovine calf serum (Invitrogen, Carlsbad, CA). Human embryonic kidney 293T cells (ATCC CRL-11268) were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (HyClone, Logan, UT). Chicken DF-1 cells (ATCC CRL-12203) were cultured in DMEM supplemented with 10% fetal calf serum (HyClone, Logan, UT). MLV pseudotyped viruses were generated as previously described (5, 21). Briefly, 293T cells were transfected by the calcium phosphate method with 4 μg of the appropriate packageable vector plasmid, 3 μg of a plasmid (pmd.oldgagpol) encoding MLV Gag/Pol, and 1 μg of plasmid encoding either the vesicular stomatitis virus glycoprotein or EnvA (pMD VSV-G or pAB6, respectively). The VSV-G-pseudotyped HIV vector was made using a Virapower kit (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. DF-1 cells were transfected using the calcium phosphate method with the subgroup A-specific avian sarcoma and leukosis virus (ASLV-A) vector, RCASBP(A)-AP, encoding alkaline phosphatase (15). Medium from transfected cells was collected 2 days posttransfection to 7 days posttransfection and filtered through a 0.45-μm bottle top filter. Virus was stored at 4°C through the collection period, combined, and then frozen at −80°C for long-term storage. Virus for use in quantitative PCR (QPCR) amplification studies was treated with DNaseI (Roche Applied Science, Indianapolis, IN) to remove contaminating plasmid DNA from the virus preps. DNaseI was added as a powder (to a final concentration of 1 μg/ml) when the virus-containing supernatants were collected. The supernatants were incubated 1 h at room temperature before filtration. The titer of VSV-G and EnvA vector stocks were determined by assaying for transduction of a marker gene following infection of either wild-type (WT) CHO-K1 cells or WT CHO-K1 cells that had been engineered to express TVA800 (described below). Calculations of the multiplicity of infection (MOI) were based on the number of transducing units observed with infection of WT CHO-K1 cells, with or without TVA800 expression.
Plasmids.
The MLV genome plasmids pMMP-nls-LacZ (encoding β-galactosidase), pCMMP-eGFP (encoding the enhanced green fluorescent protein [eGFP]), and pCMMP-IRES-GFP and the subgroup A ASLV (ASLV-A) genome plasmid RCASBP(A)-AP (encoding alkaline phosphatase) have been previously described (5, 15, 29). To construct the pCMMP-CD4-eGFP vector, a NotI-to-PstI fragment with the CD4 gene deleted for the cytoplasmic tail from pMACS4.1 (Miltenyi Biotec Inc., Auburn, CA) was inserted into the NotI/PstI sites downstream of the viral long terminal repeat (LTR) and upstream of the encephalomyocarditis virus internal ribosome entry site (IRES) in pCMMP-IRES-GFP. The HIV-1 vectors were derived from the pLenti6/V5-GW/lacZ vector (Invitrogen, Carlsbad, CA). The HIV-HcRED vector was constructed by inserting into the BamHI/EcoRV sites of pLenti6/V5-GW/lacZ the multiple cloning site and IRES from pCMMP-IRES-GFP as a BamHI/BspEI fragment upstream of an AgeI/StuI fragment containing the HcRED coding sequence from pHcRED1 (Clontech, Palo Alto, CA). To generate the pHIV-TVA800-hcRED vector, the coding sequence of TVA800 was excised from pCMMP-TVA800 (30) as a PmlI/SalI fragment and inserted into the EcoRV/SalI sites of the HIV-HcRED vector upstream of the IRES.
Mutagenesis of CHO-K1 cells.
Ten pools of 1 × 106 CHO-K1 cells were mutagenized by ICR-191 treatment (10 μg/ml) as described previously (6). Each pool was subjected to three rounds of mutagenesis before screening.
Isolation of MLV-resistant cells by fluorescence-activated cell sorting (FACS).
Two pools (numbers 1 and 4) of 1 × 106 mutagenized CHO-K1 cells were infected with CMMP-eGFP[VSV-G] at an approximate MOI of 3 to 5 GFP transducing units (GTU) for 2 h at 37°C in the presence of 4 μg/ml Polybrene (Sigma-Aldrich, Inc., St. Louis, MO). The virus-containing medium was then removed and replaced with fresh medium. Forty-eight hours postinfection (hpi), the cells were trypsinized and resuspended in medium, and GFP-negative cells were isolated using a FACSvantage high-speed cell sorter (Becton Dickinson, San Jose, CA). Cells were allowed to recover (typically for 24 to 48 h) and expanded as necessary to a level that was greater than or equal to 1 × 106 cells. The viral challenge and sorting was repeated until there was an observable resistance to infection in the sorted pools based on the level of GFP fluorescence obtained relative to a control population of unmutagenized cells. This required seven rounds of selection for pool 1 and six rounds of selection for pool 4. Once resistance was observed, a final infection and sorting was performed, and GFP-negative cells were single-cell cloned. Each of these clonal cell lines was then divided into two aliquots; one was uninfected, and the other was infected with CMMP-EGFP[VSV-G] at an approximate MOI of 3 to 5 GTU. The virus-resistant clones were identified as GFP negative following viral challenge, and the corresponding cell clone in the uninfected plate was expanded for further characterization.
Isolation of MLV-resistant cells by magnetic cell sorting (MACS).
Eight pools (pools 2, 3, and 5 to 10) of 2 × 107 mutagenized CHO-K1 cells were infected with CMMP-CD4-EGFP[VSV-G] at an approximate MOI of 1 GTU for 2 h at 37°C in the presence of 4 μg/ml Polybrene. Unbound viruses were then removed, and fresh medium was added. At 48 h postinfection, the cells were removed from the plate with phosphate-buffered saline (PBS) containing 5 mM EDTA. Cells were pelleted (200 × g, 5 min), resuspended in 500 μl of PBS containing 2 mM EDTA and 2% bovine serum albumin (BSA) (Sigma-Aldrich, Inc., St. Louis, MO), and incubated with anti-human CD4 iron-conjugated antibody (Miltenyi Biotec Inc., Auburn, CA) at 20 μg/107 cells for 15 min at 4°C. Large-cell columns (Miltenyi Biotec Inc., Auburn, CA) were applied to a magnetic field and washed with 2 ml PBS containing 2 mM EDTA and 2% BSA. Cells were filtered through a 30-μm mesh (Miltenyi Biotec Inc., Auburn, CA) and applied to the large-cell column. Cells were washed twice with 2 ml PBS containing 2 mM EDTA and 2% BSA. Column flowthrough and washes were collected, and the cells were pelleted, resuspended in medium, and replated. Cells were allowed to recover for at least 16 h before the next viral challenge. When necessary, the cells were expanded between each round of virus infection to a minimum of 5 × 105 cells per sort. The infection and selections were repeated until there was an observable resistance to infection based on eGFP fluorescence in the selected cells relative to a control population of unmutagenized CHO-K1 cells. This varied from five to seven rounds depending on the pool. Once resistance in a pool was observed, the population was infected a final time with CMMP-CD4-EGFP[VSV-G], and the GFP-negative cells were single-cell cloned after high-speed FACS.
