Abstract
Apaf-1 facilitates the proteolytic activation of procaspase-9 and maintains the hyperactive state of the processed caspase-9. The underlying molecular mechanisms for these activities remain poorly characterized. Here we report that the isolated Apaf-1 caspase recruitment domain (CARD) forms a large hetero-oligomer with the active caspase-9. The catalytic activity of caspase-9 is significantly enhanced in this complex, demonstrating that Apaf-1 CARD allosterically up-regulates caspase-9 activity. Point mutations that inactivate the interactions between Apaf-1 CARD and the prodomain of caspase-9 also abolished the formation of this complex. Based on these observations, we discuss the implications of this complex on the observed Apaf-1 function.
Apoptosis plays a central role in the development and homeostasis of metazoans (1–3). The Apaf-1/caspase-9 pathway mediates a variety of apoptotic stimuli including those initiated by the activation of tumor suppressor proteins and oncogenes. The central importance of this pathway is manifest by the observation that Apaf-1 is frequently inactivated in cancers such as malignant melanoma (4). In mice, Apaf-1 knockout leads to embryonic lethality (5, 6).
Apoptosis is executed by a cascade of caspase activation (7–9). The activation of an effector caspase, such as caspase-3, is performed by an initiator caspase, such as caspase-9, through proteolytic cleavage at specific Asp residues. Once activated, the effector caspases cleave numerous cellular targets, ultimately leading to cell death. Although the activation of effector caspases is structurally characterized (10), how the initiator caspases are activated and regulated remains poorly understood.
Recent biochemical studies have delineated the Apaf-1-mediated apoptosis in significant detail (11–13). During apoptosis, cytochrome c is released from mitochondria into the cytoplasm, a process controlled by the Bcl-2 family of proteins (12, 14). Once in the cytosol, cytochrome c induces the oligomerization of Apaf-1 in the presence of dATP or ATP, forming the so-called “apoptosome” (15–17), and then the apoptosome recruits and facilitates the proteolytic processing of procaspase-9. Unlike most other caspases, the processed caspase-9 remains associated with the apoptosome as a holo-enzyme to maintain its catalytic activity, as caspase-9 in isolation is only marginally active (17–20). In this respect, the apoptosome serves as an allosteric regulator for the enzymatic activity of caspase-9.
Apaf-1 contains three domains, an N-terminal caspase recruitment domain [CARD (21)], a central CED-4 homology domain, and 12–13 repeats of WD40 at the C-terminal half. The WD-40 repeats are thought to interact with cytochrome c whereas the CED-4 homology domain mediates the oligomerization of Apaf-1 in the presence of dATP.
Apaf-1 exhibits three distinct biochemical properties. The most extensively studied property is to recruit procaspase-9 through its CARD domain. Indeed, Apaf-1 CARD and the prodomain of caspase-9 form a heterodimer by using two complementary surfaces (22). The second property of Apaf-1 is to homo-oligomerize in the presence of cytochrome c and dATP, thus bringing procaspase-9 molecules into close proximity of one another and presumably facilitating their autoprocessing (15–17, 23–25). In support of this model, the unprocessed procaspase-9 exhibits a basal level of enzymatic activity (18). The third property of Apaf-1 is to maintain the catalytic activity of the processed caspase-9 in the holoenzyme because caspase-9 in isolation is marginally active (17–19).
Although the recognition of procaspase-9 by Apaf-1 is well characterized, how Apaf-1 mediates caspase-9 activation and helps maintain caspase-9 activity is unclear. In this manuscript, we report that the isolated Apaf-1 CARD forms a large hetero-oligomer (300–400 kDa) with the active caspase-9. Compared to the isolated caspase-9, the catalytic activity is significantly enhanced in this Apaf-1 CARD/caspase-9 complex. The formation of this complex is specific and depends on the CARD-prodomain interactions. These observations reveal mechanistic insights into the biochemical properties of Apaf-1.
Materials and Methods
Protein Preparation.
All constructs were generated by using a standard PCR-based cloning strategy. Recombinant Apaf-1 CARD (residues 1–97) was expressed and purified as described (22). Caspase-9 prodomain (1–132) and XIAP-BIR3 (238–358) were purified as described (22, 26). Caspase-9 and procaspase-3 (C163A) were expressed in Escherichia coli strain BL21(DE3) as C-terminally His-6-tagged proteins by using pET-21b (Novagen), purified over a Ni-NTA (Qiagen) column, and further fractionated by anion-exchange (Source-15Q) and gel filtration (Superdex-200).
