Abstract
Replication of the ∼30-kb plus-strand RNA genome of coronaviruses and synthesis of an extensive set of subgenome-length RNAs are mediated by the replicase-transcriptase, a membrane-bound protein complex containing several cellular proteins and up to 16 viral nonstructural proteins (nsps) with multiple enzymatic activities, including protease, polymerase, helicase, methyltransferase, and RNase activities. To get further insight into the replicase gene-encoded functions, we characterized the coronavirus X domain, which is part of nsp3 and has been predicted to be an ADP-ribose-1"-monophosphate (Appr-1"-p) processing enzyme. Bacterially expressed forms of human coronavirus 229E (HCoV-229E) and severe acute respiratory syndrome-coronavirus X domains were shown to dephosphorylate Appr-1"-p, a side product of cellular tRNA splicing, to ADP-ribose in a highly specific manner. The enzyme had no detectable activity on several other nucleoside phosphates. Guided by the crystal structure of AF1521, an X domain homolog from Archaeoglobus fulgidus, potential active-site residues of the HCoV-229E X domain were targeted by site-directed mutagenesis. The data suggest that the HCoV-229E replicase polyprotein residues, Asn 1302, Asn 1305, His 1310, Gly 1312, and Gly 1313, are part of the enzyme's active site. Characterization of an Appr-1"-pase-deficient HCoV-229E mutant revealed no significant effects on viral RNA synthesis and virus titer, and no reversion to the wild-type sequence was observed when the mutant virus was passaged in cell culture. The apparent dispensability of the conserved X domain activity in vitro indicates that coronavirus replicase polyproteins have evolved to include nonessential functions. The biological significance of the novel enzymatic activity in vivo remains to be investigated.
Coronaviruses feature the largest genomes and the most complex genetic organization of all plus-strand RNA viruses. Their replicase genes encode an exceptionally large number of nonstructural protein domains, which mediate the key functions required for genomic RNA synthesis (replication) and subgenomic RNA synthesis (transcription) (29, 41, 47, 59). The replicase gene is comprised of two open reading frames (ORFs), ORF1a and ORF1b. ORF1a encodes the replicative polyprotein (pp) 1a, and ORFs 1a and 1b together encode pp1ab (59). Expression of the ORF1b-encoded part of pp1ab requires a −1 ribosomal frameshift during translation, which occurs just upstream of the ORF1a stop codon (5). The two polyproteins pp1a and pp1ab are extensively processed by virus-encoded proteases to produce up to 16 end products called nonstructural proteins (nsps) 1 to 16 and multiple processing intermediates (59, 62). While the N-proximal regions are processed at two or three sites by one or two papain-like protease(s) termed PL1pro and PL2pro (Fig. 1), the central and C-proximal regions of the polyproteins are processed at as many as 11 conserved sites by the viral main protease (Mpro) which, due to its distant relationship to the picornavirus 3C proteases, is also called 3C-like protease (3CLpro) (Fig. 1). Mpro (3CLpro) features a pronounced substrate specificity whose structural basis has recently been elucidated by X-ray crystallography (2, 3, 56, 57). Because of their key role in proteolytic processing and strong conservation of their active site structures and substrate specificities, the viral main proteases are currently being considered the most promising targets for anticoronaviral drug design (2, 3, 52, 56, 57, 60).
FIG. 1.
Coronavirus replicase gene-encoded X domains reside in nonstructural protein 3. (A) Structures of the 5′ capped and 3′ adenylated plus-strand RNA genomes of SARS-CoV, MHV-A59, and HCoV-229E are shown. Functional ORFs in the genome are expressed from the genomic RNA and an extensive set of subgenomic mRNAs. ORFs encoding the coronavirus structural proteins, that is, the spike (S), envelope (E), membrane (M), nucleocapsid (N), and (in MHV-A59) hemagglutinin-esterase (HE) proteins are indicated in gray. MHV-A59 ORF2a has been predicted to encode a cyclic phosphodiesterase activity that is not conserved in SARS-CoV and HCoV-229E (48). The 5′ terminal replicase gene, which is comprised of ORF1a and ORF1b, encodes the replicative polyproteins pp1a and, by −1 ribosomal frameshifting, a C-terminally extended version of pp1a, which is called pp1ab. The two polyproteins are cleaved by viral proteases to yield the 16 processing end products nsps 1 to 16. Cleavage sites processed by the papain-like proteases, PL1pro and PL2pro, are indicated by open triangles, and cleavage sites processed by the 3C-like main protease, 3CLpro, are indicated by filled triangles. Conserved coronavirus replicase gene-encoded domains are illustrated for the HCoV-229E pp1a and pp1ab (48, 59). Details on the nsp3 subdomain organizations of HCoV-229E and SARS-CoV, whose X domains were characterized in this study, are given below. Please note that SARS-CoV nsp3 lacks a counterpart of PL1pro and encodes an extra domain downstream of X, the SARS-CoV unique domain, which is not conserved in other coronaviruses. The HCoV-229E X domain single-residue substitutions characterized in this study are indicated. RdRp, RNA-dependent RNA polymerase; ExoN, putative 3′-to-5′ exoribonuclease; NendoU, nidoviral uridylate-specific endoribonuclease (23); MT, putative ribose-2′-O methyltransferase; Ac, acidic domain; Y, domain containing conserved Cys and His residues and stretches of hydrophobic residues presumed to anchor nsp3 to intracellular membranes (63). (B) Sequence comparison of the X domains of HCoV-229E and SARS-CoV with cellular homologs from A. fulgidus (AF1521) (1) and S. cerevisiae (YMX7) (27). The presented set was derived from an alignment of dozens of virus and cellular homologs that was generated using the ClustalX program (versions 1.64 and 1.8) (6) with manual adjustment and cross-verification with an alignment of coronavirus nsp3 proteins (63) (A. E. Gorbalenya and J. Ziebuhr, unpublished data). The secondary structure information for AF1521 was derived from the published crystal structure (1) (Protein Data Bank accession no. 1VHU) and, together with the alignment, was used as input for the ESPript program, version 2.2 (http://prodes.toulouse.inra.fr/ESPript/cgi-bin/ESPript.cgi). Sequences of the proteins were derived from the DDBJ/EMBL/GenBank database accession numbers O28751 (for AF1521), NP_013805 (for YMX7), NC_002645 (HCoV-229E X domain, pp1a/pp1ab residues Glu 1265 to Leu 1435), and AY291315 (SARS-CoV X domain, pp1a/pp1ab residues Glu 1000 to Asp 1170). The positions of HCoV-229E X domain residues that were substituted in this study are indicated by arrowheads (see also panel A).
In contrast to most of the nonstructural proteins, which are expressed from the genome RNA, the coronavirus structural proteins and a number of virus-specific accessory proteins are expressed from an extensive nested set of 3′ coterminal subgenome-length mRNAs that possess a common 5′ leader sequence derived from the 5′ end of the genome (49). The available evidence suggests that these leader-containing subgenome-length mRNAs are generated from subgenome-length minus-strand templates containing antileader sequences at their 3′ ends (39, 40, 64).
It appears that the unique features of coronavirus (nidovirus) RNA synthesis are linked to viral proteomes that involve enzymatic activities that are rare or absent in other RNA viruses (48). Most nidovirus replicases share a conserved array of functional domains that are ordered from the N to C terminus as follows: papain-like cysteine protease(s), two-β-barrel-fold (picornavirus 3C-like) main protease, RNA polymerase, zinc-binding domain-containing helicase, and endoribonuclease (15, 48, 59, 60). In large nidoviruses, such as corona-, toro-, and roniviruses, the conserved replicase backbone may be extended by additional enzymatic activities, including putative exoribonuclease and ribose-2′-O methyltransferase activities (48, 59) (Fig. 1). Other enzymatic activities, such as putative cyclic nucleotide phosphodiesterase (CPDase) and ADP-ribose-1"-monophosphate (Appr-1"-p) processing activities are only conserved in (some) members of the family Coronaviridae (48, 59) (see below).