Screening MLV-resistant clones.
Single-cell clones from pools 2, 3, and 5 to 10 were plated on duplicate plates and incubated for 2 h with pMMP-nls-LacZ [VSV-G] at an approximate MOI of 1 LacZ transducing unit (LTU) in the presence of 4 μg/ml Polybrene. Unbound virus was then removed, and fresh medium was added. At 48 h postinfection, one plate was assayed for β-galactosidase activity with a Galacto-Star chemiluminescent kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions, and the other plate was assayed for cell number and cell viability by using CellTiter-Glo reagent (Promega, Madison, WI) following the manufacturer's instructions.
Chemiluminescent assay of viral infection.
Eight wells of a 96-well plate were seeded at 1 × 104 cells/well for each cell line tested. The cells were incubated for 2 h with an approximate MOI of 1 LTU of either MMP-nls-LacZ[VSV-G], MMP-nls-LacZ[EnvA], or Lenti6/V5-GW/lacZ [VSV-G], or an MOI of 1 alkaline phosphatase transducing unit of RCASBP(A)-AP, in the presence of 4 μg/ml Polybrene. Unbound virions were removed, and fresh medium was added. At 48 h postinfection, four wells were assayed for either β-galactosidase activity as described above or for alkaline phosphatase activity [in the case of RCASBP(A)-AP infections] by using a Phospha-Light kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The other four wells were assayed for cell number and cell viability using CellTiter-Glo reagent (Promega, Madison, WI) as described above. In these experiments, the ratio of β-galactosidase or alkaline phosphatase to luciferase activity was calculated in each case and compared to that seen with a control population of unmutagenized CHO-K1 cells to determine level of the resistance to viral infection. Under these conditions, the control cells exhibited an approximately 50-fold increase in β-galactosidase:luciferase or alkaline phosphatase:luciferase ratios following infection.
To determine the absolute level of resistance to viral infection, X-Gal (5-bromo-4-chloro-3-indoyl-β-d[scap]-galactopyranoside) staining was performed with cells that were infected with serial dilutions of viruses. For these experiments, cells were seeded at 2 × 104 cells/well in triplicate rows for each cell line tested. The cells were then infected for 2 h with 10-fold serial dilutions of MMP-nls-LacZ[VSV-G] in the presence of 4 μg/ml Polybrene as described before, and the cells were subsequently stained with X-Gal as previously described (1). The blue cells contained in wells that had between 20 and 200 β-galactosidase-positive cells were counted to give an accurate measure of the viral titer.
TVA-expressing cell lines.
Cells (1 × 106) were seeded in a 6-cm dish and infected with HIV-1-TVA800-HcRED[VSV-G] at an approximate MOI of 0.5 hcRED transducing units for 2 h. Cells infected with this virus express TVA800, HcRED, and the blasticidin S resistance gene. Infected cells were selected for 2 weeks in the presence of 3 μg/ml blasticidin (Invitrogen, Carlsbad, CA). Blasticidin-resistant clones were pooled, and TVA800 expression was confirmed by staining cells with an ASLV-A SU-immunoglobulin G immunoadhesin followed by flow cytometric analysis as previously described (47).
Real-time QPCR.
To measure the amounts of reverse transcription intermediates in infected cells, cells were seeded in triplicate wells at 5 × 105/well in a six-well plate and then infected at 4°C on a rocking platform at an MOI of 1 GTU for 2 h with an MLV vector (pLEGFP; Clontech, Palo Alto, CA) pseudotyped with VSV-G that was treated with DNaseI as described above. DNA was harvested from infected cells at 24 hpi by using a DNeasy kit (QIAGEN, Valencia, CA). For the nuclear fractionation studies, nuclei were harvested from infected cells 24 hpi using a Nuclei EZ Prep kit (Sigma-Aldrich, Inc., St. Louis, MO) following the manufacturer's instructions, and DNA was isolated from nuclei as described above. To measure integrated proviral DNA copy number, cells were seeded and infected as described above and then passaged for 10 days. DNA was then harvested from 1 × 106 infected cells as described above. To measure the number of two-long terminal repeat (2LTR) circles, 1 × 106 cells were infected as described above at a MOI of 10 GTU. DNA was harvested 24 h postinfection. DNA concentration was calculated by measuring the A260 on a SPECTRAmax Plus 96-well UV spectrophotometer (Molecular Devices, Sunnyvale, CA). Quantitative, real-time PCR (QPCR) was performed on an ABI 9600 (Applied Biosystems, Foster City, CA) using the standard cycling conditions of 50°C for 10 min and 40 cycles of 95°C for 30 s and 60°C for 2 min. DNA (10 μl/25-μl reaction) was amplified in TaqMan Universal PCR Mastermix (Applied Biosystems, Foster City, CA) with 1 μM concentrations of each primer and 0.1 μM 5′ 6-carboxyfluorescein (6-FAM), 3′ 6-carboxytetramethylrhodamine (TAMRA)-labeled probe. Each primer probe set was tested on each cell line in a minimum of three independent experiments. The number of molecules in each reaction was determined by a comparison to standard curves generated from amplification of plasmid DNA containing the target sequence. The primer probe set used to determine if the screen virus had integrated into cells is OJWB7 (5′-GAACAGATGGTCCCCAGATGC-3′), OJWB8 (CGGTGGAACCTCCAAATGAA), and OJWB21 (5′-6-FAM-AAAAGAGCCCACAACCCCTCACTCGG-TAMRA-3′). Since the 3′ LTR is replaced with the cytomegalovirus promoter in pCMMP-based vectors, this primer probe set will detect only reverse-transcribed viral genomes and does not detect pCMMP genome plasmids or reverse-transcribed viral genomes derived from pLEGFP. Primers and probes used to identify specific reverse transcription intermediates are given in Table 1.
