Abstract
Proteolytic processing of paramyxovirus fusion (F) proteins is essential for the generation of a mature and fusogenic form of the F protein. Although many paramyxovirus F proteins are proteolytically processed by the cellular protease furin at a multibasic cleavage motif, cleavage of the newly emerged Hendra virus F protein occurs by a previously unidentified cellular protease following a single lysine at residue 109. We demonstrate here that the cellular protease cathepsin L is involved in converting the Hendra virus precursor F protein (F0) to the active F1 + F2 disulfide-linked heterodimer. To initially identify the class of protease involved in Hendra virus F protein cleavage, Vero cells transfected with pCAGGS-Hendra F or pCAGGS-SV5 F (known to be proteolytically processed by furin) were metabolically labeled and chased in the absence or presence of serine, cysteine, aspartyl, and metalloprotease inhibitors. Nonspecific and specific protease inhibitors known to decrease cathepsin activity inhibited proteolytic processing of Hendra virus F but had no effect on simian virus 5 F processing. We next designed shRNA oligonucleotides to cathepsin L which dramatically reduced cathepsin L protein expression and enzyme activity. Cathepsin L shRNA-expressing Vero cells transfected with pCAGGS-Hendra F demonstrated a nondetectable amount of cleavage of the Hendra virus F protein and significantly decreased membrane fusion activity. Additionally, we found that purified human cathepsin L processed immunopurified Hendra virus F0 into F1 and F2 fragments. These studies introduce a novel mechanism for primary proteolytic processing of viral glycoproteins and also suggest a previously unreported biological role for cathepsin L.
Paramyxoviruses are enveloped, single-stranded, negative-sense RNA viruses which include a number of major human pathogens, such as measles virus, mumps virus, human respiratory syncytial virus, and the newly emergent henipaviruses, namely, Hendra virus and Nipah virus (15). The high mortality rates associated with henipaviruses, as well as the ability of these viruses to cause systemic infections in a number of different hosts (12), have resulted in the classification of Hendra and Nipah viruses as biosafety level 4 agents. Infection by paramyxoviruses is initiated by fusion between the viral envelope and a cellular membrane (15). This mode of entry is promoted by the receptor-binding (H, HN, or G) and fusion (F) viral glycoproteins. Maturation of the paramyxovirus F proteins to a fusion-active form requires posttranslational proteolytic processing of the F0 precursor protein to the F1 + F2 disulfide-linked heterodimer. F proteins of previously characterized paramyxoviruses are activated either by the ubiquitous proprotein convertase furin at a multibasic cleavage site during exocytic transport (9) or by a tissue-specific extracellular protease following a monobasic residue (28). Although Hendra virus is efficiently propagated in many different cell lines (1, 18), the Hendra virus F protein is not activated by furin or by an extracellular protease, as intracellular cleavage of this important viral protein occurs after a single basic residue (18).
Several recent studies have examined the proteolytic activation of Hendra virus and the closely related Nipah virus F proteins. N-terminal sequencing of the F1 subunit of Hendra virus F and Nipah virus F identified residue 110 as the first F1 amino acid, suggesting that cleavage occurs after lysine 109 and arginine 109, respectively (18, 19). However, since the Hendra and Nipah viruses are newly discovered, the sequence conservation of the monobasic cleavage motif for henipaviruses remains to be determined. In contrast to the case for other paramyxoviruses, proteolytic processing of Hendra virus F is maintained when this basic residue or any of the eight residues upstream of lysine is either mutated individually or in aggregate (5a). Single amino acid substitutions also did not disrupt Nipah virus F protein processing (19), suggesting that no single amino acid immediately upstream of the cleavage site is required for the cleavage of henipavirus F proteins. Our previous studies on the proteolytic processing of Hendra virus F demonstrated that cleavage did not require exogenous proteases and occurred after transport to secretory vesicles (25). Proteolytic activation of Hendra virus F had a decreased Ca2+ requirement compared to that for furin-mediated activation, and cleavage was sensitive to increases in intracellular pH (25). These studies highlight the fact that proteolytic processing of Hendra virus F (and Nipah virus F) is different from that of other viral fusion proteins. The protease responsible for cleavage, however, has not previously been determined.
Here we describe the use of protease inhibitors and small hairpin RNA (shRNA) to identify the protease involved in proteolytic processing of Hendra virus F. We show that the endosomal enzyme cathepsin L is able to activate Hendra virus F in vitro and that a knockdown of cathepsin L expression and activity affects cleavage and thus the fusion activity of Hendra virus F. Therefore, cleavage of the Hendra virus fusion protein by cathepsin L introduces a novel mechanism for the proteolytic maturation of viral glycoproteins.
MATERIALS AND METHODS
Cell culture and reagents.
Vero and BSR (provided by Karl-Klaus Conzelman, Pettenkofer Institut) cells were maintained in Dulbecco's modified Eagle's media (DMEM; Gibco Invitrogen) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Plasmids containing the Hendra virus F and G genes were provided by Lin-fa Wang (Australian Animal Health Laboratory). pCAGGS-Hendra F and -Hendra G, as well as antipeptide antibodies to the Hendra virus F cytoplasmic tail, have been previously described (25). pCAGGS-SV5 F and antipeptide antibodies to simian virus 5 (SV5) F2 (amino acids [aa] 82 to 96) were a kind gift from Robert Lamb (HHMI, Northwestern University). The protease inhibitors phenylmethylsulfonyl fluoride (PMSF), E-64, and E-64d were purchased from Sigma, CA-074Me was purchased from Peptides International, and Complete Mini without EDTA protease tablets were purchased from Roche. The protease inhibitors aprotinin, antipain, chymostatin, GM6001, 1,10-phenanthroline, calpeptin, PD150606, caspase III, caspase 3 III, MG132, lactacystin, cathepsin I, and cathepsin LIII and human liver cathepsin L were obtained from Calbiochem.
Expression of Hendra virus and SV5 F proteins, metabolic labeling, and immunoprecipitation.