Recombinant Apaf-1 protein was expressed in baculovirus-infected insect cells with a C-terminal His-6-tag. The soluble fraction was purified over a Ni-NTA column and further fractionated by anion-exchange chromatography. Horse heart cytochrome c was purchased from Sigma and purified by gel filtration.
Caspase-9 Assay.
The reaction was performed at 37°C in an assay buffer containing 20 mM HEPES (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, and 1 mM DTT. The substrate (procaspase-3, C163A) was at ≈35 μM. All components were diluted to specified concentrations by using the assay buffer. Reactions were stopped by using two times SDS-loading buffer. After SDS/PAGE, the results were visualized by Coomassie staining. The extent of procaspase-3 (C163A) cleavage was determined by densitometry using an EagleEye still video system (Stratagene).
Gel Filtration Analysis.
Gel filtration was performed by using a Superdex-200 column (10/30, Pharmacia) at 4°C. The column was calibrated with molecular weight standards. The buffer contains 10 mM HEPES (pH 7.5), 100 mM NaCl, and 2 mM DTT. To ensure complex formation, the individual components were incubated for 30 min before gel filtration.
Crystallization and Data Collection.
Crystals of caspase-9 prodomain were grown by the hanging-drop vapor-diffusion method by using a reservoir solution containing 1.5 M (NH4)2HPO4. The crystals are in the space group I23, with unit cell dimensions a = b = c = 86.4 Å, and contain one molecule per asymmetric unit. Crystals were equilibrated in a cryoprotectant buffer containing well buffer plus 20% glycerol, and were frozen in a −170°C nitrogen stream. The native data set was processed by using denzo and scalepack (27). The native data have a 4.5-fold overall redundancy, an Rsym of 0.067, and 97.3% completeness to 2.1 Å resolution.
Structure Determination.
The structure was determined by using the software amore (28), using the atomic coordinates of Apaf-1 CARD (PDB ID code 3ygs). The model was examined by using o (29), and the Apaf-1 CARD side chains were replaced with those of caspase-9 prodomain. Refinement by the program cns (30) resulted in 20.6% and 25.5% for the final R-factor and R-free, respectively. The final atomic model contains caspase-9 residues 1–99 and 39 water molecules at 2.1 Å resolution. The rms deviation for bonds and angles are 0.005 Å and 1.145°, respectively.
Results
Formation of a Heterodimer Between Apaf-1 CARD and Caspase-9 Prodomain.
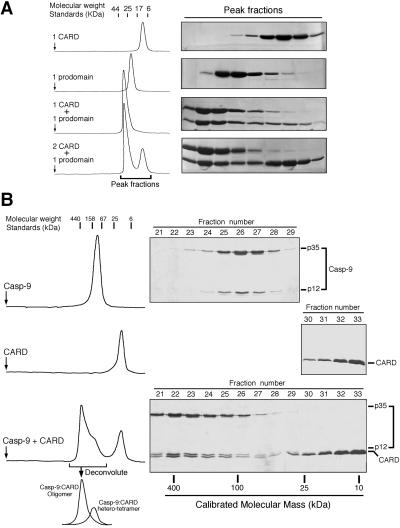
In the crystal structure, the isolated Apaf-1 CARD forms a heterodimer with the isolated prodomain of caspase-9 (22). To confirm this observation in solution, we devised an in vitro interaction assay, using size exclusion chromatography. The elution volume of Apaf-1 CARD corresponds to a monomer (11 kDa) (Fig. 1A). The elution volume of caspase-9 prodomain corresponds to ≈23 kDa (Fig. 1A), larger than that expected for a monomer (15 kDa). This difference is likely caused by the enlarged radius of hydration because of the C-terminal flexible sequence of the caspase-9 prodomain. When equimolar amounts of Apaf-1 CARD and caspase-9 prodomain were used, there was little protein at the elution volume for either of the individual components, indicating a 1:1 stoichiometry (Fig. 1A). In addition, the elution volume for the complex corresponds to an apparent molecular weight of 34 kDa, consistent with a 1:1 complex of Apaf-1 CARD and caspase-9 prodomain (Fig. 1A).