TABLE 1.
Quantitative real-time PCR primer and probe sets for the detection of reverse transcription intermediates
| RT step detecteda | Oligonucleotide nameb | Sequencec | Binding positiond |
|---|---|---|---|
| Minus-strand strong stop | OJWB45 | GCGCCAGTCTTCCGATAGACTG | R (589-610) |
| OJWB47 | GTCGTGGGTAGTCAATCACTCAG | R and U5 (697-719) | |
| OJWB38 | 6-FAM-ATCCGAATCGTGGTCTCGCTGTTC-TAMRA | R (657-680) | |
| Minus-strand synthesis | OJWB32 | CCACAAGTTCAGCGTGTCC | GFP (3148-3166) |
| OJWB33 | CGTCGTCCTTGAAGAAGATGG | GFP (3366-3386) | |
| OJWB34 | 6-FAM-ATGTGGTCGGGGTAGCGGCTGAAGCAC-TAMRA | GFP (3283-3309) | |
| Plus-strand strong stop | OJWB39 | CAGTTCGCTTCTCGCTTCTGTTC | U3 (513-535) |
| OJWB40 | TGGGTAGTCAATCACTCAG | R and U5 (697-715) | |
| OJWB38 | 6-FAM-ATCCGAATCGTGGTCTCGCTGTTC-TAMRA | U5 (657-680) | |
| Plus-strand extension | OJWB45 | GCGCCAGTCTTCCGATAGACTG | R (589-610) |
| OJWB48 | ACAATCGGACAGACACAGATAAGTTG | Gag (807-832) | |
| OJWB38 | 6-FAM-ATCCGAATCGTGGTCTCGCTGTTC-TAMRA | R (657-680) | |
| 2LTR circles | OJWB45 | GCGCCAGTCTTCCGATAGACTG | R (589-610) |
| OJWB46 | GGGCTCTTTTATTGAGCTCGGAG | U3 (549-571) | |
| OJWB38 | 6-FAM-ATCCGAATCGTGGTCTCGCTGTTC-TAMRA | R (657-680) |
The DNA intermediate of the MLV reverse transcription process that the primer/probe set detects.
Designation given to each primer in a primer/probe set. Sets are listed in the following order: plus-strand primer, minus-strand primer, probe. The OJWB38 probe binds on the plus strand, and the OJWB34 probe binds to the minus strand.
Sequence of the oligonucleotide written in the 5′-to-3′ orientation. Oligonucleotides used as Taqman probes are modified with a 6-FAM fluorescent probe at the 5′ end and a TAMRA quencher at the 3′ end.
The functional elements of the viral genome the primer binds are denoted along with base pair position in the viral genome plasmid pLEGFP-C1. U3 is the unique 3′ region of the viral LTR, U5 is the unique 5′ region of the viral LTR, R is the repeat region of the LTR, and GFP correlates to the green fluorescent protein marker gene.
RESULTS
Isolation of cell lines resistant to retroviral infection.
FACS and MACS approaches were employed to isolate cells that are resistant to retroviral infection. Both approaches involved subjecting chemically mutagenized CHO-K1 cells to infection with a VSV-G-pseudotyped, replication-defective MLV vector. The use of VSV-G-pseudotyped virions reduced the possibility of identifying mutations that abolish receptor expression, since VSV-G has been reported to bind to a ubiquitous nonproteinaceous receptor, potentially phosphatidylserine (32, 33). Ten independent pools of CHO-K1 cells (pools 1 to 10) were mutagenized with the alkylating agent ICR-191. This mutagen induces frameshift and small deletions which have a low reversion rate relative to point mutations (17, 26). CHO-K1 cells were employed because they have been shown to be functionally hypodiploid, so that in most cases, only one copy of a gene needs to be disrupted to exhibit a mutant phenotype (20).
Pools 1 and 4 of the mutagenized cells were challenged with VSV-G-pseudotyped pCMMP-eGFP (CMMP-eGFP[VSV-G]), an MLV vector that encodes the eGFP, and the eGFP-negative population (enriched for uninfected cells) was then collected by FACS (Fig. 1A). This procedure was repeated until there was an observable resistance to virus infection, and single cell clones were isolated as described in Materials and Methods. The most resistant clone from pool 1 (designated mutant cell line 1 [MCL1]) and the most resistant clone from pool 4 (designated MCL4) were characterized further (see below).
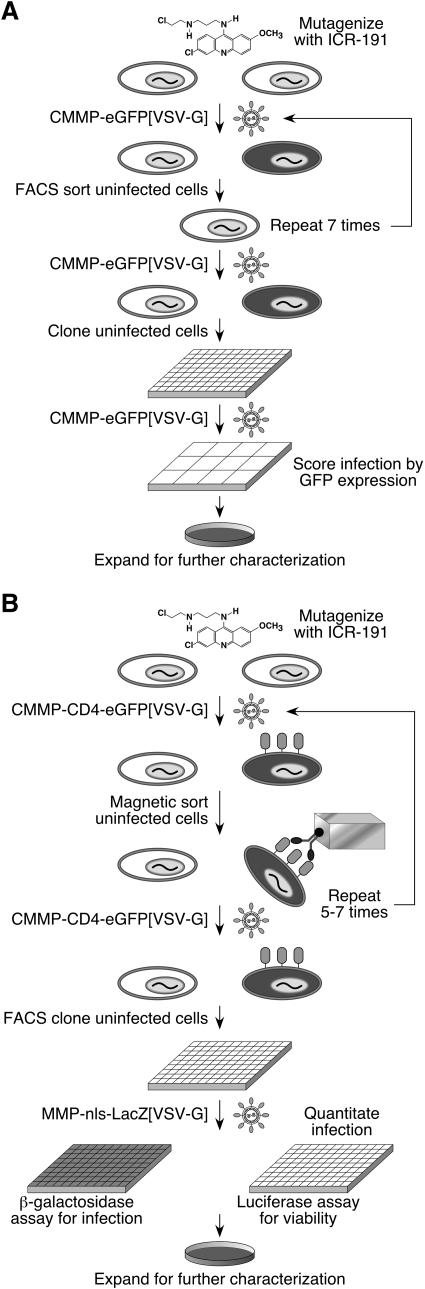
FIG. 1.