Vero cells were transiently transfected with pCAGGS expression vectors using the Lipofectamine Plus reagent (25). At 24 h posttransfection, cells were starved in methionine- and cysteine-deficient DMEM for 45 min and then metabolically labeled with Tran35S label (100 μCi/ml; MP Biomedicals) for 30 min. After being washed twice, cells were either immediately lysed in radioimmunoprecipitation assay buffer containing protease inhibitors and 20 mM iodoacetamide or incubated for 2 h at 37°C in fresh DMEM in the absence or presence of specific protease inhibitors before cell lysis. Immunoprecipitations were performed (25), and samples were electrophoresed in 15% reducing polyacrylamide gels (sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) and analyzed by storage phosphor autoradiography on a Typhoon imaging system (Amersham).
Cathepsin L shRNA.
Cathepsin L-specific shRNA oligonucleotides were designed using the Oligoengine toolset. Sense and antisense Cath L98 (GATCCCCGGCGATGCACAACAGATTATTCAAGAGATAATCTGTTGTGCATCGCCTTTTTA) and Cath L662 (GATCCCCTGACACCGGCTTTGTGGACTTCAAGAGAGTCCACAAAGCCGGTGTCATTTTTA) DNA oligonucleotides were ligated into pSUPER-GFP/Neo and sequenced to confirm the insertion of oligonucleotides (underlined and italicized sequences denote shRNA target regions). pSuper plasmids were transiently transfected into Vero cells as described above. At 24 h posttransfection, cells were trypsinized, collected in DMEM-10% FBS, and centrifuged at 1,500 rpm for 5 min at 4°C. Cells were washed twice and resuspended in a basic sorting buffer (phosphate-buffered saline without Ca2+ and Mg2+, 1 mM EDTA, 25 mM HEPES, and 1% FBS; pH 7.2), and green fluorescent protein (GFP)-expressing cells were sorted from non-GFP-expressing cells (MoFlo Dako Cytomation cell sorter). GFP-expressing cells were plated in 35-mm dishes (5 × 105 cells/dish) in DMEM-10% FBS with 5 mg/ml G418 and incubated overnight at 37°C in a 5% CO2 incubator. The pSuper-Scramble plasmid (GATCCCCTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTA), generously provided by Doug Andres (University of Kentucky), was used as a control for all experiments. shRNA-expressing cells were analyzed for cathepsin L protein expression and enzyme activity. pCAGGS-Hendra F, -Hendra G, -SV5 F, and empty vector were expressed in shRNA-expressing cells by transient transfection using the Lipofectamine 2000 reagent according to Invitrogen's instructions.
Immunoblot analysis of cathepsin L.
shRNA-expressing Vero cells were lysed as described above and then scraped, and the collected lysate was centrifuged at 13,000 × g for 20 min at 4°C. Five micrograms of total protein was resolved by 15% SDS-PAGE and transferred to a nitrocellulose membrane, and cathepsin L was detected with rabbit anti-human cathepsin L antisera (Athens Research and Technology). Equal protein concentrations were determined by the detection of β-actin with a monoclonal antibody (Sigma).
Cathepsin B and L enzymatic activity assays.
shRNA-expressing Vero cells were washed three times with PBS and lysed in 50 mM Tris (pH 7.4), 150 mM NaCl, 10% glycerol, and 1% NP-40. Scraped cell lysates were centrifuged at 4°C for 20 min at 13,000 × g and then placed on ice. InnoZyme cathepsin L and B activity kits (Calbiochem) were used to measure the enzyme activity of 10 μg of total protein. Fluorescence was measured at excitation and emission wavelengths of 360 nm and 480 nm, respectively (SpectraMAX Gemini XS; Molecular Devices), and analysis was performed using Softmax Pro software. For each sample, the enzyme activity of triplicate samples was measured in duplicate.
Viral fusion assays.
Luciferase T7 control DNA (1 μg), pCAGGS-Hendra F (1 μg), and pCAGGS-Hendra G (3 μg) were transfected into shRNA-expressing Vero cells as described above. At 24 h posttransfection, BSR cells were overlaid onto the Vero cells and incubated for 3 h at 37°C. Cells were lysed and analyzed for luciferase activity per the manufacturer's protocol (Promega). Since Vero cells lacking shRNA expression plasmids could not be sorted, control background values for Scramble-expressing Vero cells transfected with pCAGGS-Hendra G alone were subtracted from the values for Hendra virus F-plus-G-expressing samples. The luciferase activity of triplicate samples was measured in duplicate, and the assay was repeated three times.
In vitro cleavage of immunoprecipitated Hendra virus F.
Hendra virus F protein was expressed in Vero cells and metabolically labeled as described above. To generate uncleaved F product, cells were chased in DMEM at 20°C for 2 h, lysed, and immunoprecipitated as described above. After the final immunoprecipitation wash, 10 nM human cathepsin L (in 50 mM sodium acetate [pH 5.5], 5 mM EDTA, and 2 mM dithiothreitol buffer; Calbiochem) was added to Hendra virus F-bound protein A-Sepharose and incubated at 37°C for 0 to 8 h. Digestion reactions were stopped by the addition of 1 mM E-64, and samples were resolved by 15% SDS-PAGE and analyzed as previously described.
RESULTS
Proteolytic processing of Hendra virus F is not decreased by general protease inhibitors.