As reverse genetics approaches for coronaviruses have become available only recently, the activities of the diverse nidovirus enzymes and, in particular, their precise functions in the viral life cycle are only slowly beginning to emerge (12, 23, 50). Previous work has mainly concentrated on the elucidation of the replicase polyprotein processing pathways as well as coronavirus proteases and helicases (2, 3, 22, 24, 25, 44, 61, 62). More recently, this work has been extended to other proteins, such as uridylate-specific replicative endoribonuclease activities, which are conserved in all nidoviruses and therefore are thought to play a key role in the life cycle of these viruses (4, 23). Furthermore, the functions in nidoviral replication and transcription of the complex Zn-binding domains associated with nidovirus helicases have been characterized (45, 54).
In this study, we addressed the activities of a coronavirus pp1a/pp1ab domain that is not conserved in arteri- and roniviruses (7, 48). The domain, which is known as “X domain,” resides in the coronavirus nsp3 and was previously predicted to be an Appr-1"-pase (also called ADRP) (48). Outside the coronaviruses, X domain homologs are conserved in only very few plus-strand RNA viruses, such as hepatitis E virus, rubella virus, and alphaviruses (16). For most of them, there is nearly no data available. By contrast, there are numerous cellular homologs and, at least for some of them, structural and functional information has recently become available (1, 27, 46). Thus, for example, crystal structure analysis of the Archaeoglobus fulgidus AF1521 and Saccharomyces cerevisiae YMX7 proteins revealed a mixed α/β fold, and YMX7 was proposed to employ a catalytic triad consisting of Asn-Asp-His (27). However, the fact that the proposed active-site residues are only partially conserved in other homologs indicates that more studies are needed to obtain conclusive evidence regarding the catalytic mechanism(s) employed by these ubiquitous proteins. In two studies (31, 46), the substrate of two different X domain homologs was confirmed to be Appr-1"-p, a downstream metabolite of cellular tRNA splicing, that is produced from ADP-ribose 1",2" cyclic phosphate (Appr>p) by the activity of a cyclic phosphodiesterase (CPDase) (8). Putative CPDase domains have also been identified in several group II coronaviruses, such as mouse hepatitis virus (MHV) and bovine coronavirus, as well as in the torovirus Berne virus. In MHV, the CPDase domain resides in the 2a protein which is expressed from a subgenomic mRNA (RNA 2.1), whereas the Berne virus homolog is encoded by the 3′ terminal region of ORF1a and, thus, expressed from the genome RNA.
To determine the properties of the conserved but largely uncharacterized coronavirus X domain and to get first insights into its potential functions in coronavirus replication, we expressed coronavirus X domains in Escherichia coli and characterized the proteins biochemically. The data provide convincing evidence that the human coronavirus 229E (HCoV-229E) and severe acute respiratory syndrome coronavirus (SARS-CoV) X domains are highly specific monophosphoesterases that act on Appr-1"-p. Furthermore, the characterization of mutant forms of the HCoV-229E X domain suggests that the pp1a/pp1ab residues, Asn 1302, Asn 1305, His 1310, Gly 1312, and Gly 1313, are part of the Appr-1"-pase active site and might be involved in catalysis or substrate binding. Finally, the study revealed that, in cultured cells, an Appr-1"-pase-deficient HCoV-229E mutant generated by reverse genetics had no detectable defect in viral RNA synthesis. The mutant grew to similar titers as the wild-type virus, and no reversion to the wild-type sequence was observed upon passaging of the mutant virus in cell culture. Together, the data argue for a regulatory rather than essential function for this enzyme in coronavirus replication. The relevant substrates and biological significance of the conserved activity remain to be identified.
MATERIALS AND METHODS
Cloning and expression of coronavirus X domains.
The coding sequences of the X domains of HCoV-229E and SARS-CoV, respectively, were amplified by reverse transcription-PCR (RT-PCR) using poly(A) RNA isolated from HCoV-229E-infected MRC-5 cells and SARS-CoV-infected Vero cells, respectively, as described previously (20, 52). Briefly, oligonucleotides JZ161 (for HCoV-229E) and S74 (for SARS-CoV), respectively, were used for reverse transcription of poly(A) RNA isolated from virus-infected cells. Subsequent PCRs were performed with the primer pairs JZ174/JZ105 (for HCoV-229E) and AP162/AP163 (for SARS-CoV) (Table 1). The reverse primers contained a translation stop codon and an EcoRI restriction site. Following treatment with T4 DNA polymerase, polynucleotide kinase, and digestion with EcoRI, the PCR products were ligated with XmnI/EcoRI-digested pMal-c2 plasmid DNA (800-64S; New England Biolabs, Schwalbach, Germany). The resulting plasmids, pMal-HCoV-X and pMal-SARS-CoV-X, encoded the respective X domains fused to the E. coli maltose-binding protein (MBP). The fusion proteins and their mutant derivatives (Table 2 and Fig. 1) were overexpressed at 18°C in E. coli TB1 cells, purified by amylose-affinity chromatography, and cleaved with factor Xa to separate the X domain from MBP using previously described protocols (18). The buffer used for protein purification and storage contained 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Virus | Sequence (5′ to 3′) | Nucleotidesa | Polarity | Use |
|---|---|---|---|---|---|
| JZ161 | HCoV-229E | TTATTCCACGCAGCATCAAGACCT | 5452-5475 | Reverse | Reverse transcription |
| JZ174 | HCoV-229E | GAATTTGTTGTTAAAGAGAAGTTG | 4085-4108 | Forward | PCR |
| JZ105 | HCoV-229E | tcccgaattcttaCACTAAACCAGACACAAAATCCTT | 4577-4600 | Reverse | PCR |
| S74 | SARS-CoV | GCTCTTGAGAGCATCTCAGTAGTG | 4116-4139 | Reverse | Reverse transcription |
| AP162 | SARS-CoV | GAAGAACCAGTTAATCAGTTTACT | 3262-3285 | Forward | PCR |
| AP163 | SARS-CoV | tttgaattcttaCTTCAGGTTATCAAGATAATCCAT | 3760-3783 | Reverse | PCR |
| AP153 | HCoV-229E | aaaggatcccgtctcaAGTGAATGTTCAAAAAGTTGAGCA | 4597-4620 | Forward | PCR |
| AP154 | HCoV-229E | tttgagctcCCCTTGTCACCTTTCATTAGTCTT | 5527-5550 | Reverse | PCR |
| AP155 | HCoV-229E | caagaattcCATGAAGGTGACTGGGATAGCTT | 3116-3138 | Forward | PCR |
| AP152 | HCoV-229E | tttctgcagcgtctctATTCTTCCTTAGGTGTTGTATCAA | 4065-4088 | Reverse | PCR |
| Oli147 | HCoV-229E | AAACCAGTCTGCTCATC | 3860-3876 | Forward | PCR |
| Oli48 | HCoV-229E | GATTACCAGAAGGTATAGC | 4853-4871 | Reverse | PCR |
| Oli180 | HCoV-229E | CATGCCACTGTGTTGTTTGA | 26857-26876 | Forward | PCR |
| 24.3 down | HCoV-229E | GATAGGTCTCAAGACAAATGACTG | 27212-27235 | Reverse | PCR |
TABLE 2.
Proteins and viruses characterized in this study
| Protein or virus | Substitution(s)a | Codon in:
|
|
|---|---|---|---|
| Wild type | Mutant | ||
| Proteinb | |||
| MBP-HCoV-X | None (wild type) | ||
| MBP-HCoV-X_N1293A | Asn1293→Ala | AAT | GCT |
| MBP-HCoV-X_N1302A | Asn1302→Ala | AAT | GCT |
| MBP-HCoV-X_N1305A | Asn1305→Ala | AAT | GCT |
| MBP-HCoV-X_N1302A/ | Asn1302→Ala | AAT | GCT |
| N1305A | Asn1305→Ala | AAT | GCT |
| MBP-HCoV-X_H1310L | His1310→Leu | CAT | CTT |
| MBP-HCoV-X_G1312A | Gly1312→Ala | GGA | GCA |
| MBP-HCoV-X_G1312V | Gly1312→Val | GGA | GTA |
| MBP-HCoV-X_G1313A | Gly1313→Ala | GGA | GCA |
| MBP-HCoV-X_G1313V | Gly1313→Val | GGA | GTA |
| MBP-SARS-CoV-X | None (wild-type) | ||
| Virusc | |||
| HCoV-229E | None (wild type) | ||
| HCoV_1ab-N1293A | Asn1293→Ala | AAU | GCU |
| HCoV_1ab-N1305A | Asn1305→Ala | AAU | GCU |
The numbering of HCoV-229E and SARS-CoV (isolate Frankfurt 1) pp1a/pplab residues was according to GenBank accession numbers X69721 (for HCoV-229E) and AY291315 (for SARS-CoV).