Figure 1.
Formation of the CARD/caspase-9 complex. (A) Apaf-1 CARD forms a heterodimer with the isolated prodomain of caspase-9. (B) Apaf-1 CARD oligomerizes the processed caspase-9. The elution volume of the primary caspase-9:CARD complex corresponds to an apparent molecular weight of 300–400 kDa (Bottom).
Apaf-1 CARD Oligomerizes Caspase-9.
Apaf-1 CARD is responsible for the recruitment of procaspase-9 (12, 22). After autoprocessing, the mature caspase-9 remains bound to the apoptosome, and the isolated caspase-9 is only marginally active (17). This observation indicates that caspase-9 must undergo some conformational change on binding to the apoptosome. One possibility is that Apaf-1 binding could induce a conformational change in the catalytic subunit of caspase-9. In this case, because CARD is the only domain of Apaf-1 known to interact with caspase-9, complex formation between the CARD and the prodomain likely creates a novel interface that facilitates interactions with the catalytic subunit of caspase-9, allosterically activating it.
To examine this scenario, we incubated Apaf-1 CARD with the processed caspase-9 and applied the complex to size exclusion chromatography (Fig. 1B Bottom). If the interactions were restricted to those between the CARD and the prodomain, the resulting complex would be a heterotetramer, comprising a caspase-9 homodimer and two bound CARD molecules. In this case, the complex is expected to display a combined molecular weight of ≈110 kDa, similar to that of the caspase-9 homodimer. Surprisingly, the elution volume of the primary complex corresponds to ≈300–400 kDa, far greater than that of the caspase-9 homodimer (≈90 kDa, Fig. 1B Top). Thus, Apaf-1 CARD induces the formation of a large multimeric complex involving caspase-9, hereafter referred to as the “CARD/caspase-9 complex.”
The formation of this CARD/caspase-9 complex is concentration dependent because the caspase-9:CARD heterotetramer (≈110 kDa) was also present in a separate peak despite excess amount of CARD (Fig. 1B Bottom). Higher concentrations of caspase-9 and excess Apaf-1 CARD drives the equilibrium toward the formation of the CARD/caspase-9 complex, as judged by the diminished peak of the caspase-9:CARD heterotetramer (data not shown).
To examine whether the formation of this CARD/caspase-9 complex is dependent on the interactions between Apaf-1 CARD and caspase-9 prodomain, we incubated caspase-9 with a mutant CARD (D27A), which does not interact with the prodomain (22). In this case, no complex formation was detected (data not shown), demonstrating that the interactions between Apaf-1 CARD and the prodomain of caspase-9 are a prerequisite for the formation of the CARD/caspase-9 complex.
Enhanced Caspase-9 Activity in the CARD/Caspase-9 Complex.
To further characterize this complex, we reconstituted an in vitro caspase-9 assay by using its physiological substrate procaspase-3. To ensure that the cleavage of this substrate is a direct readout of caspase-9 activity, we introduced a point mutation in the substrate, changing its catalytic residue Cys-163 to an Ala (C163A).
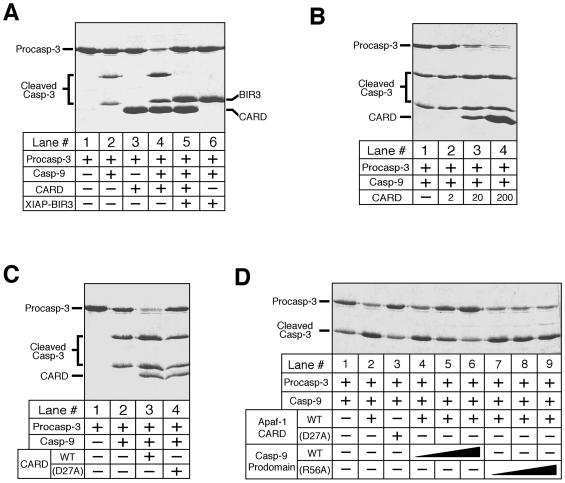
The isolated caspase-9 exhibits a basal level of activity, as judged by the cleavage of procaspase-3 precursor into the p20 and p12 subunits (Fig. 2A, lane 2). Interestingly, this activity is significantly enhanced in the presence of Apaf-1 CARD (lane 4) whereas CARD by itself does not have any enzymatic activity (lane 3). The addition of the inhibitory XIAP-BIR3 completely blocked the cleavage of procaspase-3 by caspase-9 (lanes 5 and 6).