To determine the class of protease involved in processing of the Hendra virus F protein, Vero cells expressing either the Hendra virus F or SV5 F (known to be cleaved by Ca2+-dependent furin) protein via the pCAGGS system were metabolically labeled in the absence or presence of different protease inhibitors. Immunoprecipitated F proteins were resolved by 15% SDS-PAGE and analyzed by the Typhoon imaging system. As observed previously, only the inactive F precursor F0 (Fig. 1, lane 1) was observed immediately after Tran35S labeling, and F0 was converted to the F1 + F2 heterodimer during the 2-h chase period (Fig. 1, lane 2) (25). Aprotinin and PMSF, representing membrane-impermeant and -permeant serine protease inhibitors, respectively, did not reduce Hendra virus F cleavage (Fig. 1, lanes 4 and 5). The impermeant agents leupeptin and antipain and the permeant agent chymostatin, inhibitors of trypsin-like serine proteases and some cysteine proteases, similarly did not inhibit processing of the Hendra virus F proteins (Fig. 1, lanes 6 to 8). Pepstatin A (Fig. 1, lane 9), a membrane-permeant inhibitor of aspartic proteases, also did not affect Hendra virus F processing. Tumor necrosis factor alpha-converting enzyme, a zinc-dependent metalloprotease, was reported to cleave the ectodomain of the Ebola virus glycoprotein (GP), resulting in GP release from the cell surface (6). Despite the suggestion that tumor necrosis factor alpha-converting enzyme might be involved in Nipah virus F protein processing (19), the addition of the metalloprotease inhibitors GM6001 and 1,10-phenanthroline (Fig. 1, lanes 10 and 11) did not inhibit processing of Hendra virus F. All of the above inhibitors also do not affect furin activity, in that cleavage of SV5 F proceeded in the presence of these inhibitors (21; data not shown). To exclude the possibility that the inhibitor concentrations were too low, we examined the effect of 10 times the recommended inhibitor concentration and found (with the exception of 1,10-phenanthroline, which was toxic to cells at concentrations of >1 mM) that the Hendra virus F protein was efficiently processed (data not shown). Therefore, Hendra virus F was not processed by a cell surface serine or cysteine protease, as aprotinin, leupeptin, and antipain did not inhibit cleavage (Fig. 1, lanes 4, 6, and 7). Similarly, inhibition of intracellular serine, aspartic, and metalloproteases did not affect Hendra virus F cleavage (Fig. 1, lanes 5 and 8 to 11). In contrast, the presence of a Complete protease tablet (without EDTA) decreased proteolytic processing of Hendra virus F (Fig. 1, lane 3). Since each Complete protease tablet contains a cocktail of serine and cysteine inhibitors but other serine protease inhibitors had no effect, we further examined the role of cysteine proteases on Hendra virus F processing.
FIG. 1.
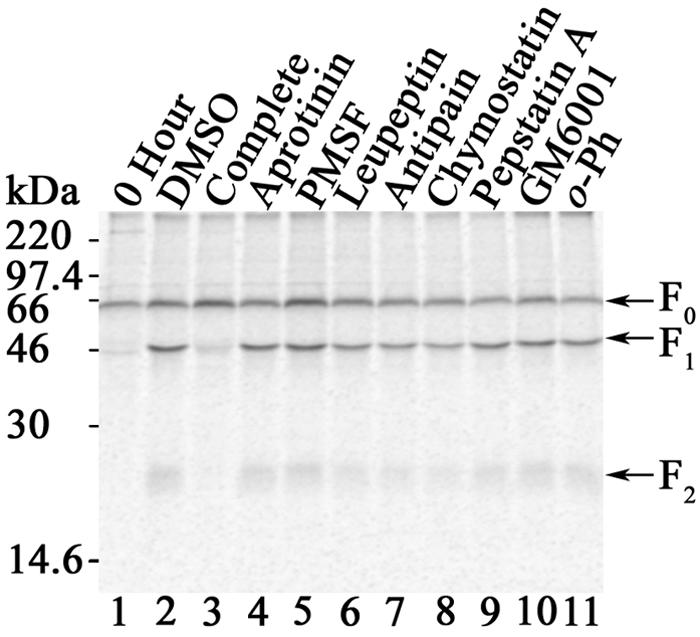
General protease inhibitors do not retard proteolytic processing of Hendra virus F. Vero cells transfected with pCAGGS-Hendra F were metabolically labeled and either lysed immediately (0 h) or chased for 2 h in DMEM in the absence (DMSO) or presence of a Complete protease tablet (without EDTA), 1 kU aprotinin, 1 mM PMSF, 100 μM leupeptin, 100 μM antipain, 100 μM chymostatin, 1 μM pepstatin A, 5 μM GM6001, or 0.1 mM 1,10-phenanthroline (o-Ph). Samples were analyzed by immunoprecipitation, 15% SDS-PAGE, and the Typhoon imaging system. Positions of uncleaved F0 and cleaved F1 and F2 subunits are indicated.
Nonspecific and specific cathepsin inhibitors block processing of Hendra virus F.
The general cysteine protease inhibitors E-64 and E-64d inhibit calpain and cathepsins B, H, and L (33). While the addition of the membrane-impermeant E-64 had no effect, membrane-permeant E-64d ablated cleavage of Hendra virus F (Fig. 2A, lanes 3 and 4), indicating that an intracellular cysteine protease is involved in processing. To examine the role of calpain in Hendra virus F processing, calpeptin (an inhibitor of calpain and other cysteine proteases [32]) and PD150606 (a more specific calpain inhibitor that binds Ca2+-binding domains of calpain versus the active site [31]) were added. Calpeptin, but not PD150606, inhibited the cleavage of Hendra virus F (Fig. 2A, lanes 5 and 6), ruling out calpain but again pointing to cleavage by a cysteine protease. Neither the broad-spectrum caspase inhibitor III nor an inhibitor of caspases 3, 6, 7, 8, and 10 (caspase 3 inhibitor III) blocked the cleavage of Hendra virus F (Fig. 2A, lanes 7 and 8). A 1 μM concentration of the reversible proteasome inhibitor MG132 significantly reduced the processing of Hendra virus F (Fig. 2A, lane 9), but the irreversible proteasome inhibitor lactacystin had no effect on F cleavage (Fig. 2A, lane 10). MG132, however, has been demonstrated to block cathepsin L activity (10). Finally, cleavage of Hendra virus F was impeded in the presence of a general cathepsin inhibitor (cathepsin inhibitor I [Cath I]) and inhibitors of cathepsin L (cathepsin L inhibitor III [Cath LIII]) and cathepsin B/L (CA-074Me) (Fig. 2A, lanes 11 to 13) (22). We subsequently verified that cathepsin L and B enzyme activity in Vero cells was decreased in the presence of the CA-074Me and Cath LIII inhibitors (data not shown). Cysteine protease inhibitors and increasing concentrations thereof did not influence the processing of SV5 F (Fig. 2B and data not shown), indicating that the effects of these inhibitors were specific to Hendra virus F processing and not due to a nonspecific block on intracellular trafficking. We further confirmed proper intracellular trafficking of the Hendra virus F protein in the presence of E-64d, calpeptin, MG132, and cathepsin inhibitors by endoglycosidase H resistance and cell surface expression (biotinylation assays) (data not shown). The inhibition of Hendra virus F processing in the presence of nonspecific and specific cathepsin inhibitors, together with the sensitivity of Hendra virus F cleavage to increases in intracellular pH (25), strongly indicates that endosomal cathepsin proteases are involved in the maturation of Hendra virus F.