The HCoV-229E and SARS-CoV X domain sequences were expressed as fusions with the E. coli MBP.
HCoV-229E mutants were derived from the full-length cDNA clone, vHCoV-inf-1, as described in Materials and Methods.
Site-directed mutagenesis of the HCoV-229E X domain sequence.
Using pMal-HCoV-X plasmid DNA as a template, site-directed codon mutagenesis was performed by a PCR-recombination method (20, 58). A summary of all the nucleotide and amino acid substitutions characterized in this study is given in Table 2 and Fig. 1.
Appr-1"-pase activity assay and substrates.
Appr-1"-p was generated enzymatically from chemically synthesized Appr>p (17) using Arabidopsis thaliana cyclic nucleotide phosphodiesterase as previously described (14). Without further purification Appr-1"-p was then incubated with the coronavirus X domain or its mutant derivatives at 30°C for the indicated time periods (2, 3, or 5 h). Reaction mixtures (5 to 10 μl) contained 2 mM Appr-1"-p and 10 ng/μl X domain, which were incubated in buffer consisting of 35 mM Tris-HCl (pH 7.5), 0.005% Triton X-100, 0.5 mM EDTA, 100 mM NaCl, and 0.5 mM dithiothreitol. Control reactions contained 0.5 U of alkaline phosphatase isolated from calf intestine (Roche, Mannheim, Germany). Following incubation at 94°C for 2 min to terminate the enzyme reactions, the reaction products were analyzed by cellulose thin-layer chromatography (TLC) in solvent I [saturated (NH4)2SO4-3 M sodium acetate-isopropyl-alcohol (80:6:2)]. Alternatively, polyethyleneimine-cellulose plates were used with solvent II (0.75 M LiCl) and solvent III (0.15 M LiCl-0.15 M HCOOH, pH 3.0), respectively, as liquid phases. Reaction products were visualized under UV light. To test whether the coronavirus X domain acts on other substrates, the nucleoside phosphates ATP, ADP, 5′ AMP, 3′ AMP, and 2′ AMP (each at 10 mM) were incubated with 10 ng/μl HCoV-229E X domain in buffer containing 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol. As a positive control for phosphohydrolase activity, calf intestine phosphatase (0.05 U/μl) was used in these experiments. The samples were incubated at 30°C for 3 h, and the reaction products were analyzed by cellulose TLC in solvent I.
Mutagenesis of the full-length HCoV-229E cDNA clone, vHCoV-inf-1.
Nucleotides 4597 to 5550 of the HCoV-229E full-length cDNA were amplified by PCR using vHCoV-inf-1 (51) as a template and oligonucleotides AP153 and AP154 as primers. The PCR product was digested with BamHI and SacI and ligated with BamHI/SacI-digested pGPT-1 plasmid DNA (21). The resulting plasmid was digested with PstI and EcoRI and ligated with a PstI/EcoRI-digested PCR product generated from the vHCoV-inf-1 template using the primer pair AP155 and AP152. The resulting plasmid, pGPT-X, contains downstream of a vaccinia virus promoter the E. coli guanine phosphoribosyltransferase gene (gpt) that is flanked by HCoV-229E nucleotides 3116 to 4088 and 4597 to 5550, respectively. To produce vHCoV-ΔX_GPT (a vHCoV-inf-1 derivative in which the X domain coding sequence was replaced with gpt), the pGPT-X plasmid DNA was transfected into vHCoV-inf-1-infected CV-1 cells, and gpt-positive vaccinia virus recombinants were selected and isolated using previously established protocols (21). The correct insertion of the gpt gene at the desired locus in the ORF1a cDNA sequence by vaccinia virus-mediated homologous recombination was verified by Southern blotting and nucleotide sequence analysis. Next, the HCoV-229E ORF1a sequence 3860 to 4871, encoding the HCoV-229E X domain together with flanking sequences of about 250 nucleotides on either side, was amplified by PCR using vHCoV-inf-1 as a template and oligonucleotide primers oli147 and oli48. The PCR product was treated with T4 DNA polymerase and polynucleotide kinase and cloned into EcoRV-digested pBluescriptII KS+ plasmid DNA (Stratagene, Heidelberg, Germany). The resultant plasmid DNA, pBS-HCoV-3860-4871, served then as a template for site-directed mutagenesis (see above) to produce two mutant derivatives, pBS-HCoV-3860-4871_N1293A and pBS-HCoV-3860-4871_N1305A, in which the HCoV-229E replicase gene codons for N1293 and N1305 were replaced with Ala codons. The pBS-HCoV-3860-4871_N1293A and pBS-HCoV-3860-4871_N1305A plasmid DNAs were then used for a second round of vaccinia virus-mediated homologous recombination, in which gpt-negative derivatives of vHCoV-ΔX_GPT were selected on D980R cells (21). The desired replacement of the gpt coding sequence with the HCoV-229E sequences containing the pp1a/pp1ab substitutions N1293A and N1305A, respectively, was confirmed by Southern blotting and sequence analysis.
Production and characterization of the HCoV-229E mutants, HCoV_1ab-N1293A and HCoV_1ab-N1305A.
The genomic DNAs of the vaccinia virus recombinants, vHCoV-inf-1 (51), vHCoV_1ab-N1293A (this study), and vHCoV_1ab-N1305A (this study) were isolated from virus-infected BHK-21 cells using previously described methods (21). Following digestion with ClaI, the DNAs were used as templates for T7 runoff transcriptions to produce genome-length (wild-type and mutant) HCoV-229E RNAs. Following phenol-chloroform extraction and ethanol precipitation, 10 μg of each of these RNAs was electroporated into BHK-21 cells expressing the HCoV-229E nucleocapsid protein [BHK-21(N) cells] (41). Viral RNA synthesis was analyzed 3 days posttransfection by Northern blotting using poly(A) RNA that was isolated with Dynabeads oligo(dT)25 (Dynal Biotech, Hamburg, Germany) according to the manufacturer's instructions. As a probe, an HCoV-229E-specific 32P-labeled DNA probe was generated using the Megaprime DNA-labeling system (Amersham, Freiburg, Germany) and template DNA that was generated by PCR from vHCoV-inf-1 genomic DNA using the oligonucleotide primers oli180 and 24.3down (Table 1).
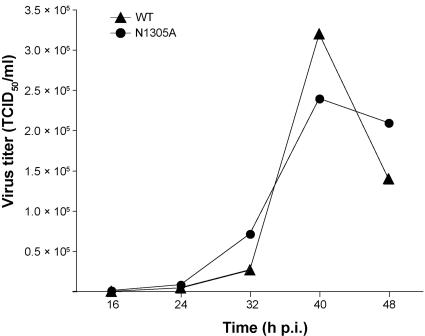
The tissue culture supernatant(s) from BHK-21(N) cells transfected with (mutant or wild-type) HCoV-229E full-length RNAs were collected at 72 h posttransfection and used to infect MRC-5 cells (CCL-171; American Type Culture Collection). The MRC-5 tissue culture supernatants (virus stocks) were collected after 72 h and (without plaque purification) passaged five times on MRC-5 cells (each passage for 72 h at 33°C in 5% CO2). Following determination of the virus titers by the method of Reed and Muench (38), the three virus stocks were then used to infect 6 × 105 MRC-5 cells at a multiplicity of infection (MOI) of 0.01 50% tissue culture infective dose (TCID50) per cell. At 72 h postinfection, poly(A) RNA was isolated and used for Northern blotting, RT-PCR, and nucleotide sequencing. To characterize the growth kinetics of the Appr-1"-pase-deficient HCoV-229E mutant, 6 × 105 MRC-5 cells were infected with HCoV_1ab-N1305A and HCoV-229E (wild type), respectively, at an MOI of 0.01, and virus titers (given as TCID50/ml) were determined at 16, 24, 32, 40, and 48 h postinfection as described previously (37).