Two approaches used to isolate mutagenized CHO-K1 cell lines resistant to retroviral infection. (A) Isolation of MLV resistant cells by FACS. CHO-K1 cells mutagenized with ICR-191 were infected with a VSV-G-pseudotyped MLV vector that encodes eGFP, and the resultant eGFP-negative cells were collected by FACS. After a minimum of five rounds of infection and sorting, the eGFP-negative cells were single-cell cloned, expanded into 12-well plates, and assayed for resistance to infection by infection by the pseudotyped MLV vector. (B) Isolation of MLV-resistant cells by MACS. CHO-K1 cells mutagenized with ICR-191 were infected with a VSV-G-pseudotyped MLV vector that encodes both CD4 and eGFP. Infected cells were depleted from the population by magnetic sorting with an iron-conjugated anti-CD4 antibody. After a minimum of five rounds of infection and sorting, the eGFP-negative cells were single-cell cloned, expanded, and seeded into duplicate assay plates. The assay plates were infected with another VSV-G-pseudotyped MLV vector that encodes β-galactosidase. One plate was then assayed with a chemiluminescent assay for β-galactosidase, and the other plate was assayed with a luciferase-based chemiluminescent viability assay.
The flow cytometric method, while useful for isolating virus-resistant cells, was time-consuming and was not suitable for screening the additional pools. Therefore, to screen the eight remaining pools (2, 3, and 5 to 10), MACS was employed (Fig. 1B). In this approach, cells in each population were infected with CMMP-CD4-eGFP[VSV-G], a VSV-G-pseudotyped MLV vector that encodes the cell surface protein CD4 and contained an IRES that allowed for translation of the eGFP. Two days postinfection, the cells were incubated with an anti-CD4 antibody that was conjugated to an iron moiety. The infected cells were removed from the population by exposure to a magnetic field. The infections and sorting were repeated for 5 to 7 rounds until there was an observable resistance to virus infection, and single cell clones of eGFP-negative cells were isolated as described in Materials and Methods. This approach gave rise to eight cell lines: MCL2, MCL3, and MCL5 through MCL10 (from pools 2, 3, and 5 through 10, respectively) (see below).
Characterization of the resistance to MLV infection in isolated clones.
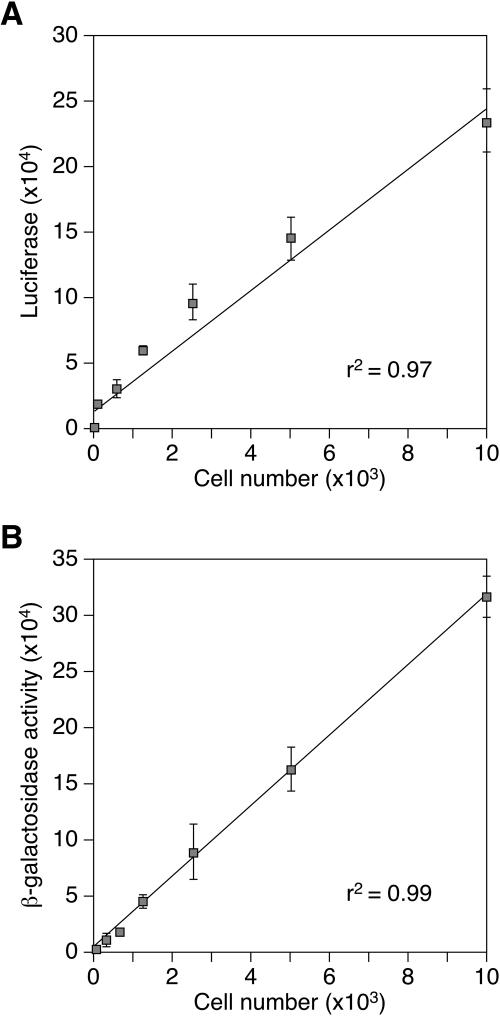
To measure the defects in infection seen with the mutant cell lines, MCL1 to MCL10 were challenged with VSV-G-pseudotyped pMMP-nls-LacZ (MMP-nls-lacZ[VSV-G], an MLV vector that encodes β-galactosidase, and assayed 48 h later for the virus-encoded β-galactosidase activity. Since these studies involved a direct comparison between the levels of virus infection seen in mutagenized versus nonmutagenized CHO-K1 cells, it was important to correct the data for any differences in either the relative plating efficiencies or the growth rates of the mutant cell types. This was accomplished by incorporating a luciferase-based cell viability assay during the analysis. Reconstruction experiments performed with increasing numbers of CHO-K1 cells showed that the β-galactosidase and luciferase signals that were obtained increased linearly with the number of input cells (Fig. 2A and 2B). Therefore, in the experiments described below, the data were normalized to the number of cells in each population by dividing the β-galactosidase signal by the luciferase signal (to give a ratio of the amount of viral infection to relative cell number).
FIG. 2.
The β-galactosidase-based infection assay and the luciferase-based cell viability assay display a linear dependence upon cell number. Cells were plated for 24 h and then challenged with 8 × 104 LTU of MMP-nls-lacZ[VSV-G]. The cells were assayed 48 h postinfection with (A) a CellTiter-Glo kit (Promega, Madison, WI), which measures the viability of infected cells by measuring cellular ATP as a substrate for luciferase, or (B) a Galacto-star chemiluminescent β-galactosidase assay (Applied Biosystems, Foster City, CA). Uninfected controls were assayed 72 h after plating. The data shown are the average mean values obtained in an experiment with quadruplicate samples and are representative of results of three independent experiments. Error bars indicate the standard deviations of the data.