FIG. 2.
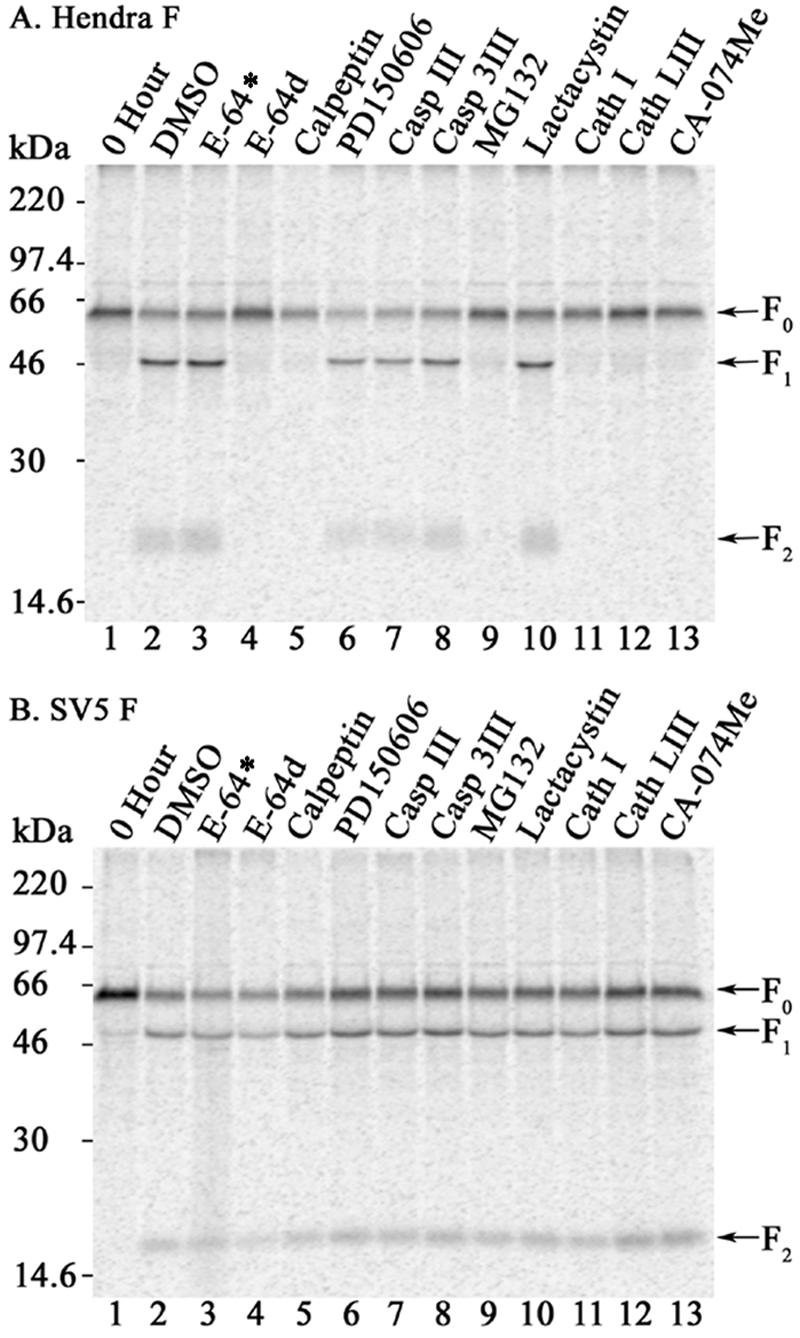
Nonspecific and specific cathepsin inhibitors block Hendra virus F cleavage. Vero cells expressing pCAGGS-Hendra F (A) or SV5 F (B) were starved, labeled with Tran35S label, and chased for 2 h in the absence (DMSO) or presence of 10 μM E-64, E-64d, calpeptin, PD150606, caspase 3 inhibitor III (Casp 3 III) or lactacystin, 1 μM MG132, 100 μM caspase inhibitor III (Casp III), or 50 μM cathepsin inhibitor I (Cath I), cathepsin L inhibitor III (Cath LIII), or CA-074Me. Samples were prepared and analyzed as described in the text. The positions of the F0, F1, and F2 proteins are indicated. All cysteine protease inhibitors, with the exception of E-64*, are membrane permeant.
Cathepsin L shRNA suppresses protein and enzyme activity in Vero cells.
Our results indicated that a cysteine cathepsin protease was likely involved in Hendra virus F processing. Cathepsins B and L are ubiquitously expressed proteases in the endosomal/lysosomal pathway and, to date, are the most well-studied cathepsins (reviewed in reference 17). Cathepsin B exhibits both carboxydipeptidase and endopeptidase activities (17). In contrast, cathepsin L displays only endopeptidase activity and has been reported to proteolytically process prohormones at both mono- and dibasic residues (11, 17). For these reasons, we chose to further examine the role of cathepsin L in the proteolytic maturation of Hendra virus F. Cathepsin L protein expression and enzyme activity in Vero cells were suppressed by the expression of two shRNA DNA oligonucleotides, Cath L98 and Cath L662, targeted to nucleotides (nt) 98 to 116 and 662 to 680, respectively, of the human cathepsin L cDNA sequence. Additionally, oligonucleotides with a scramble sequence were included as a control for all assays. Oligonucleotides were cloned into the pSuper expression vector and transiently transfected into Vero cells. The presence of both GFP and neomycin selection markers within pSuper permitted efficient sorting and maintenance of only shRNA-expressing Vero cells.
Cathepsin L is synthesized as a proprotein (∼39 kDa; ProL). The single-chain form of cathepsin L (∼30 kDa; SCL) is generated following the removal of the prodomain. SCL is further processed to a double-chain form of the enzyme (∼25 and 5 kDa; DCL) (reviewed in reference 13). SCL and DCL are the mature forms of the protein, although most cell types predominantly harbor DCL (14). An immunoblot of untransfected Vero cells using anti-cathepsin L antibodies demonstrated a majority of the DCL form (Fig. 3A, lane 1). DCL and SCL were identified in Vero cells transfected with pSuper-Scramble (Fig. 3A, lane 2). At the protein detection level, cathepsin L was completely suppressed when pSuper-Cath L98 was expressed in Vero cells (Fig. 3A, lane 3). However, a faint band corresponding to DCL was visible in Vero cells expressing pSuper-Cath L662 (Fig. 3A, lane 4). Equivalent amounts of protein were demonstrated by Western blotting using β-actin monoclonal antibodies (Fig. 3A, bottom panel).