RESULTS
The conserved coronavirus replicase gene-encoded X domain is a highly specific phosphatase that converts Appr-1"-p to Appr.
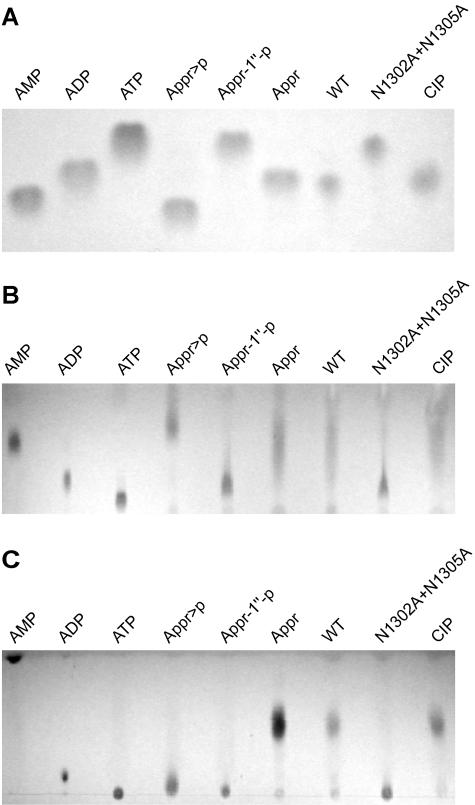
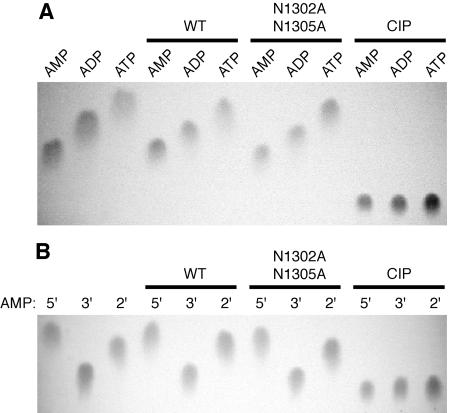
Recent data obtained for the yeast protein Poa1p (previously called YBR022w) suggested that viral X domains (and their cellular homologs) might be phosphohydrolases that specifically convert Appr-1"-p to Appr (31, 46, 48). To test this hypothesis, the HCoV-229E pp1a/pp1ab residues Glu 1265 to Val 1436 were expressed as a fusion with the E. coli MBP. The borders of the HCoV-229E X domain, which in all coronaviruses is part of nsp3 (Fig. 1A) (59), were selected on the basis of previous sequence alignments with other coronavirus X domains (63) and numerous cellular homologs including an archaeal protein, A. fulgidus AF1521, for which a crystal structure was available when this study was started (1) (Fig. 1B). As a (presumed) negative control in these experiments, two conserved Asn residues (pp1a/pp1ab residues 1302 and 1305) in the X domain sequence were replaced with Ala. Following purification of the MBP-X fusion proteins on amylose columns and subsequent release of the X domain from MBP by factor Xa cleavage, the proteins were incubated with Appr-1"-p, which was generated from chemically synthesized ADP-ribose 1",2" cyclic phosphate (Appr>p) using A. thaliana CPDase as described previously (14, 17). Analysis of the reaction products by TLC revealed that incubation with the wild-type HCoV-229E X domain but not with the negative control protein generated a reaction product that comigrated with ADP-ribose (Appr) (Fig. 2A). The same processing product was observed when the substrate was incubated with a nonspecific nucleoside phosphohydrolase, alkaline phosphatase from calf intestine (CIP). The fact that the reaction product consistently comigrated with Appr (one of our markers) using different chromatography media and liquid phases (Fig. 2) strongly suggests that the coronavirus X domain is a phosphatase that converts Appr-1"-p to Appr. To investigate the specificity of the coronavirus X domain in more detail and to answer the question of whether or not the enzyme also acts on other nucleoside (adenosine) phosphate derivatives, a range of adenosine phosphates were incubated with the HCoV-229E X domain and the two control proteins, HCoV-X_N1302A/N1305A and CIP, respectively. Analysis of the reaction products by TLC revealed that none of the substrates was processed by the coronavirus enzyme, whereas the positive control protein, CIP, dephosphorylated all these substrates, including 5′ AMP, 3′ AMP, 2′ AMP, ADP, and ATP (Fig. 3A and B). Furthermore, the HCoV-229E X domain had no detectable activity on α-NAD, β-NAD, α-NADP, and β-NADP (data not shown). Taken together, the data lead us to conclude that the HCoV-229E X domain (and probably other viral X domains) are highly specific for Appr-1"-p.
FIG. 2.
Bacterially expressed HCoV-229E X domain converts Appr-1"-p to Appr. (A) The HCoV-229E X domain (pp1a/pp1ab residues 1265 to 1436) was expressed and purified as an MBP fusion protein (see Materials and Methods) and subsequently cleaved with factor Xa. This protein and a mutant derivative carrying two substitutions of conserved Asn residues were incubated for 3 h at 30°C with Appr-1"-p that was generated from Appr>p using A. thaliana CPDase (see Materials and Methods). Lanes: WT, incubation with HCoV-229E X (wild-type sequence); N1302A+N1305A, incubation with a mutant form of HCoV-229E X in which the pp1a/pp1ab Asn residues 1302 and 1305 were each substituted with Ala; CIP, incubation with alkaline phosphatase isolated from calf intestine. Reaction products were separated on cellulose TLC plates using solvent I and visualized under UV light. Marker nucleotides: AMP, ADP, ATP, Appr, Appr-1"-p, and Appr>p. (B and C) The reactions described in panel A were analyzed on polyethyleneimine-cellulose TLC plates using solvent II (B) and III (C), respectively.
FIG. 3.
The phosphatase activity of the HCoV-229E X domain is highly specific for Appr-1"-p. (A) The adenosine phosphates, AMP, ADP, and ATP, respectively, were incubated for 3 h at 30°C with the following proteins: WT, HCoV-229E X (wild-type sequence); N1302A+N1305A, mutant form of HCoV-229E X in which the pp1a/pp1ab Asn residues 1302 and 1305 were each replaced by Ala; CIP, alkaline phosphatase isolated from calf intestine. (B) The adenosine monophosphates, 5′ AMP (5′),3′ AMP (3′), and 2′ AMP (2′), respectively, were incubated with HCoV-229E X (WT; wild-type sequence),a mutant form of HCoV-229E X (N1302A+N1305A), and CIP. In both experiments, the reaction products were separated on cellulose TLC plates using solvent I as liquid phase. Nucleotide markers: AMP, ADP, ATP, 5′ AMP, 3′ AMP, and 2′ AMP.
Mutagenesis data support predictions on potential active-site residues of the HCoV-229E X domain.