Figure 2.
Caspase-9 exhibits significantly enhanced activity in the CARD/caspase-9 complex. The concentrations for caspase-9 and procaspase-3 (C163A) were 0.2 and 35 μM, respectively. (A) Increased caspase-9 activity in the presence of Apaf-1 CARD. The concentrations are: CARD ≈ 200 μM; XIAP-BIR3 ≈ 70 μM. (B) Dependence of enhanced caspase-9 activity on the concentrations of Apaf-1 CARD. CARD: 2, 20, and 200 μM for lanes 2, 3, and 4, respectively. (C) Dependence of enhanced caspase-9 activity on the CARD-prodomain interactions. CARD ≈ 20 μM; CARD (D27A) ≈ 20 μM. (D) Disruption of the CARD/caspase-9 complex by the wt prodomain of caspase-9. The concentrations for the wt (lanes 4–6) and R56A (lanes 7–9) prodomain are 10, 20, and 40 μM, respectively.
Because the formation of the CARD/caspase-9 complex is concentration dependent, the increased activity of caspase-9 is also expected to depend on the concentrations of the CARD. Indeed, increasing amounts of CARD progressively elevated the enzymatic activity of caspase-9 (Fig. 2B). To demonstrate that the elevated caspase-9 activity is due to the formation of the CARD/caspase-9 complex, we showed that a mutant CARD (D27A), which cannot form this complex, has no impact on the basal-level activity of caspase-9 (Fig. 2C).
Because the formation of the CARD/caspase-9 complex depends on the interactions between Apaf-1 CARD and caspase-9 prodomain, the prodomain by itself should antagonize the formation of this complex by competing with caspase-9 for binding to the CARD. Indeed, the wild-type (wt) prodomain is able to abolish the formation of this complex in a concentration-dependent manner (Fig. 2D, lanes 4–6). In contrast, a mutant prodomain (R56A), which does not interact with CARD (22), exhibited no effect (Fig. 2D, lanes 7–9).
The Prodomain of Caspase-9 Is Important for Its Activity.
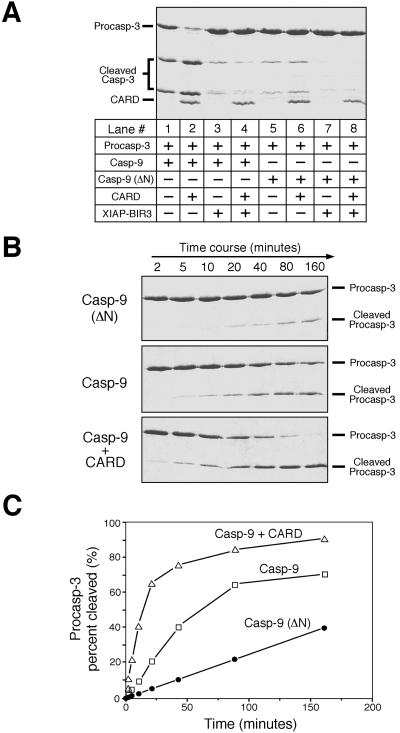
One possible explanation for the CARD-mediated caspase-9 activation is that the prodomain of caspase-9 may inhibit its catalytic activity and binding by CARD could relieve this inhibition. To examine this possibility, we generated a caspase-9 (residues 148–416, termed “caspase-9 (ΔN)”) with the prodomain removed. During over-expression in bacteria, this variant undergoes the same proteolytic activation as the wt caspase-9.
We first compared the basal-level activity of caspase-9 (ΔN) to that of the wt caspase-9 (Fig. 3A). Surprisingly, removal of the prodomain renders the resulting caspase-9 (ΔN) much less active (Fig. 3A, lanes 1 and 5), indicating that the prodomain is important for the maintenance of caspase-9 basal-level activity. In contrast to the wt caspase-9, caspase-9 (ΔN) does not interact with Apaf-1 CARD and can no longer form the CARD/caspase-9 complex; thus its activity remains the same regardless of CARD (lanes 5 and 6). Because the catalytic site is intact in both cases, XIAP-BIR3 still potently inhibits both the wt and the prodomain-deleted caspase-9 (lanes 3, 4, 7, and 8).