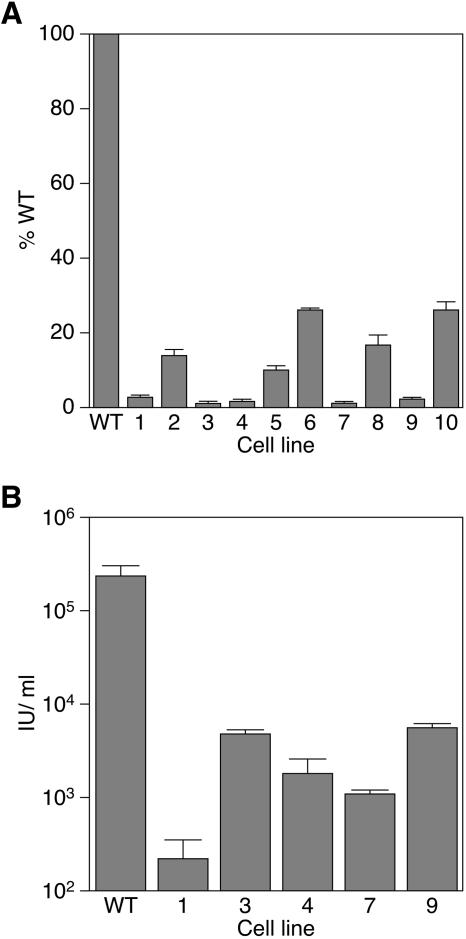
Infection of unmutagenized CHO-K1 cells gave rise to a 40-fold increase in the ratio of β-galactosidase to luciferase (defined as 100%) (Fig. 3A). Each of the clones tested was judged to be at least fivefold resistant to infection with MMP-nls-LacZ[VSV-G] relative to WT CHO-K1 cells (Fig. 3A). MCLs 1, 3, 4, 7, and 9 were ≥40-fold resistant to infection (defined as background levels for this assay) and were characterized further. MCL5 was 10-fold resistant to MLV, infection, but this phenotype spontaneously reverted to WT after several weeks in culture and therefore, given its instability, this cell line was not further characterized.
FIG. 3.
Resistance of isolated cell lines to infection by a VSV-G-pseudotyped MLV vector. (A) Cells (1 × 104/well of a 96-well plate) of the indicated cell lines were infected with 5 × 104 IU of MMP-nls-lacZ[VSV-G] and assayed 48 h postinfection with chemiluminescent assays for β-galactosidase and viability as described for Fig. 2. (B) Cells (1 × 104/well) of the indicated cell lines were infected with different concentrations of MMP-nls-lacZ[VSV-G] and stained 48 h postinfection with X-Gal. The data shown are the average mean values obtained in an experiment with triplicate samples and are representative of results of three independent experiments. Error bars indicate the standard deviations of the data.
In order to precisely determine the level of resistance in the highly resistant cell lines, they were infected with serial dilutions of MMP-nls-LacZ[VSV-G] and stained 48 hpi with X-Gal. The resultant β-galactosidase-positive cells were counted, and the relative infectious units per ml of virus stock were calculated for each cell line (Fig. 3B). The average resistance to MLV infection for each cell line was 1,100-fold for MCL1, 50-fold for MCL3, 130-fold for MCL4, 230-fold for MCL7, and 43-fold for MCL9 (Fig. 3B).
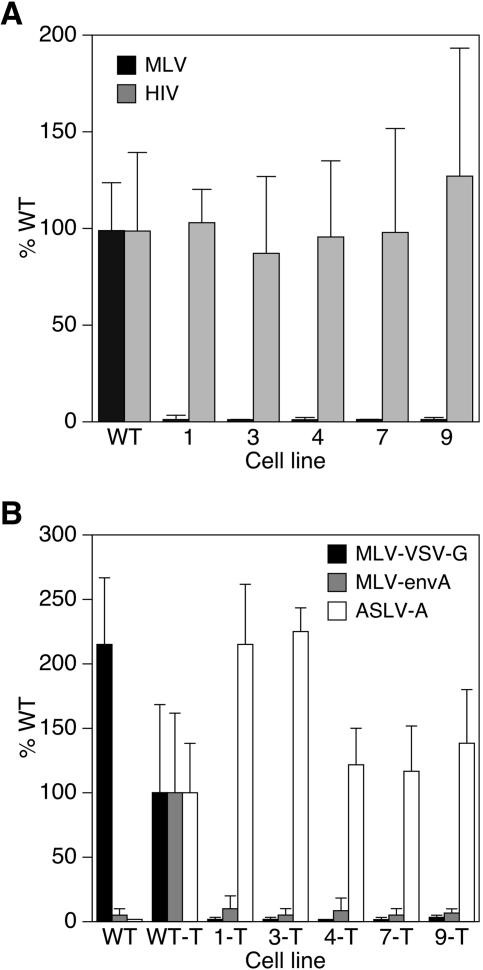
Resistance to infection is specific to the MLV core.
We next asked if the resistance was specific to MLV or common to other retroviruses. To address this question, cells were infected with MMP-nls-LacZ[VSV-G] or with Lenti6/V5-GW/lacZ[VSV-G], an HIV-1 vector encoding β-galactosidase. As expected, MCLs 1, 3, 4, 7, and 9 were at least 40-fold resistant to MLV infection, as seen before (Fig. 4A). However, each of these cell lines showed no obvious defect in their susceptibility to infection by the HIV-1 vector (Fig. 4A). Since both the MLV and the HIV-1 vectors used in these experiments contained VSV-G, these data demonstrated that the defects in these cells are not linked to the viral glycoprotein but instead map specifically to the core of the MLV vector. To address this issue more thoroughly, the MCLs were engineered to express TVA800, the cellular receptor for ASLV-A (2, 43). The TVA800-expressing cells were resistant to infection by MLV vectors pseudotyped with VSV-G or with the ASLV-A Env (EnvA) (Fig. 4B). However, the TVA-expressing cells were fully susceptible to infection with an ASLV-A vector that encodes heat-stable alkaline phosphatase (Fig. 4B). These data confirmed that the resistance in these cell lines is independent of viral envelope/receptor interactions and is specific for MLV cores.
FIG. 4.
Resistance to retroviral infection is specific to the MLV core. (A) Cells (1 × 104/well of a 96-well plate) of the indicated cell lines were infected with either 5 × 104 IU (LTU) of MMP-nls-lacZ[VSV-G] or 5 × 104 IU of a VSV-G-pseudotyped HIV-1 vector that encodes β-galactosidase (Lenti6/V5-GW/lacZ [VSV-G]) and assayed 48 h postinfection with chemiluminescent assays for β-galactosidase and for viable cells as described for Fig. 2. (B) Cells (1 × 104/well of a 96-well plate) of WT CHO-K1 or the indicated cell lines engineered to express TVA800 were infected with either 5 × 104 IU (LTU) of the MLV vectors MMp-nls-LacZ[VSV-G], 5 × 104 IU of pMMp-nls-LacZ[envA], or the ASLV-A vector RCASBP(A)-AP and assayed 48 h postinfection with chemiluminescent assays for cell viability and either β-galactosidase or alkaline phosphatase. TVA-expressing cell lines are designated with a T after the line name. The data shown are the average mean values obtained in an experiment with quadruplicate samples and are representative of results of three independent experiments. Error bars indicate the standard deviations of the data.