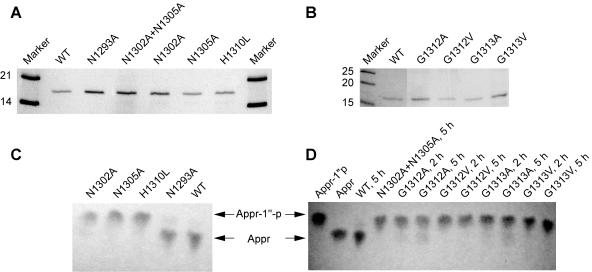
To gain insight into potential active-site residues of the coronavirus Appr-1"-pase, we replaced a series of HCoV-229E Appr-1"-pase residues whose equivalents in AF1521 are involved in the formation of a cleft that, in the published crystal structure (1), accommodated a 2-morpholinoethanesulfonic acid buffer molecule, suggesting that this solvent-exposed region on the molecule's surface may be part of the active site. The relevant AF1521 segment interacting with the 2-morpholinoethanesulfonic acid molecule included a His residue followed by a Gly-rich sequence that forms part of the loop connecting β-strand 3 and α-helix 1 as well as two Asn residues of β-strand 3. Despite the low degree of sequence conservation between the cellular and viral homologs shown in Fig. 1B, all these residues proved to be conserved in the HCoV-229E X domain (Asn 1302, Asn 1305, His 1310, and Gly residues 1311 to 1313), strongly supporting their critical role (Fig. 1B). To obtain further evidence for a potentially essential function of these residues, a total of 6 residues were replaced with (mainly) Ala residues (Fig. 1B and Table 2). These included two conserved Asn residues (1302 and 1305), His 1310, and two Gly residues (1312 and 1313). The purity and stability of the bacterially expressed proteins were confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 4A and B), and the Appr-1"-pase activities were determined using equal amounts of protein (Fig. 4C and D).
FIG. 4.
Activities of mutant forms of the HCoV-229E Appr-1"-pase. (A and B) Coomassie brilliant blue-stained sodium dodecyl sulfate-polyacrylamide gels showing the bacterially expressed HCoV-229E X domain (wild-type sequence) and the mutant forms characterized in this study (substitutions indicated) after purification and factor Xa-mediated release from the MBP fusion protein. (C and D) Appr-1"-pase activities of HCoV-229E Appr-1"-pases containing the indicated substitutions of conserved and nonconserved residues (Fig. 1 and Table 2). Incubation of the substrate Appr-1"-p with HCoV-229E Appr-1"-pase (wild-type sequence [WT]) served as a positive control, and for each of the reactions an equal amount of purified protein was used (for details, see Material and Methods). Reactions were performed at 30°C for 3 h and the reaction products were analyzed by cellulose TLC using solvent I as liquid phase. The positions of nucleotide markers (Appr-1"-p and Appr) are indicated by arrowheads.
The analysis of the AF1521 crystal structure (1) suggested to us that the above-mentioned conserved Gly residues of coronavirus (and other) Appr-1"-pases may contribute to substrate binding by contacts of their main chain atoms with the substrate. To test this idea, we replaced the Gly residues 1311 and 1312 with Ala (small side chain) and Val (larger and branched side chain). If the Gly residues were involved in the binding of the substrate, we expected to see a significant reduction of enzymatic activity, which should be more pronounced in the case of the Gly-to-Val than in the Gly-to-Ala substitutions. As Fig. 4D shows, the Gly substitutions resulted in strongly reduced (but still detectable) enzymatic activities. As predicted, the Gly-to-Val substitutions reduced the activities more strongly than the corresponding Gly-to-Ala substitutions, which became particularly evident after longer reaction times (Fig. 4D). The data support the idea that the Gly-rich segment of the loop connecting β-strand 3 and α-helix 1 (Fig. 1) is involved in substrate binding rather than playing an essential role in catalysis. By contrast, the Asn 1302, Asn 1305, and His 1310 residues proved to be essential for activity since their substitution abolished the enzymatic activities completely in our assay (Fig. 4C, N1302A, N1305A, and H1310L). As expected, the substitution of an upstream nonconserved Asn residue, Asn1293, that was located outside of the presumed active site and therefore used as a control in this experiment, had no significant effect on enzymatic activity.
Appr-1"-pase activity is dispensable for HCoV-229E replication in tissue culture.
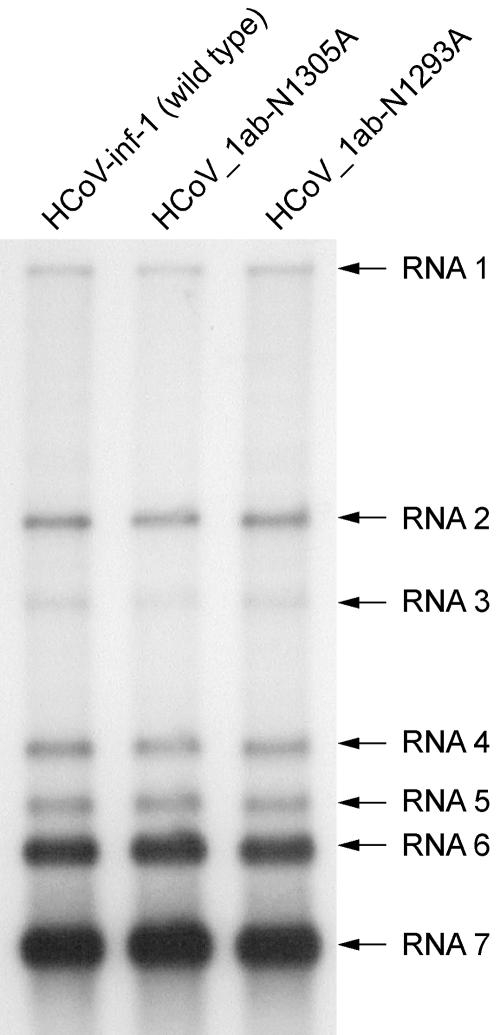
The conservation of the X domain in coronaviruses and several other plus-strand RNA viruses suggested an important function for this enzyme in the viral life cycle. To test whether the Appr-1"-pase activity is essential for coronavirus RNA synthesis and/or production of virus progeny, two single-point mutations were introduced into the X domain-coding sequence of the full-length HCoV-229E cDNA clone, replacing the Asn 1305 and Asn 1293 codons, respectively, with Ala. The N1305A substitution was chosen because this Asn belongs to the most conserved residues in viral X domains (and their cellular homologs) and because this replacement inactivated the HCoV-229E Appr-1"-pase activity in vitro (Fig. 4C). By contrast, the N1293A substitution had been shown to preserve the Appr-1"-pase activity (Fig. 4C), and, therefore, it was used as a negative control in subsequent experiments. In passage 0, that is, in BHK-21(N) cells transfected with each of the three full-length RNAs, we observed no difference in viral genomic and subgenomic RNA synthesis as determined by Northern blotting (data not shown). To produce virus stocks suitable for the determination of single-cycle growth curves, the cell culture supernatants collected from BHK-21(N) cells were used to infect human embryonic lung fibroblast (MRC-5) cells, which [in contrast to BHK-21(N) cells] are susceptible to infection by HCoV-229E. After five passages, the obtained high-titer stocks of HCoV-1ab-N1305A, HCoV-1ab-N1293A, and wild-type HCoV-229E were used to infect MRC-5 cells at an equal MOI. The subsequent characterization of viral RNA synthesis in MRC-5 cells at 72 h postinfection and the determination of virus titers revealed no obvious differences between the wild-type virus and the mutants (Fig. 5 and data not shown). To further corroborate this conclusion, we sought to characterize and compare the growth kinetics of the wild-type and Appr-1"-pase-deficient coronaviruses in more detail. To do this, we infected MRC-5 cells with HCoV-229E and HCoV-1ab-N1305A, respectively, at an equal MOI. and monitored the release of infectious virus progeny over time. As Fig. 6 shows, only very minor differences could be observed between the titers of the two viruses. To test the stability of the introduced mutations, we passaged the recombinant virus stocks six times in MRC-5 cells and subsequently amplified the X domain and flanking sequences by RT-PCR using poly(A) RNA isolated from virus-infected cells. The determination of the consensus sequence revealed that the introduced mutations were still present and that no additional changes had occurred in the X domain. We think that this lack of rapidly emerging revertants (or their failure to overgrow the mutant virus) argues against a superior fitness of the wild-type over an Appr-1"-pase-deficient virus in vitro. However, even though we did not observe differential growth kinetics of the mutant virus at early versus late passages, it remains to be formally excluded that compensatory mutations outside the X domain have emerged to restore the wild-type-like fitness observed for our X domain mutant. Taken together, the data lead us to suggest that the X domain-associated Appr-1"-pase activity is dispensable for HCoV-229E replication, transcription, and the production of virus progeny in tissue culture.