Figure 3.
The prodomain of caspase-9 is required to maintain its basal-level activity. The concentrations for caspase-9, caspase-9 (ΔN), and procaspase-3 (C163A) were 0.2, 0.2, and 35 μM, respectively. (A) Caspase-9 (ΔN) exhibits a decreased level of activity. CARD ≈ 20 μM; XIAP-BIR3 ≈ 2 μM. (B) A time course of procaspase-3 cleavage by caspase-9 (ΔN) and wt caspase-9 in the presence or absence of Apaf-1 CARD. CARD ≈ 20 μM. (C) Quantitation of substrate cleavage by caspase-9.
To quantitatively compare caspase-9 activity in the absence and presence of Apaf-1 CARD, we measured the percentage of cleaved substrates (Fig. 3 B and C). This analysis reveals that caspase-9 exhibits ≈5-fold higher activity in the presence of 20 μM CARD than in its absence, whereas caspase-9 (ΔN) exhibits ≈20% activity of the wt caspase-9. As formation of the CARD/caspase-9 complex is concentration dependent, higher concentrations of CARD further improved the catalytic activity of caspase-9 (data not shown).
Effect of Full-Length Apaf-1.
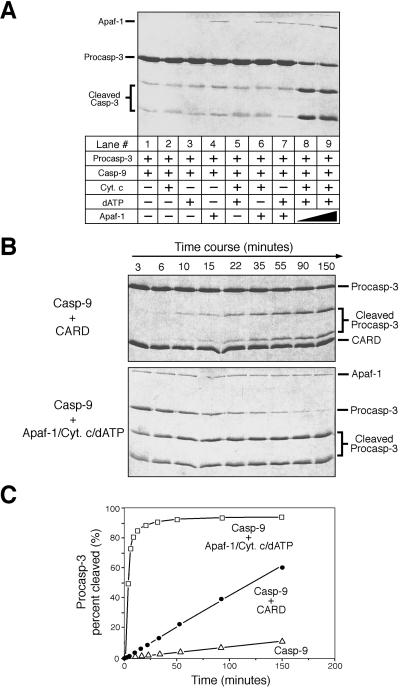
Our observations demonstrate that Apaf-1 CARD is sufficient for the maintenance of the hyperactive state of caspase-9 in a concentration-dependent manner. To compare the effect of Apaf-1 CARD to that of the full-length Apaf-1, we reconstituted another caspase-9 assay by using purified full-length Apaf-1, cytochrome c, dATP, and the physiological substrate procaspase-3 (C163A) (Fig. 4A). Caspase-9 by itself exhibits a basal-level activity (Fig. 4A, lane 1). This activity was not improved upon incubation with 10 μM cytochrome c (lane 2), 1 mM dATP (lane 3), 0.2 μM full-length Apaf-1 (lane 4), or any two of the three components (lanes 5, 6, and 7). In contrast, incubation with Apaf-1 in the presence of dATP and cytochrome c significantly enhanced caspase-9 activity (lanes 8 and 9). This result confirms that the holoenzyme comprises three proteins (Apaf-1, caspase-9, and cytochrome c) and one critical cofactor (dATP) and that the absence of any component abrogates the formation of the holoenzyme.
Figure 4.
Comparison of caspase-9 activity in the presence of Apaf-1 CARD or the full-length Apaf-1. The concentrations for caspase-9 and procaspase-3 (C163A) are 0.2 and 35 μM, respectively. (A) Assembly of an apoptosome holoenzyme by using recombinant proteins. Cytochrome c ≈ 10 μM; dATP ≈ 1 mM; Apaf-1 ≈ 0.2 and 0.6 μM for lanes 8 and 9, respectively. (B) A time course of caspase-9 activity in the CARD/caspase-9 complex or the holoenzyme. CARD ≈ 20 μM; Cytochrome c ≈ 10 μM; dATP ≈ 1 mM; Apaf-1 ≈ 0.6 μM. (C) Quantitation of substrate cleavage by caspase-9.
Next, we compared caspase-9 activity in the presence of the full-length Apaf-1 or the CARD (Fig. 4 B and C). Both promoted caspase-9 activity; but the full-length Apaf-1 appears to be much more efficient. Although this observation appears to argue that caspase-9 in the holoenzyme has much higher activity than that in the CARD/caspase-9 complex, it in fact does not and the discrepancy can be explained by the observed differences in association affinity.