FIG. 3.
Effect of cathepsin L shRNA on protein and enzymatic activity in Vero cells. (A) Cathepsin L protein expression in untransfected and shRNA-expressing Vero cells. Five micrograms of total cell extract was subjected to SDS-PAGE and analyzed by Western blotting with anti-cathepsin L antibodies (top panel) and anti-β-actin monoclonal antibodies (bottom panel). The positions of procathepsin L (ProL), single-chain cathepsin L (SCL), and double-chain cathepsin L (DCL) are indicated. (B) Cathepsin L enzyme activity levels in untransfected and shRNA-expressing Vero cells. (C) Cathepsin B enzyme activity levels in untransfected and pSuper-Scramble-, -Cath L98-, and Cath L662-expressing Vero cells.
To confirm the Western blot results, the cathepsin L enzyme activity in both untransfected and shRNA-expressing Vero cells was determined (Fig. 3B). Untransfected and pSuper-Scramble shRNA-expressing cells demonstrated high levels of cathepsin L activity, although control shRNA-expressing cells exhibited a lower activity than untransfected cells. Cathepsin L activity in Cath L98-expressing cells was 10× less than that in cells expressing Scramble, while Cath L662-expressing cells maintained a moderate enzyme activity (2.5× less than that with Scramble), consistent with the immunoblot results. The cathepsin B activity in Vero cells expressing pSuper-Scramble, -Cath L98, and -Cath L662 was not significantly different from that in untransfected cells, indicating that shRNA reduction of cathepsin L does not inhibit cathepsin B activity (Fig. 3C).
Suppression of cathepsin L activity inhibits proteolytic processing of Hendra virus F and fusion activity.
Cath L98- and Cath L662-expressing Vero cells provide two levels of cathepsin L activity with which to study the effect of cathepsin L on the proteolytic processing of Hendra virus F. Twenty-four hours after cell sorting, shRNA-expressing Vero cells were transiently transfected with pCAGGS-Hendra F or -SV5 F. Metabolic labeling and immunoprecipitation were then used to examine the expression and proteolytic processing of the Hendra virus and SV5 F proteins. In agreement with our earlier inhibitor data, a shRNA knockdown of cathepsin L did not influence cleavage of SV5 F by furin (Fig. 4A, lanes 6 to 10). However, Hendra virus F cleavage was almost completely inhibited (Fig. 4A, lane 4) in Cath L98-expressing Vero cells exhibiting undetectable cathepsin L protein levels and minimal enzyme activity (Fig. 3A and B). Although only a small amount of cathepsin L protein expression was observed in cells transfected with pSuper-Cath L662 (Fig. 3, lane 4), the amount of cathepsin L activity (Fig. 3B) remaining was sufficient to proteolytically process Hendra virus F (Fig. 4A, lane 5), though to a reduced extent compared to that in untransfected and Scramble shRNA-expressing cells (Fig. 4A, lanes 2 and 3). These results strongly argue that cathepsin L is involved in proteolytic activation of the Hendra virus F protein.
FIG. 4.
Proteolytic processing of Hendra virus F and membrane fusion in shRNA-expressing Vero cells. (A) Expression and proteolytic processing of Hendra virus F and SV5 F in shRNA-expressing Vero cells were examined by metabolic labeling and immunoprecipitation as described previously. (B) Luciferase reporter gene assay to examine the ability of Hendra virus F and G to promote membrane fusion in shRNA-expressing Vero cells. The results presented are representative of three separate experiments.
Inhibition of Hendra virus F proteolytic processing clearly correlates with reductions in cathepsin L activity. We therefore examined the ability of Hendra virus F to mediate cell-cell fusion when expressed in Vero cells with shRNA to cathepsin L using a quantitative luciferase reporter gene assay. shRNA-expressing Vero cells were transfected with pCAGGS-Hendra F, -Hendra G, and a plasmid containing the luciferase gene under the control of a T7 promoter. These cells were overlaid with BSR cells expressing the T7 polymerase (2), and luciferase activity was assessed. It should be noted that these target cells express endogenous cathepsin L. Membrane fusion in Cath L98 shRNA-expressing cells was significantly debilitated (Fig. 4B), in agreement with the negligible amount of cleaved Hendra virus F in these cells. The moderate amounts of Hendra virus F cleavage observed following expression in Cath L662-expressing cells was reflected by increased fusion compared that in Cath L98-expressing cells. However, the fusion activity by Hendra virus F and G in Cath L662-expressing cells was not equivalent to that in Vero cells expressing pSuper-Scramble. The importance of cleavage for fusion was further confirmed by syncytium formation assays. We found that the size and extent of syncytium formation in Scramble and Cath L662 shRNA-expressing Vero cells appeared equivalent, but fewer and smaller syncytia were observed in Cath L98-expressing cells (data not shown). Therefore, suppression of cathepsin L inhibited the cleavage of Hendra virus F, leading to a loss of fusogenic activity.
Cathepsin L can cleave immunoprecipitated uncleaved Hendra virus F.
Our results indicate that cathepsin L is involved in the processing of Hendra virus F in cells. To confirm the ability of cathepsin L to process the Hendra virus F protein, we examined the ability of purified cathepsin L to cleave Hendra virus F0 to F1 and F2 subunits. Incubation of Hendra virus F-transfected Vero cells at 20°C inhibits the budding of secretory vesicles from the trans-Golgi network and restricts Hendra virus F processing (Fig. 5, lane 3), while cleavage may be restored by returning cells to 37°C (Fig. 5, lane 4) (25). Uncleaved Hendra virus F, generated by metabolic labeling at 20°C followed by immunoprecipitation, was used as a substrate for in vitro digestion with human cathepsin L. In the absence of enzyme, no processing of Hendra virus F was observed (Fig. 5, lane 5). Following a 2-h incubation (Fig. 5, lane 6), cathepsin L proteolytically processed Hendra virus F0 to several products, including two that corresponded to the F1 and F2 subunits. With increasing times of digestion, the amount of F0 decreased and the amount of F1 plus F2 increased (Fig. 5, lanes 6 to 9). Recently, proteolytic processing of Ebola virus GP was shown to require cathepsin B and L to facilitate infection (4). We found that immunopurified uncleaved Hendra virus F was processed by cathepsin B into multiple bands within 2 hours, including those corresponding to F1 and F2. However longer incubations resulted in Hendra virus F degradation to nonspecific products (data not shown). Similarly, cathepsin B and L rapidly digested uncleaved SV5 F to nonspecific products. Thus, cathepsin L is able to specifically convert the precursor Hendra virus F0 to stable products corresponding to those present in the fusogenically active F protein.