The mutant cell lines exhibit defects either at pre- or post-reverse transcription steps.
A real-time QPCR analysis revealed that, despite multiple rounds of challenge with an MLV vector during the screening process, MCLs 1, 3, 4, 7, and 9 did not contain integrated viral DNA (Fig. 5A). Therefore, these data indicated that these mutant cell lines were deficient at supporting an early step of MLV replication prior to integration. To determine whether the defects in these cells lie upstream or downstream of specific stages during reverse transcription, additional QPCR experiments were performed.
FIG. 5.
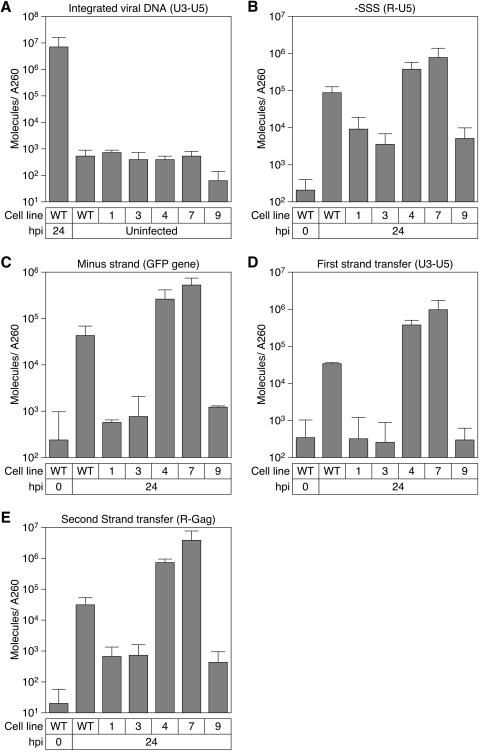
Production of reverse transcription intermediates in resistant cell lines. (A) DNA was harvested from infected and uninfected cells (5 × 105/well of a six-well plate) of the indicated cell lines. QPCR was performed using a primer/probe set that recognizes the first-strand transfer (+SSS) reverse transcription intermediate of the MLV genome that was used in the initial screen. (B to E) Cells (5 × 105/well of a six-well plate) of the indicated cell lines were infected with 5 × 105 IU of LEGFP[VSV-G] at 4°C for 2 h. Total DNA was harvested at 0 hpi and 24 hpi. DNA concentration was quantitated by A260, and QPCR was performed using primer/probe sets that recognize the (B) minus-strand strong stop, (C) minus strand, (D) first-strand transfer (+SSS) products, and (E) second-strand transfer (plus strand) reverse transcription intermediates. The numbers of DNA molecules were determined by comparison to a standard curve generated from serial dilutions of plasmid DNA. The data shown are the average mean values obtained in an experiment with triplicate samples and are representative of results of three independent experiments. Error bars indicate the standard deviations of the data.
The primer/probe sets used in these experiments recognize (in temporal order) the minus-strand strong stop (-SSS), elongated minus strand (minus strand), plus-strand strong stop (+SSS), and the plus-strand extension product after the second-strand transfer event (plus strand). Each primer/probe set recognizes the specific temporal reverse transcription step and will also amplify reverse transcription products from subsequent steps. Primer sets were chosen to amplify the specific reverse transcription intermediates based on hybridization to different regions of the viral genome (19, 42). Primers used to detect -SSS products hybridized to the viral repeat (R) and unique 5′ (U5) regions, those that detected minus strand elongation hybridized to the GFP coding sequence, those that detected products after the first-strand transfer (+SSS) hybridized to the viral unique 3′ (U3) and U5 regions, and those that detected products after the second strand transfer hybridized to the viral R and Gag regions (Table 1). To reduce the detection of plasmid DNA from the viral stocks, the virus stocks were treated with DNaseI, and cells were washed twice after infection. MCLs 1, 3, and 9 exhibited 10-fold, 20-fold, and 20-fold fewer -SSS products, respectively, than WT CHO-K1 cells (Fig. 5B). Minus-strand (Fig. 5C) and +SSS (Fig. 5D) reverse transcription products were at background levels in cell lines 1, 3, and 9. The final reverse transcription products accumulated to levels 50-fold lower than WT in these three cell lines (Fig. 5E). These data indicate that MLV does not undergo efficient reverse transcription in MCLs 1, 3, and 9. In contrast, MCL4 and MCL7 contained WT or greater levels of reverse transcription products with all four primer/probe sets, indicating that there is no defect in their ability to support reverse transcription (Fig. 5).
MCL4 and MCL7 support movement of the MLV reverse transcription complex to the nucleus but fail to integrate MLV.
To determine if viral DNA is localized to the nucleus for either cell line, MCL4 or MCL7, infections were performed, nuclei were isolated, DNA was extracted, and QPCR was performed. Late reverse transcription products were detected in the nuclei of these cells at levels equal to or greater than the levels detected for WT CHO-K1 cells (Fig. 6A). By contrast, no nuclear reverse transcription products were detected with MCL1, as predicted, since this cell line does not give rise to reverse transcription products. The reason for the increase in reverse transcription products in MCL4 and MCL7, which was observed with all primer/probe sets (Fig. 5 and 6A), is unknown, but the increase is highly reproducible, suggesting that it is caused by the mutation which blocks MLV infection. It may be that the incoming genome is trapped in the nucleus in a form that is slightly more stable than in WT cells.
FIG. 6.