The association affinity for the CARD/caspase-9 complex is relatively low. At ≈100 μM concentrations of the CARD and caspase-9, the formation of the CARD/caspase-9 complex was still incomplete (Fig. 1B). In the in vitro assays, the caspase-9 and CARD concentrations were 0.2 and 20 μM, respectively (Fig. 4 B and C). At these concentrations, the vast majority of caspase-9 (>98%) is estimated to remain unbound to this complex and thus exhibits only the basal-level activity. In the case of the full-length Apaf-1, the association affinity for the holoenzyme is very high. At ≈1 μM concentration each, free caspase-9 was undetectable. Presumably, this increased association affinity is due to the homo-oligomeric interactions of the CED-4 homology domain in Apaf-1.
A Model of the CARD/Caspase-9 Complex.
We demonstrated that the interactions between Apaf-1 CARD and the prodomain of caspase-9 are indispensable to the formation of the CARD/caspase-9 complex. Because the isolated CARD only forms a heterodimer with the isolated prodomain, an additional interface between the CARD and caspase-9 has to be involved to facilitate the formation of this multimeric complex.
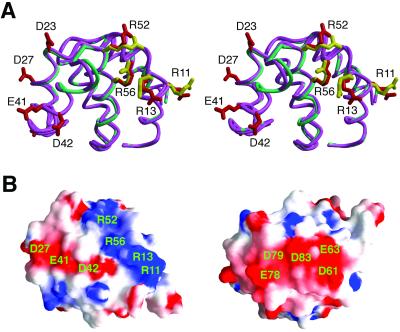
Where is this novel interface located on caspase-9? To help answer this question, we determined the crystal structure of caspase-9 prodomain by itself (Fig. 5A). Comparison analysis reveals that the structures of caspase-9 prodomain by itself and in complex with the CARD are nearly identical to each other, with 0.36 Å rms deviation for all 97 aligned Cα atoms (Fig. 5A). Similar to the CARD, the surface of the prodomain is enriched with charged residues, which form two acidic and one basic patches (Fig. 5B). In the heterodimer, one acidic surface of Apaf-1 CARD is recognized by a basic surface on the prodomain of caspase-9 (22). This arrangement leaves the other charged surfaces intact on the complex.
Figure 5.
Structure of the prodomain of caspase-9. (A) Stereo superposition of the structure of the prodomain in isolation (green) with that in complex with Apaf-1 CARD (pink). For the prodomain in isolation, some of the charged residues are highlighted in red. For the prodomain in complex with CARD, the basic residues involved in CARD recognition are colored yellow. (B) Surface representation of the caspase-9 prodomain highlighting three highly charged surface patches.
During characterization of the CARD/caspase-9 complex, we found that oligomerization of caspase-9 by the CARD is abolished at elevated ionic strength, such as 500 mM NaCl. Under this condition, caspase-9 exhibits only a basal level of activity despite the fact that caspase-9 retained formation of a 2:2 heterotetramer with the CARD. These observations suggest that the interactions responsible for caspase-9 oligomerization are predominantly electrostatic in nature. Thus it is likely that the other charged surfaces on the CARD:prodomain heterodimer are involved in oligomerizing caspase-9 through electrostatic interactions (Fig. 6). These interactions may in turn rearrange the active site of caspase-9 to better accommodate substrates, thus enhancing activity.
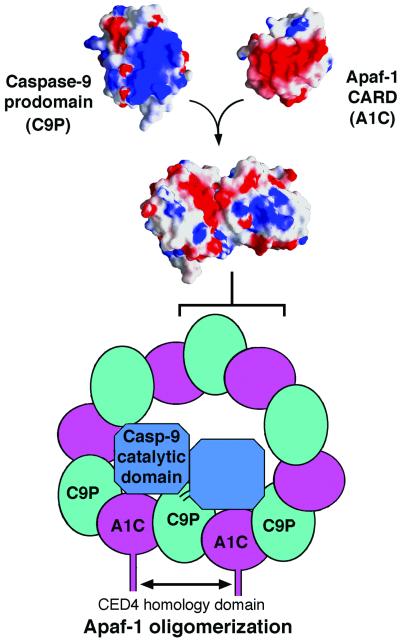
Figure 6.