FIG. 5.
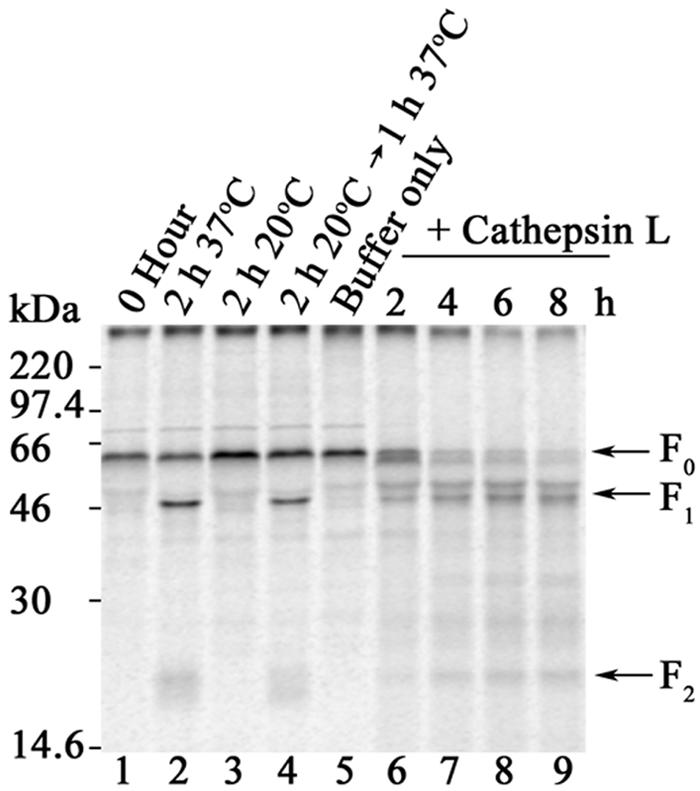
Cathepsin L is able to cleave Hendra virus F0 to F1-plus-F2 heterodimers. Immunopurified uncleaved Hendra virus F was digested with 10 nM human cathepsin L at 37°C for 2 to 8 h. An uncleaved Hendra virus F control sample was incubated in buffer without enzyme for 8 h at 37°C. Samples were analyzed as described in the other figure legends. F0, F1, and F2 are designated by arrows on the right.
DISCUSSION
Our data here demonstrate that the endosomal/lysosomal protease cathepsin L is involved in proteolytic maturation of the Hendra virus F protein. The incubation of Vero cells expressing Hendra virus F in the presence of nonspecific and specific cathepsin inhibitors reduced the cleavage of Hendra virus F (Fig. 2). shRNA to cathepsin L suppressed cellular protein levels and enzymatic activity (Fig. 3) and gave dramatically reduced proteolytic processing of the Hendra virus F protein (Fig. 4A). The low fusogenic activity of Hendra virus F in shRNA-expressing cells paralleled negligible amounts of cleaved Hendra virus F (Fig. 4B). Finally, we demonstrated that purified cathepsin L can digest precursor Hendra virus F0 to corresponding F1 and F2 subunits (Fig. 5). These results fit with our previous characterization of Hendra virus F processing (25), as cathepsin L does not require Ca2+ but is highly sensitive to increases in intracellular pH.
Lysosomal cysteine cathepsin proteases historically are thought of as destructive enzymes involved in the degradation of intracellular and endocytosed proteins (17). Despite the adverse association of cathepsin L with tumor metastasis (29) and osteoporosis (26), cathepsin L plays a number of important physiological roles in the cell, including major histocompatibility complex class II antigen presentation in cortical epithelial cells of the thymus (24), epidermal homeostasis (26), hair differentiation (27), and proteolytic processing of the CDP/CUX transcription factor (10) and prohormones (34). Cathepsins have also been reported to affect some viral processes. Following receptor-mediated uptake of the nonenveloped reovirus, cathepsins B and L digest the outer capsid proteins to form infectious subvirion particles which are then able to enter cells (8). Proteolytic digestion of Ebola virus GP by endosomal proteases was recently described (4). Viral entry following a secondary proteolytic event of Ebola GP was described to depend primarily upon cathepsin B digestion, with cathepsin L cleavage augmenting viral entry. Our finding of a role for cathepsin L in primary proteolytic processing of a viral glycoprotein suggests a novel activity for this important cellular protease. Unlike many cellular proteases, cathepsin L has no distinctive substrate recognition site, although the endoprotease is thought to favor hydrophobic amino acids at P2 and P3 sites (reviewed in reference 17). The cleavage of Hendra virus or Nipah virus F protein is thought to occur after a monobasic residue in the G-D-V-K/R sequence (18, 19). However, single mutations of the basic residue as well as of amino acids upstream of the cleavage site to alanine failed to eliminate henipavirus F processing (5a, 19). Only when six residues immediately upstream of arginine 109 in Nipah virus F were deleted was the cleavage of Nipah virus F ablated (19). These mutagenesis studies therefore demonstrate that no single amino acid is essential for processing of these F proteins. The apparent lack of specificity for henipavirus F cleavage aligns with the absence of a specific cleavage motif for cathepsin L.