FIG. 5.
RNA synthesis of the HCoV-229E Appr-1"-pase mutants HCoV_1ab-N1293A and HCoV_1ab-N1305. Shown is a Northern blot analysis of viral RNA synthesis at 72 h postinfection using poly(A) RNA from 6 × 105 MRC-5 cells that had been infected with HCoV-229E (wild type [WT]), HCoV_1ab-N1293A, and HCoV_1ab-N1305A, respectively, at an MOI of 0.01 TCID50 per cell. The 32P-labeled probe used in this experiment was specific for the HCoV-229E 3′ terminal nucleotides 26857 to 27235.
FIG. 6.
Growth kinetics in MRC-5 cells of an HCoV-229E mutant with an Appr-1"-pase active-site substitution. Virus titers (given as mean values of two independent experiments) of wild-type HCoV-229E (WT) and HCoV_1ab-N1305A (N1305A) were determined at the indicated times points as described in Materials and Methods. p.i., postinfection.
Appr-1"-pase activity of the SARS-CoV X domain.
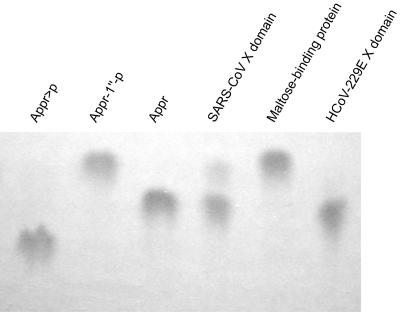
The N-terminal pp1a/pp1ab region represents the most divergent region in coronavirus replicase polyproteins (15, 59), and it has been noted previously that the subdomain organization of the SARS-CoV nsp3 differs from that of other coronaviruses in that it employs only one papain-like protease to process three cleavage sites (48, 52). On top of that, a domain called SARS-CoV unique domain with currently unknown functions that is not conserved in other coronaviruses was identified in this region (48) (Fig. 1). To address the question of whether, in this very divergent sequence context, the SARS-CoV X domain has retained its Appr-1"-pase activity, we expressed the SARS-CoV pp1a/pp1ab residues 1000 to 1173 as an E. coli MBP fusion protein using the protocols established for the expression of the HCoV-229E X domain homolog. The SARS-CoV protein was purified by amylose-affinity chromatography along with the negative and positive control proteins, MBP and HCoV-229E X domain, respectively. Following factor Xa cleavage, equal amounts of the three proteins were incubated with Appr-1"-p, and the reaction products were analyzed by TLC. As Fig. 7 shows, the bacterially expressed SARS-CoV X domain converted Appr-1"-p to a product that comigrated with the product of the HCoV 229E Appr-1"-pase reaction, which has been identified as Appr in the experiments described above. As expected, the negative control, MBP, had no Appr-1"-p processing activity. These data show that, despite a significantly divergent evolution of the coronavirus nsp3 homologs, the Appr-1"-pase activities have been preserved in HCoV-229E and SARS-CoV, suggesting that the activity is conserved in most (if not all) coronaviruses. At the present stage, the conservation of the X domain in coronaviruses is difficult to reconcile with the observed dispensability of the activity in HCoV-229E replication. One possibility to explain these somewhat contradictory results could be that the X domain activities might only be advantagous (or even essential) to the virus under specific (in vivo) conditions that remain to be investigated (see below).
FIG. 7.
Appr-1"-pase activity of the SARS-CoV X domain. The X domain of SARS-CoV (pp1a/pp1ab residues 1000 to 1173) was overexpressed as an E. coli MBP fusion protein, purified by amylose-affinity chromatography and released from MBP by factor Xa treatment. Following incubation of the Appr-1"-p substrate with the SARS-CoV X domain for 3 h at 30°C, the reaction products were separated by cellulose TLC using solvent I and visualized under UV light. As positive and negative controls, respectively, the HCoV-229E X domain and MBP were used. Appr-1"-p, Appr, and Appr>p were used as markers.
DISCUSSION
The study identifies a novel RNA virus phosphoesterase activity that is conserved in a limited number of plus-strand RNA viruses, including coronaviruses, hepatitis E virus, rubella virus, and animal alphaviruses (16, 48, 59). Interestingly, homologs of these viral enzymes exist in a variety of organisms, suggesting that they are involved in a very basic mechanism that is employed by all kingdoms of life (35). This protein family also includes the large C-terminal (non-histone) domain of macroH2A histones and, therefore, is often referred to as the macrodomain family. Despite their widespread occurrence, only very few of these proteins have been characterized to date, and their physiological role is far from being understood.
The data presented in this paper provide conclusive evidence that the nsp3-associated X domains of HCoV-229E, SARS-CoV, and probably other (corona)viruses are phosphoesterases that act to convert Appr-1"-p to Appr. The fact that no activity was detectable when a range of other adenosine phosphates were used as substrates suggests that coronavirus X domains are very specific for this particular substrate which, in eukaryotic cells, is produced as a downstream metabolite of tRNA splicing (8). Consistent with our data, it was recently reported that also Poa1p (previously called YBR022w), a macrodomain homolog from S. cerevisiae, converts Appr-1"-p to Appr in a very specific manner, and no activity on several other substrates could be detected (46). Despite its slow activity (kcat of 1.7 min−1), Poa1p was estimated to convert as much as 90% of the intracellular Appr-1"-p pool. The HCoV-229E Appr-1"-pase activity proved to have a higher (but still modest) kcat, which we estimated to be about 20 min−1 under the experimental conditions used in our assay. Since the HCoV-229E X domain is part of nsp3, a large multidomain protein of ∼177 kDa, which, in turn, is part of the multisubunit viral replicase, it remains to be studied whether (and to what extent) the presence of other nsp3 domains (or even other replicase subunits) will affect the activities of coronavirus X domains.
Information on the active site of macrodomain proteins is extremely limited and, to our knowledge, restricted to crystal structure information (1, 27). This lack of information is mainly due to the fact that there is no significant sequence similarity between macrodomain-related phosphatases and other (often quite well characterized) phosphatases. The AF1521 crystal structure, as well as previously made predictions on potential active-site residues derived from this structure (1), and our own sequence comparisons (Fig. 1 and data not shown) led us to predict that the HCoV-229E pp1a/pp1ab residues Asn 1302, Asn 1305, His 1310, Gly 1312, and Gly 1313 are part of the Appr-1"-pase active site, which was tested by experiments. As Fig. 4 shows, the data obtained for bacterially expressed mutant forms of the HCoV-229E X domain strongly support our predictions and suggest that the active site cleft has been localized correctly for the AF1521 and YMX7 homologs (1, 27). Specifically, our data are in agreement with the previous prediction that YMX7 Asn80 (which corresponds to Asn1305 in the HCoV-229E pp1a/pp1ab) has a catalytic function. With respect to the other two residues of the predicted Asn-Asp-His catalytic triad of YMX7, additional support would be highly desirable, in particular, as the Asp and His residues are only partly conserved in other macrodomain family members (including the coronavirus X domains).