A model of the CARD/caspase-9 complex. In this model, the building block is the CARD:prodomain heterodimer, which creates a novel surface onto which the catalytic subunit of caspase-9 docks. This docking stabilizes the interactions between two units of the CARD:prodomain heterodimer, which forms a donut-shaped structure. This docking also rearranges the active site of caspase-9 for higher activity. The CARD/caspase-9 complex may resemble the holoenzyme, in which oligomerization is further stabilized by the CED-4 homology domain of Apaf-1.
To support this model, we looked for missense mutations in the catalytic subunits of caspase-9 that would disrupt the CARD/caspase-9 complex but not a heterotetramer between caspase-9 and the CARD. Interestingly, we found that C287A mutation in the active site of caspase-9 completely disrupted this complex while retaining the formation of a heterotetramer with the CARD. In contrast, a number of other mutations in the vicinity of the active site (such as D315A or D293A/D315A) exhibited no impact on the formation of this complex. These observations demonstrate that the CARD/caspase-9 complex is formed through highly specific interactions. In the crystal structure of caspase-9 (31), Cys-287 interacts with surrounding residues through van der Waals contacts, which are likely important for the CARD-mediated oligomerization. The C287A mutation presumably disrupts these local interactions and prevents the CARD-mediated oligomerization of caspase-9.
Discussion
The processed caspase-9 and Apaf-1 CARD together form a ≈300–400-kDa concentration-dependent complex. Several lines of evidence indicate that this complex is highly specific and may recapitulate some aspects of the holoenzyme. First, the formation of this CARD/caspase-9 complex depends on the interaction between Apaf-1 CARD and caspase-9 prodomain. Mutations that inactivate a CARD-prodomain hetero-dimer also abrogated the formation of this complex. Second, caspase-9 exists in a hyperactive state in this complex. Although caspase-9 was only 5-fold more active in the presence of 20 μM CARD, higher levels of activity were recorded with higher concentrations of Apaf-1 CARD. When taking into account the differences in association affinities, the activity of caspase-9 in this complex is comparable to that in the holoenzyme. Third, we identified at least one missense mutation in the catalytic site of caspase-9 (C287A) that abolished formation of this complex but not the heterotetramer with Apaf-1 CARD.
If this CARD/caspase-9 complex indeed recapitulates the holoenzyme (with the omission of the bulk of Apaf-1 and cytochrome c), it can explain the observed biochemical properties of Apaf-1. For example, the CED-4 homology domain of Apaf-1 is known to form a homo-oligomer (23, 24). This activity is expected to greatly strengthen the CARD/caspase-9 complex by providing additional energy, explaining the higher association affinity for the holoenzyme. In the CARD/caspase-9 complex, the catalytic subunit of caspase-9 is likely to be in direct contact with the CARD/prodomain complex through electrostatic interactions, which rearrange the catalytic site of caspase-9 for higher activity.
Our observations also have significant implications for the activation process of procaspase-9. Because the bound procaspase-9 in the holoenzyme presumably rearranges its active site before autoprocessing, it is possible that the elevated enzymatic activity of the rearranged procaspase-9 is the true cause of proteolytic autoactivation, rather than that predicted by “induced proximity.”
If Apaf-1 CARD forms a large complex with caspase-9, then why is the full-length Apaf-1 by itself insufficient for this activity? (Fig. 4A, lane 4). The reason is that the full-length Apaf-1 by itself does not interact with the processed caspase-9. The association with cytochrome c and dATP changes the conformation of Apaf-1, enabling it to recruit caspase-9 (12, 19, 25, 32).
Using dATP-activated cell lysates, two different sizes (700 and 1,400 kDa) of the Apaf-1-containing apoptosome were recently identified and only the 700-kDa complex was shown to be active (33). These results are in contrast to other studies that revealed only one size for the apoptosome (≈1,400 kDa) by using purified recombinant proteins (15–17). It is unclear at present how to reconcile this discrepancy. In addition, the CARD/caspase-9 complex described here is not related to the reported “microaposome” that mainly contains active caspase-3 and -7 but little Apaf-1 or caspase-9 (34). More recently, Apaf-1 was reported to undergo proteolytic cleavage during apoptosis (35, 36). In one case, caspase-3 was found to cleave Apaf-1 in the CED-4 homology domain to generate a 30-kDa N-terminal fragment. Although this fragment is unable to promote caspase-9 maturation (35), it may be able to maintain the processed caspase-9 in a hyperactive state to promote apoptosis, much the same way as Apaf-1 CARD.