Cathepsin L is localized primarily to the endosomal/lysosomal pathway and recently was reported to be in the secretory granules of the regulated secretory pathway (17, 34). A functional endocytosis motif is present within the cytoplasmic tails of the Hendra virus and Nipah virus F proteins (17a, 30). Mutagenesis of this endocytosis motif did not affect the intracellular trafficking of Hendra virus F to the plasma membrane; however, the rate of endocytosis and proteolytic processing of Hendra virus F was significantly decreased (17a). This indicates a novel mechanism of proteolytic maturation of Hendra virus F compared to those of many viral fusion proteins, which are either processed during exocytic trafficking (e.g., Ebola virus GP, human immunodeficiency virus gp160, and Lassa virus GP [16, 20]) or later cleaved by an extracellular protease (28). We propose that proteolytic processing of Hendra virus F by cathepsin L occurs during recycling of the F protein between the endosome and the cell surface and not in the secretory pathway during initial transport to the cell surface. Interestingly, purified Hendra virions are reported to contain both cleaved and uncleaved fusion proteins (18), suggesting that a percentage of Hendra virus F protein has undergone at least one round of recycling prior to budding of the virion from the cell surface.
The proteolytic processing of Hendra virus F by an endosomal protease and the presence of both uncleaved and cleaved F proteins on the surfaces of Hendra virions suggest that Hendra virus can utilize several mechanisms of entry. Enveloped viruses are thought to enter cells by one of two well-characterized pathways (7). The first, pH-independent entry occurs following fusion of the viral envelope with the plasma membrane. The second pathway requires endocytosis of the virus, where the low pH in the endosome serves to trigger conformational changes in the fusion protein, which stimulate fusion of the viral envelope with the endosomal membrane. As with other members of the paramyxovirus family, the presence of cleaved F protein on the surfaces of Hendra virions may permit fusion at the plasma membrane, especially since the Hendra virus F protein can promote fusion at neutral pH (1, 3). However, the Hendra virus could also enter cells via the endocytic pathway, where uncleaved F protein on the virion surface could be processed by cathepsin L, leading to fusion of the viral envelope with the endosomal membrane promoted by newly cleaved F protein. In this case, the pH of the environment would not be required to trigger membrane fusion, but rather the acidic environment would allow efficient enzymatic processing of the F protein. The potential ability of the virus to enter cells at the plasma membrane and/or the endocytic system would ensure viral infectivity and efficient propagation in the host. Future experiments examining the role of cathepsin L in Hendra virus infection as well as investigations into the mode(s) of Hendra virus entry will address these important possibilities.
Hendra virus was first identified as the etiological agent causing the deaths of 14 horses and 1 human due to severe respiratory illness during an outbreak in Australia in 1994 (23). The fatal viral encephalitis outbreak attributed to Nipah virus in Malaysia in 1999 caused 105 fatalities out of 265 reported cases (5). Currently, no antiviral therapies are available. Our finding that cathepsin L carries out a crucial processing event on the Hendra virus F protein, combined with knowledge of the cathepsin L crystal structure, the availability of cathepsin inhibitors, and studies examining the effectiveness of these inhibitors on tumor invasion (29) and osteoporosis prevention (35), suggests the possible use of cathepsin L inhibitors as therapy for Hendra virus infection.
Acknowledgments
We thank Lin-Fa Wang of the Australian Animal Health Laboratory for the Hendra virus F and G plasmids, Robert Lamb (HHMI, Northwestern University) for the pCAGGS-SV5 F expression vector and SV5 F-specific antibodies, and Doug Andres (University of Kentucky) for the pSuper-Scramble plasmid. Karl-Klaus Conzelman (Max Pettenkofer Institut) kindly provided the BSR cells. Furthermore, our thanks go to Wally Whiteheart, Doug Andres, and Louis Hersh for important suggestions and discussions during the course of this work. Technical advice from Brian Finlan and Martin Chow as well as assistance from Jennifer Strange in the UK Flow Cytometry Core Facility is greatly appreciated. We are grateful to members of the Dutch lab for critically reviewing the manuscript.
C.T.P. is a recipient of an AHA Ohio Valley Affiliate predoctoral fellowship. This study was supported by NIAID grant AI063052 to R.E.D.
REFERENCES
- 1.Bossart, K. N., L.-F. Wang, B. Eaton, and C. C. Broder. 2001. Functional expression and membrane fusion tropism of the envelope glycoproteins of Hendra virus. Virology 290:121-135. [DOI] [PubMed] [Google Scholar]
- 2.Buchholz, U. J., S. Finke, and K.-K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter, J. R., C. T. Pager, S. D. Fowler, and R. E. Dutch. 2005. Role of N-linked glycosylation of the Hendra virus fusion protein. J. Virol. 79:7922-7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandran, K., N. J. Sullivan, U. Felbor, S. P. Whelan, and J. M. Cunningham. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 5a.Craft, W. W., and R. E. Dutch. Sequence motif of the Hendra virus fusion protein is not sufficient to promote efficient protolytic processing. Virology, in press. [DOI] [PubMed]
- 6.Dolnik, O., V. Volchkov, W. Garten, C. Carbonelle, S. Becker, J. Kahnt, U. Ströher, H.-D. Klenk, and V. Volchkov. 2004. Ectodomain shedding of the glycoprotein GP of Ebola virus. EMBO J. 23:2175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2004. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285:25-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebert, D. H., J. Deussing, C. Peters, and T. S. Dermody. 2002. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 277:24609-24617. [DOI] [PubMed] [Google Scholar]
- 9.Garten, W., S. Hallenberger, D. Ortmann, W. Schäfer, M. Vey, H. Angliker, E. Shaw, and H. D. Klenk. 1994. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie 76:217-225. [DOI] [PubMed] [Google Scholar]