To gain insight into possible functions of the Appr-1"-pase activity in coronavirus replication, we used the information derived from site-directed mutagenesis of the HCoV-229E X domain (see below) to generate an Appr-1"-pase-deficient HCoV-229E mutant. The characterization of this mutant, HCoV_1ab-N1305A, revealed no significant defect in viral RNA synthesis. Also, the production of infectious virus progeny and the growth kinetics of the mutant in cultured cells proved to be virtually indistinguishable from that of the wild-type virus. Furthermore, the fact that the introduced mutation remained stable for (at least) six passages suggests that, in cell culture, there is no strong selection pressure to restore the Appr-1"-pase activity. The data lead us to conclude that the Appr-1"-pase activity is dispensable for coronaviral replication under in vitro conditions. This finding is consistent with previously published S. cerevisiae data, demonstrating that POA1 deletion mutants had no obvious growth defects, although Appr-1"-p is produced in appreciable quantities in the cell and Poa1p was revealed to be the key enzyme in the processing of the intracellular Appr-1"-p pool. The yeast data were interpreted to suggest a regulatory (rather than essential) function for this enzyme (46). As previously noted for the yeast homolog and now corroborated for the coronavirus X domain, the presumed regulatory function gains further support by the observed high specificities of these Appr-1"-pases, which resemble those of typical regulatory phosphatases, such as fructose-2,6-bisphosphatase (36), and stands in clear contrast to alkaline, acidic, and even the more specific metabolic phosphatases which, in many cases, act on a wider range of substrates (19, 33, 34). Evidence for a regulatory rather than essential role of Appr-1"-p and its precursor, Appr>p, also came from an elegant study in yeast, showing that the functional replacement of the endogenous tRNA ligase with the T4 phage enzymes RNA ligase 1 and polynucleotide kinase/phosphatase had no effect on cell growth and viability although, under these conditions, no Appr>p and Appr-1"-p were produced (43). It is thus tempting to believe that Appr-1"-pases might act to regulate (or modulate) the concentrations and molecular ratios of several intracellular metabolites, such as Appr-1"-p, Appr, and poly(ADP-ribose), which under specific conditions might have consequences for the metabolism of the cell, including chromatin remodeling, DNA repair, genomic stability, and transcription (9, 10, 28, 53). It is also worth mentioning that several structurally related molecules including cyclic Appr and NAADP (nicotinic acid adenine dinucleotide phosphate) are key signaling molecules that regulate the mobilization of calcium ions in a variety of biological systems (13, 55), once again pointing to the regulatory functions of Appr and its derivatives.
The conditions under which the conserved Appr-1"-pase activities might provide a selective advantage in viral replication or favorably affect host cell functions to the benefit of the virus remain to be investigated by further experiments including in vivo studies. This also applies to the (putative) CPDases encoded by toroviruses and several group II coronaviruses (48) (Fig. 1). By analogy to previously characterized cellular CPDases (8, 14, 32), the nidovirus CPDases are thought to convert Appr>p to Appr-1"-p which, in turn, could be processed to Appr by the X domain-associated Appr-1"-pase. The observed dispensability of the HCoV-229E Appr-1"-pase activity in vitro provides an interesting link to previous MHV studies, which have shown that the CPDase residing in the 2a protein (Fig. 1) is also not essential for replication (42) and also that point mutations in the MHV-A59 CPDase sequence proved to be compatible with efficient viral replication in cell culture (50). The fact that the CPDase and Appr-1"-pase activities, mediating consecutive reactions of the same metabolic pathway, are (i) only partly conserved in coronaviruses (nidoviruses) (48) and (ii) dispensable for coronavirus replication in vitro leads us to suggest that these activities might provide a selective advantage only under very specific conditions. The idea that these nonessential activities might be linked to viral pathogenicity has received support from recent MHV data showing that a range of mutations in the ORF2a (CPDase) and replicase genes were well tolerated in vitro but proved to be attenuating in mice (50). Finally, it should be mentioned that Sindbis virus temperature-sensitive mutants with insertions and substitutions at (or very close to) the putative active site of the nsP3-associated X domain proved to replicate to high titers in tissue culture, even though some of them had partial defects in minus-strand RNA synthesis and nsP3 phosphorylation at the restrictive temperature (11, 30). Also these Sindbis virus data would argue for a regulatory rather than essential function of the X domain. Taken together, the available data seem to support our previous hypothesis (48) that the coronavirus Appr-1"-pase (and CPDase) activities might interfere with or modulate a yet-to-be-characterized regulatory pathway of the host, whereas a critical role for these enzymes in viral RNA synthesis seems less likely.
In conclusion, our study identified a coronavirus-wide conserved, highly specific phosphoesterase activity. The conservation in several plus-strand RNA viruses and many more cellular systems implicate this activity in a ubiquitous metabolic pathway whose biological significance is only slowly beginning to emerge (26). More experiments, including studies on the pathogenesis of genetically engineered Appr-1"-pase-deficient animal coronaviruses in their natural hosts and studies aimed at identifying possible effects of Appr-1"-pase activities on the metabolism of the host, will be required to get a detailed understanding of the biological role of this activity.
Acknowledgments
We gladly acknowledge the generous gift of the recombinant vaccinia virus vHCoV-inf-1 from Volker Thiel and Stuart G. Siddell.
The work was supported by grants from the Deutsche Forschungsgemeinschaft (EGK 587, ZI 618/2-3, and SFB 479-TPA) and the European Commission (SP22-CT-2004-511064).
REFERENCES
- 1.Allen, M. D., A. M. Buckle, S. C. Cordell, J. Lowe, and M. Bycroft. 2003. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 330:503-511. [DOI] [PubMed] [Google Scholar]
- 2.Anand, K., G. J. Palm, J. R. Mesters, S. G. Siddell, J. Ziebuhr, and R. Hilgenfeld. 2002. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 21:3213-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. [DOI] [PubMed] [Google Scholar]
- 4.Bhardwaj, K., L. Guarino, and C. C. Kao. 2004. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 78:12218-12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brierley, I., M. E. Boursnell, M. M. Binns, B. Bilimoria, V. C. Blok, T. D. Brown, and S. C. Inglis. 1987. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 6:3779-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowley, J. A., C. M. Dimmock, K. M. Spann, and P. J. Walker. 2000. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 81:1473-1484. [DOI] [PubMed] [Google Scholar]
- 8.Culver, G. M., S. A. Consaul, K. T. Tycowski, W. Filipowicz, and E. M. Phizicky. 1994. tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose 1",2"-cyclic phosphate. J. Biol. Chem. 269:24928-24934. [PubMed] [Google Scholar]
- 9.D'Amours, D., S. Desnoyers, I. D'Silva, and G. G. Poirier. 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342:249-268. [PMC free article] [PubMed] [Google Scholar]
- 10.Dantzer, F., V. Schreiber, C. Niedergang, C. Trucco, E. Flatter, G. De La Rubia, J. Oliver, V. Rolli, J. Menissier-de Murcia, and G. de Murcia. 1999. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie 81:69-75. [DOI] [PubMed] [Google Scholar]
- 11.Dé, I., C. Fata-Hartley, S. G. Sawicki, and D. L. Sawicki. 2003. Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus. J. Virol. 77:13106-13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denison, M. R., B. Yount, S. M. Brockway, R. L. Graham, A. C. Sims, X. Lu, and R. S. Baric. 2004. Cleavage between replicase proteins p28 and p65 of mouse hepatitis virus is not required for virus replication. J. Virol. 78:5957-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galione, A., and G. C. Churchill. 2002. Interactions between calcium release pathways: multiple messengers and multiple stores. Cell Calcium 32:343-354. [DOI] [PubMed] [Google Scholar]
- 14.Genschik, P., J. Hall, and W. Filipowicz. 1997. Cloning and characterization of the Arabidopsis cyclic phosphodiesterase which hydrolyzes ADP-ribose 1",2"-cyclic phosphate and nucleoside 2′,3′-cyclic phosphates. J. Biol. Chem. 272:13211-13219. [DOI] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E. 2001. Big nidovirus genome. When count and order of domains matter. Adv. Exp. Med. Biol. 494:1-17. [PubMed] [Google Scholar]