Nuclear localization and integration of reverse transcription intermediates in resistant cell lines. (A) Cells (5 × 105/well of a six-well plate) of the indicated cell lines were infected as described for Fig. 5, and nuclear DNA was harvested at 0 hpi and 24 hpi. QPCR was performed using a primer/probe set that recognizes the second-strand transfer reverse transcription intermediate. (B) Cells (5 × 106/6-cm dish) of the indicated cell lines were infected with 5 × 106 IU of LEGFP[VSV-G]. DNA was harvested 24 hpi, and QPCR was performed with a primer/probe set that recognizes 2LTR circles. (C) Cells (5 × 105/well of a six-well plate) of the indicated cell lines were infected with 5 × 105 IU of pLEGFP[VSV-G]and passaged for 10 days postinfection. Total DNA was isolated, and QPCR was performed using a primer/probe set that recognizes the first-strand transfer (+SSS) and subsequent reverse transcription intermediates. DNA concentration was quantitated by A260, and number of DNA molecules was determined by comparison to a standard curve generated from serial dilutions of plasmid DNA. The data shown are the average mean values obtained in an experiment with triplicate samples and are representative of results of three independent experiments. Error bars indicate the standard deviations of the data.
To determine if the viral DNA in MCL4 and MCL7 is accessible to the interior of the nucleus, we exploited the fact that viral DNA that is localized to the nucleus can be circularized by cellular enzymes into integration incompetent products known as 2LTR circles (34, 35). The formation of 2LTR circles has been used as a marker for the exposure of viral reverse transcription products to the interior of the nucleus (10, 17, 25). To test if the reverse transcription products in MCL4 and MCL7 get imported to the interior of the nucleus, cells were infected and QPCR amplification was used to measure the amount of viral 2LTR circular DNA that was produced. Cell lines 4 and 7 had close to WT levels of 2LTR circles, while cell line 1, which does not make reverse transcription products, had only 6% of the WT levels of 2LTR circles (Fig. 6B). These data suggest that reverse transcription products synthesized in cell lines 4 and 7 are transported to the interior of the nucleus. To determine if the cell lines could support integration, cells were infected and then passaged for 10 days. Since episomal retroviral DNA is not maintained with cell division, only integrated viral DNA would be predicted to remain after 10 days (41, 46). All of the cell lines had at least 50-fold fewer molecules of integrated viral DNA than WT cells (Fig. 6C). These data suggest that the majority of resistance to MLV in cell lines 4 and 7 is due to the failure of nuclear localized viral DNA to integrate into the host genome. Taken together, the data strongly suggest that the resistance to infection in cell lines 4 and 7 is due to a failure of MLV to integrate into the genomes of these cells.
DISCUSSION
We have employed a high-throughput screening method with chemically mutagenized CHO-K1 cells that has led to the identification of mutant cell lines that exhibit novel blocks to the early steps of retroviral infection. These effects were specific for the MLV core and were not noted with HIV-1 or ASLV vectors. These cell lines fall into two distinct phenotypic groups.
Members of the first group, consisting of MCL1, MCL3, and MCL9, appear to have a defect in uncoating, since they are impaired in supporting the earliest steps of MLV-specific reverse transcription. Indeed, the impairment in initiating reverse transcription is sufficient to explain the MLV resistance in MCL3 and MCL9. In the case of MCL1, the cell line exhibits an ∼1,100-fold resistance to MLV infection, and at least 50-fold of this effect is due to impaired reverse transcription. The discrepancy between results of the reporter gene assay used to determine infectivity and the QPCR results may be due, in part, to an approximately sevenfold defect in this cell line in transcription from the MLV LTR (data not shown). This transcriptional defect, while not specific—it was also noted with the cytomegalovirus promoter—likely contributes to the MCL1 resistance phenotype.
The second group, consisting of MCL4 and MCL7, contained even higher levels (three- to fivefold) of reverse transcription products relative to WT CHO-K1 cells. In these cells, viral DNA was associated with the nucleus and 2LTR circular viral DNA forms were produced, indicative of nuclear translocation. However, the viral DNA genomes failed to integrate in these cells. The defects observed in the two distinct classes of mutant CHO-K1 cells are also clearly different from those described for two Rat-2 cell lines (R3-2 and R4-7) (17). Like MCL1, MCL3, and MCL9, the R4-7 cell line exhibits a block to initiation of reverse transcription. However, this defect is not MLV specific, since a similar block to infection was seen with an HIV-1 vector (17). Unlike MCL4 and MCL7, the R3-2 cell line exhibits a block to nuclear translocation of viral DNA-containing complexes, and again this effect is not MLV-specific, since it was also observed with an HIV-1 vector (17). It is unlikely that the MLV-specific defects in MCL4 and MCL7 can be explained by mutations in the cellular factors barrier-to-autointegration factor or HMGa1, since these factors support MLV and HIV-1 DNA integration (7, 22-24, 27, 37, 39, 40). Given these novel features, we anticipate that further characterization of the mutant cell lines that are described in this study will provide new insights into the role of viral-host cell factor interactions in promoting the early steps of retroviral replication. Moreover, the high-throughput MACS assay that is described in this report should prove useful for screening for cellular factors that help facilitate the early replication events for a number of different viruses.
Acknowledgments
We thank Jessica Berneström for the screening of Pool 4 cells.
This work was supported by NIH training grant T32 CA009075 and by NIH grants CA70810 (J.A.T.Y.) and CA22443 (P.A.)