- 10.Goulet, B., A. Baruch, N.-S. Moon, M. Poirier, L. L. Sansregret, A. Erickson, M. Bogyo, and A. Nepveu. 2004. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell 14:207-219. [DOI] [PubMed] [Google Scholar]
- 11.Hook, V., S. Yasothornsrikul, D. Greenbaum, K. F. Medzihradszky, K. Troutner, T. Toneff, R. Bundey, A. Logrinova, T. Reinheckel, C. Peters, and M. Bogyo. 2004. Cathepsin L and Arg/Lys aminopeptidase: a distinct prohormone processing pathway for the biosynthesis of peptide neurotransmitters and hormones. Biol. Chem. 385:473-480. [DOI] [PubMed] [Google Scholar]
- 12.Hooper, P., S. R. Zaki, P. Daniels, and D. Middleton. 2001. Comparative pathology of the disease caused by Hendra and Nipah viruses. Microbes Infect. 3:315-322. [DOI] [PubMed] [Google Scholar]
- 13.Ishidoh, K., and E. Kominami. 2002. Processing and activation of lysosomal proteinases. Biol. Chem. 383:1827-1831. [DOI] [PubMed] [Google Scholar]
- 14.Kominami, E., T. Tsukahara, K. Hara, and N. Katunuma. 1988. Biosyntheses and processing of lysosomal cysteine proteinases in rat macrophages. FEBS Lett. 231:225-228. [DOI] [PubMed] [Google Scholar]
- 15.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 16.Lenz, O., J. ter Meulen, H.-D. Klenk, N. G. Seidah, and W. Garten. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. USA 98:12701-12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGrath, M. E. 1999. The lysosomal cysteine proteases. Annu. Rev. Biophys. Biomol. Struct. 28:181-204. [DOI] [PubMed] [Google Scholar]
- 17a.Meulendyke, K. A., M. A. Wurth, R. O. McCann, and R. E. Dutch. 2005. Endocytosis plays a critical role in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79:12643-12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michalski, W. P., G. Crameri, L.-F. Wang, B. J. Shiell, and B. Eaton. 2000. The cleavage activation and sites of glycosylation in the fusion protein of Hendra virus. Virus Res. 69:83-93. [DOI] [PubMed] [Google Scholar]
- 19.Moll, M., S. Diederich, H.-D. Klenk, M. Czub, and A. Maisner. 2004. Ubiquitous activation of the Nipah virus fusion protein does not require a basic amino acid at the cleavage site. J. Virol. 78:9705-9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molloy, S. S., E. D. Anderson, F. Jean, and G. Thomas. 1999. Bi-cycling the furin pathway: from TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 9:28-35. [DOI] [PubMed] [Google Scholar]
- 21.Molloy, S. S., P. A. Bresnahan, S. H. Leppla, K. R. Klimpel, and G. Thomas. 1992. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J. Biol. Chem. 267:16396-16402. [PubMed] [Google Scholar]
- 22.Montaser, M., G. Lalmanach, and L. Mach. 2002. CA-074, but not its methyl ester CA-074Me, is a selective inhibitor of cathepsin B within living cells. Biol. Chem. 383:1305-1308. [DOI] [PubMed] [Google Scholar]
- 23.Murray, K., P. Selleck, P. Hooper, A. Hyatt, A. Gould, L. Gleeson, H. Westbury, L. Hiley, L. Selvey, B. Rodwell, and P. Ketterer. 1995. A morbillivirus that caused fatal disease in horses and humans. Science 268:94-97. [DOI] [PubMed] [Google Scholar]
- 24.Nakagawa, T., W. Roth, P. Wong, A. Nelson, A. Farr, J. Deussing, J. A. Villadangos, H. Ploegh, C. Peters, and A. Y. Rudensky. 1998. Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science 280:450-453. [DOI] [PubMed] [Google Scholar]
- 25.Pager, C. T., M. A. Wurth, and R. E. Dutch. 2004. Subcellular localization and calcium and pH requirements for proteolytic processing of the Hendra virus fusion protein. J. Virol. 78:9154-9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potts, W., J. Bowyer, H. Jones, D. Tucker, A. J. Freemont, A. Millest, C. Martin, W. Vernon, D. Neerunjun, G. Slynn, F. Harper, and R. Maciewicz. 2004. Cathepsin L-deficient mice exhibit abnormal skin and bone development and show increased resistance to osteoporosis following ovariectomy. Int. J. Exp. Pathol. 85:85-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth, W., J. Deussing, V. A. Botchkarev, M. Pauly-Evers, P. Saftig, A. Hafner, P. Schmidt, W. Schmahl, J. Scherer, I. Anton-Lamprecht, K. Von Figura, R. Paus, and C. Peters. 2000. Cathepsin L deficiency as molecular defect of furless: hyperproliferation of keratinocytes and perturbation of hair follicle cycling. FASEB J. 14:2075-2086. [DOI] [PubMed] [Google Scholar]
- 28.Scheid, A., and P. W. Choppin. 1974. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology 57:475-490. [DOI] [PubMed] [Google Scholar]
- 29.Sever, N., M. Filipic, J. Brzin, and T. T. Lah. 2002. Effect of cysteine proteinase inhibitors on murine B16 melanoma cell invasion in vitro. Biol. Chem. 383:839-842. [DOI] [PubMed] [Google Scholar]
- 30.Vogt, C., M. Eickmann, S. Diederich, M. Moll, and A. Maisner. 2005. Endocytosis of the Nipah virus glycoproteins. J. Virol. 79:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, K. K. W., R. Nath, A. Posner, K. J. Raser, M. Buroker-Kilgore, I. Hajimohammadreza, A. W. Probert, Jr., F. W. Marcoux, Q. Ye, E. Takano, M. Hatanaka, M. Maki, H. Caner, J. L. Collins, A. Fergus, K. S. Lee, E. A. Lunney, S. J. Hays, and P.-W. Yuen. 1996. An alpha-mercaptoacrylic acid derivative is a selective nonpeptide cell-permeable calpain inhibitor and is neuroprotective. Proc. Natl. Acad. Sci. USA 93:6687-6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, K. K. W., and P.-W. Yuen. 1994. Calpain inhibition: an overview of its therapeutic potential. Trends Pharmacol. Sci. 15:412-419. [DOI] [PubMed] [Google Scholar]
- 33.Wilcox, D., and R. W. Mason. 1992. Inhibition of cysteine proteinases in lysosomes and whole cells. Biochem. J. 285:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasothornsrikul, S., D. Greenbaum, K. F. Medzihradszky, T. Toneff, R. Bundey, R. Miller, B. Schilling, I. Petermann, J. Dehnert, A. Logvinova, P. Goldsmith, J. M. Neveu, W. S. Lane, B. Gibson, T. Reinheckel, C. Peters, M. Bogyo, and V. Hook. 2003. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc. Natl. Acad. Sci. USA 100:9590-9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yasuma, T., S. Oi, N. Choh, T. Nomura, N. Furuyama, A. Nishimura, Y. Fujisawa, and T. Sohda. 1998. Synthesis of peptide aldehyde derivatives as selective inhibitors of human cathepsin L and their inhibitory effect on bone resorption. J. Med. Chem. 41:4301-4308. [DOI] [PubMed] [Google Scholar]