- 16.Gorbalenya, A. E., E. V. Koonin, and M. M. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, J., P. Genschik, and W. Filipowicz. 1996. Synthesis and characterization of the (5′→5")-ester of adenosine 5′-diphosphate with α-d-ribofuranose cyclic 1",2"-phosphate: a NAD derivative produced during tRNA splicing. Helv. Chim. Acta 79:1005-1010. [Google Scholar]
- 18.Hegyi, A., A. Friebe, A. E. Gorbalenya, and J. Ziebuhr. 2002. Mutational analysis of the active centre of coronavirus 3C-like proteases. J. Gen. Virol. 83:581-593. [DOI] [PubMed] [Google Scholar]
- 19.Heppel, L. A., D. R. Harkness, and R. J. Hilmoe. 1962. A study of the substrate specificity and other properties of the alkaline phosphatase of Escherichia coli. J. Biol. Chem. 237:841-846. [PubMed] [Google Scholar]
- 20.Herold, J., S. Siddell, and J. Ziebuhr. 1996. Characterization of coronavirus RNA polymerase gene products. Methods Enzymol. 275:68-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hertzig, T., E. Scandella, B. Schelle, J. Ziebuhr, S. G. Siddell, B. Ludewig, and V. Thiel. 2004. Rapid identification of coronavirus replicase inhibitors using a selectable replicon RNA. J. Gen. Virol. 85:1717-1725. [DOI] [PubMed] [Google Scholar]
- 22.Heusipp, G., U. Harms, S. G. Siddell, and J. Ziebuhr. 1997. Identification of an ATPase activity associated with a 71-kilodalton polypeptide encoded in gene 1 of the human coronavirus 229E. J. Virol. 71:5631-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov, K. A., T. Hertzig, M. Rozanov, S. Bayer, V. Thiel, A. E. Gorbalenya, and J. Ziebuhr. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA 101:12694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov, K. A., and J. Ziebuhr. 2004. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 78:7833-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karras, G. I., G. Kustatscher, H. R. Buhecha, M. D. Allen, C. Pugieux, F. Sait, M. Bycroft, and A. G. Ladurner. 2005. The macro domain is an ADP-ribose binding module. EMBO J. 24:1911-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumaran, D., S. Eswaramoorthy, F. W. Studier, and S. Swaminathan. 2005. Structure and mechanism of ADP-ribose-1"-monophosphatase (Appr-1"-pase), a ubiquitous cellular processing enzyme. Protein Sci. 14:719-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ladurner, A. G. 2003. Inactivating chromosomes: a macro domain that minimizes transcription. Mol. Cell. 12:1-3. [DOI] [PubMed] [Google Scholar]
- 29.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviridae: the viruses and their replication, p. 1163-1185. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 30.LaStarza, M. W., J. A. Lemm, and C. M. Rice. 1994. Genetic analysis of the nsP3 region of Sindbis virus: evidence for roles in minus-strand and subgenomic RNA synthesis. J. Virol. 68:5781-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martzen, M. R., S. M. McCraith, S. L. Spinelli, F. M. Torres, S. Fields, E. J. Grayhack, and E. M. Phizicky. 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 286:1153-1155. [DOI] [PubMed] [Google Scholar]
- 32.Nasr, F., and W. Filipowicz. 2000. Characterization of the Saccharomyces cerevisiae cyclic nucleotide phosphodiesterase involved in the metabolism of ADP-ribose 1",2"-cyclic phosphate. Nucleic Acids Res. 28:1676-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neumann, H. 1968. Substrate selectivity in the action of alkaline and acid phosphatases. J. Biol. Chem. 243:4671-4676. [PubMed] [Google Scholar]
- 34.Nigam, V. N., H. M. Davidson, and W. H. Fishman. 1959. Kinetics of hydrolysis of the orthophosphate monoesters of phenol, p-nitrophenol, and glycerol by human prostatic acid phosphatase. J. Biol. Chem. 234:1550-1554. [PubMed] [Google Scholar]
- 35.Pehrson, J. R., and R. N. Fuji. 1998. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 26:2837-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilkis, S. J., J. Pilkis, M. R. el-Maghrabi, and T. H. Claus. 1985. The sugar phosphate specificity of rat hepatic 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. J. Biol. Chem. 260:7551-7556. [PubMed] [Google Scholar]
- 37.Raabe, T., B. Schelle-Prinz, and S. G. Siddell. 1990. Nucleotide sequence of the gene encoding the spike glycoprotein of human coronavirus HCV 229E. J. Gen. Virol. 71:1065-1073. [DOI] [PubMed] [Google Scholar]
- 38.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 39.Sawicki, S. G., and D. L. Sawicki. 1995. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 380:499-506. [DOI] [PubMed] [Google Scholar]
- 40.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-219. [DOI] [PubMed] [Google Scholar]
- 41.Schelle, B., N. Karl, B. Ludewig, S. G. Siddell, and V. Thiel. 2005. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 79:6620-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarz, B., E. Routledge, and S. G. Siddell. 1990. Murine coronavirus nonstructural protein ns2 is not essential for virus replication in transformed cells. J. Virol. 64:4784-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwer, B., R. Sawaya, C. K. Ho, and S. Shuman. 2004. Portability and fidelity of RNA-repair systems. Proc. Natl. Acad. Sci. USA 101:2788-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seybert, A., A. Hegyi, S. G. Siddell, and J. Ziebuhr. 2000. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA 6:1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seybert, A., C. C. Posthuma, L. C. van Dinten, E. J. Snijder, A. E. Gorbalenya, and J. Ziebuhr. 2005. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J. Virol. 79:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shull, N. P., S. L. Spinelli, and E. M. Phizicky. 2005. A highly specific phosphatase that acts on ADP-ribose 1"-phosphate, a metabolite of tRNA splicing in Saccharomyces cerevisiae. Nucleic Acids Res. 33:650-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siddell, S. G., J. Ziebuhr, and E. J. Snijder. 2005. Coronaviruses, toroviruses, and arteriviruses, p. 823-856. In B. W. J. Mahy and V. ter Meulen (ed.), Topley and Wilson's microbiology and microbial infections, 10th ed., vol. 1. Hodder Arnold, London, United Kingdom. [Google Scholar]
- 48.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spaan, W., H. Delius, M. Skinner, J. Armstrong, P. Rottier, S. Smeekens, B. A. van der Zeijst, and S. G. Siddell. 1983. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 2:1839-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sperry, S. M., L. Kazi, R. L. Graham, R. S. Baric, S. R. Weiss, and M. R. Denison. 2005. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 79:3391-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82:1273-1281. [DOI] [PubMed] [Google Scholar]
- 52.Thiel, V., K. A. Ivanov, Á. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 53.Tulin, A., D. Stewart, and A. C. Spradling. 2002. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 16:2108-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Dinten, L. C., H. van Tol, A. E. Gorbalenya, and E. J. Snijder. 2000. The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. J. Virol. 74:5213-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamasaki, M., J. M. Thomas, G. C. Churchill, C. Garnham, A. M. Lewis, J. M. Cancela, S. Patel, and A. Galione. 2005. Role of NAADP and cADPR in the induction and maintenance of agonist-evoked Ca2+ spiking in mouse pancreatic acinar cells. Curr. Biol. 15:874-878. [DOI] [PubMed] [Google Scholar]
- 56.Yang, H., W. Xie, X. Xue, K. Yang, J. Ma, W. Liang, Q. Zhao, Z. Zhou, D. Pei, J. Ziebuhr, R. Hilgenfeld, K. Y. Yuen, L. Wong, G. Gao, S. Chen, Z. Chen, D. Ma, M. Bartlam, and Z. Rao. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biology, in press. [DOI] [PMC free article] [PubMed]
- 57.Yang, H., M. Yang, Y. Ding, Y. Liu, Z. Lou, Z. Zhou, L. Sun, L. Mo, S. Ye, H. Pang, G. F. Gao, K. Anand, M. Bartlam, R. Hilgenfeld, and Z. Rao. 2003. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA 100:13190-13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yao, Z., D. H. Jones, and C. Grose. 1992. Site-directed mutagenesis of herpesvirus glycoprotein phosphorylation sites by recombination polymerase chain reaction. PCR Methods Appl. 1:205-207. [DOI] [PubMed] [Google Scholar]
- 59.Ziebuhr, J. 2005. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 287:57-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ziebuhr, J. 2004. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 7:412-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ziebuhr, J., G. Heusipp, and S. G. Siddell. 1997. Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J. Virol. 71:3992-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
- 63.Ziebuhr, J., V. Thiel, and A. E. Gorbalenya. 2001. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 276:33220-33232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zúñiga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78:980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]