REFERENCES
- 1.Adkins, H. B., S. C. Blacklow, and J. A. Young. 2001. Two functionally distinct forms of a retroviral receptor explain the nonreciprocal receptor interference among subgroups B, D, and E avian leukosis viruses. J. Virol. 75:3520-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, P., J. A. Young, and H. E. Varmus. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043-1051. [DOI] [PubMed] [Google Scholar]
- 3.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109-1115. [DOI] [PubMed] [Google Scholar]
- 5.Boerger, A. L., S. Snitkovsky, and J. A. Young. 1999. Retroviral vectors preloaded with a viral receptor-ligand bridge protein are targeted to specific cell types. Proc. Natl. Acad. Sci. USA 96:9867-9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley, K. A., J. Mogridge, M. Mourez, R. J. Collier, and J. A. Young. 2001. Identification of the cellular receptor for anthrax toxin. Nature 414:225-229. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H., and A. Engelman. 1998. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc. Natl. Acad. Sci. USA 95:15270-15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin, J. M., S. H. Hughes, and H. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N. Y. [PubMed]
- 9.Dvorin, J. D., and M. H. Malim. 2003. Intracellular trafficking of HIV-1 cores: journey to the center of the cell. Curr. Top. Microbiol. Immunol. 281:179-208. [DOI] [PubMed] [Google Scholar]
- 10.Ellis, J., and A. Bernstein. 1989. Retrovirus vectors containing an internal attachment site: evidence that circles are not intermediates to murine retrovirus integration. J. Virol. 63:2844-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelman, A. 2003. The roles of cellular factors in retroviral integration. Curr. Top. Microbiol. Immunol. 281:209-238. [DOI] [PubMed] [Google Scholar]
- 12.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 73:8919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fassati, A., D. Gorlich, I. Harrison, L. Zaytseva, and J. M. Mingot. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 22:3675-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Federspiel, M. J., P. Bates, J. A. Young, H. E. Varmus, and S. H. Hughes. 1994. A system for tissue-specific gene targeting: transgenic mice susceptible to subgroup A avian leukosis virus-based retroviral vectors. Proc. Natl. Acad. Sci. USA 91:11241-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, G., and S. P. Goff. 2004. Isolation of suppressor genes that restore retrovirus susceptibility to a virus-resistant cell line. Retrovirology 1:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, G., and S. P. Goff. 1999. Somatic cell mutants resistant to retrovirus replication: intracellular blocks during the early stages of infection. Mol. Biol. Cell 10:1705-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goff, S. P. 2001. Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J. Gene Med. 3:517-528. [DOI] [PubMed] [Google Scholar]
- 19.Gotte, M., X. Li, and M. A. Wainberg. 1999. HIV-1 reverse transcription: a brief overview focused on structure-function relationships among molecules involved in initiation of the reaction. Arch. Biochem. Biophys. 365:199-210. [DOI] [PubMed] [Google Scholar]
- 20.Gupta, R. S., D. Y. Chan, and L. Siminovitch. 1978. Evidence for functional hemizygosity at the Emtr locus in CHO cells through segregation analysis. Cell 14:1007-1013. [DOI] [PubMed] [Google Scholar]
- 21.Landau, N. R., and D. R. Littman. 1992. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J. Virol. 66:5110-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, L., C. M. Farnet, W. F. Anderson, and F. D. Bushman. 1998. Modulation of activity of Moloney murine leukemia virus preintegration complexes by host factors in vitro. J. Virol. 72:2125-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, L., K. Yoder, M. S. Hansen, J. Olvera, M. D. Miller, and F. D. Bushman. 2000. Retroviral cDNA integration: stimulation by HMG I family proteins. J. Virol. 74:10965-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, C. W., and A. Engelman. 2003. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J. Virol. 77:5030-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobel, L. I., J. E. Murphy, and S. P. Goff. 1989. The palindromic LTR-LTR junction of Moloney murine leukemia virus is not an efficient substrate for proviral integration. J. Virol. 63:2629-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacInnes, M. A., U. Friedrich, T. van Daalen Wetters, and P. Coffino. 1982. Quantitative forward-mutation specificity of mono-functional alkylating agents, ICR-191, and aflatoxin B1 in mouse lymphoma cells. Mutat. Res. 95:297-311. [DOI] [PubMed] [Google Scholar]
- 27.Mansharamani, M., D. R. Graham, D. Monie, K. K. Lee, J. E. Hildreth, R. F. Siliciano, and K. L. Wilson. 2003. Barrier-to-autointegration factor BAF binds p55 Gag and matrix and is a host component of human immunodeficiency virus type 1 virions. J. Virol. 77:13084-13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melikyan, G. B., R. J. Barnard, R. M. Markosyan, J. A. Young, and F. S. Cohen. 2004. Low pH is required for avian sarcoma and leukosis virus Env-induced hemifusion and fusion pore formation but not for pore growth. J. Virol. 78:3753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narayan, S., R. J. Barnard, and J. A. Young. 2003. Two retroviral entry pathways distinguished by lipid raft association of the viral receptor and differences in viral infectivity. J. Virol. 77:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narayan, S., and J. A. Young. 2004. Reconstitution of retroviral fusion and uncoating in a cell-free system. Proc. Natl. Acad. Sci. USA 101:7721-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlegel, R., T. S. Tralka, M. C. Willingham, and I. Pastan. 1983. Inhibition of VSV binding and infectivity by phosphatidylserine: is phosphatidylserine a VSV-binding site? Cell 32:639-646. [DOI] [PubMed] [Google Scholar]
- 33.Schlegel, R., M. C. Willingham, and I. H. Pastan. 1982. Saturable binding sites for vesicular stomatitis virus on the surface of Vero cells. J. Virol. 43:871-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shank, P. R., S. H. Hughes, H. J. Kung, J. E. Majors, N. Quintrell, R. V. Guntaka, J. M. Bishop, and H. E. Varmus. 1978. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell 15:1383-1395. [DOI] [PubMed] [Google Scholar]
- 35.Shank, P. R., and H. E. Varmus. 1978. Virus-specific DNA in the cytoplasm of avian sarcoma virus-infected cells is a precursor to covalently closed circular viral DNA in the nucleus. J. Virol. 25:104-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 37.Shumaker, D. K., K. K. Lee, Y. C. Tanhehco, R. Craigie, and K. L. Wilson. 2001. LAP2 binds to BAF · DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J. 20:1754-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki, Y., and R. Craigie. 2002. Regulatory mechanisms by which barrier-to-autointegration factor blocks autointegration and stimulates intermolecular integration of Moloney murine leukemia virus preintegration complexes. J. Virol. 76:12376-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki, Y., H. Yang, and R. Craigie. 2004. LAP2α and BAF collaborate to organize the Moloney murine leukemia virus preintegration complex. EMBO J. 23:4670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weller, S. K., A. E. Joy, and H. M. Temin. 1980. Correlation between cell killing and massive second-round superinfection by members of some subgroups of avian leukosis virus. J. Virol. 33:494-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitcomb, J. M., and S. H. Hughes. 1992. Retroviral reverse transcription and integration: progress and problems. Annu. Rev. Cell Biol. 8:275-306. [DOI] [PubMed] [Google Scholar]
- 43.Young, J. A., P. Bates, and H. E. Varmus. 1993. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J. Virol. 67:1811-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yueh, A., and S. P. Goff. 2003. Phosphorylated serine residues and an arginine-rich domain of the Moloney murine leukemia virus p12 protein are required for early events of viral infection. J. Virol. 77:1820-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 47.Zingler, K., and J. A. Young. 1996. Residue Trp-48 of Tva is critical for viral entry but not for high-affinity binding to the SU glycoprotein of subgroup A avian leukosis and sarcoma viruses. J. Virol. 70:7510-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]