While this paper was in review, caspase-9 structure was reported (31). In this study, caspase-9 was found to exist in an equilibrium between monomers and dimers and only the dimeric form was active (31). Thus the activation of caspase-9 was attributed to dimer formation (31). Under our experimental conditions, we have only observed one form of caspase-9, which is active. This dimeric form of caspase-9 can be further activated by Apaf-1 or the CARD (Figs. 3 and 4). We do not yet know the precise stoichiometry for the constituents of the CARD/caspase-9 complex. The relatively low association affinity for this complex makes determination of its precise molecular weight difficult.
In summary, we demonstrate the formation of a multimeric complex between Apaf-1 CARD and caspase-9 in which the enzymatic activity of caspase-9 is significantly elevated. Although formation of the CARD/caspase-9 complex may lack physiological significance, our observations reveal important insights into the mechanism of caspase-9 activation by Apaf-1, a biologically significant process. The biochemical nature of this complex remains speculative. Nevertheless, these findings suggest a link between the observed CARD/caspase-9 complex and the holoenzyme.
Acknowledgments
We thank N. Hunt for administrative assistance. This research is supported by the National Institutes of Health (R01-CA90269). Y.S. is a Searle Scholar and a Rita Allen Foundation Scholar.
Abbreviations
- CARD
caspase recruitment domain
- Apaf-1
apoptotic protease-activating factor 1
- wt
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Steller H. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson M D, Weil M, Raff M C. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 3.Horvitz H R. Cancer Res. 1999;59:1701–1706. [PubMed] [Google Scholar]
- 4.Soengas M S, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, McCombie R, Herman J G, Gerald W L, Lazebnik Y A, et al. Nature (London) 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida H, Kong Y-Y, Yoshida R, Elia A J, Hakem A, Hakem R, Penninger J M, Mak T W. Cell. 1998;94:739–750. doi: 10.1016/s0092-8674(00)81733-x. [DOI] [PubMed] [Google Scholar]
- 6.Cecconi F, Alvarez-Bolado G, Meyer B I, Roth K A, Gruss P. Cell. 1998;94:727–737. doi: 10.1016/s0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]
- 7.Thornberry N A, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 8.Budihardjo I, Oliver H, Lutter M, Luo X, Wang X. Annu Rev Cell Dev Biol. 1999;15:269–290. doi: 10.1146/annurev.cellbio.15.1.269. [DOI] [PubMed] [Google Scholar]
- 9.Earnshaw W C, Martins L M, Kaufmann S H. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 10.Chai J, Wu Q, Shiozaki E, Srinivasula S M, Alnemri E S, Shi Y. Cell. 2001;107:399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 11.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 12.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y. Nat Struct Biol. 2001;8:394–401. doi: 10.1038/87548. [DOI] [PubMed] [Google Scholar]
- 14.Adams J M, Cory S. Trends Biochem Sci. 2001;26:61–66. doi: 10.1016/s0968-0004(00)01740-0. [DOI] [PubMed] [Google Scholar]
- 15.Zou H, Li Y, Liu X, Wang X. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 16.Saleh A, Srinivasula S M, Acharya S, Fishel R, Alnemri E S. J Biol Chem. 1999;274:17941–17945. doi: 10.1074/jbc.274.25.17941. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez J, Lazebnik Y. Genes Dev. 1999;13:3179–3184. doi: 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stennicke H R, Deveraux Q L, Humke E W, Reed J C, Dixit V M, Salvesen G S. J Biol Chem. 1999;274:8359–8362. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- 19.Jiang X, Wang X. J Biol Chem. 2000;275:31199–31203. doi: 10.1074/jbc.C000405200. [DOI] [PubMed] [Google Scholar]
- 20.Srinivasula S M, Saleh A, Hedge R, Datta P, Shiozaki E, Chai J, Robbins P D, Fernandes-Alnemri T, Shi Y, Alnemri E S. Nature (London) 2001;409:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann K, Bucher P. Trends Biochem Sci. 1997;22:155–156. doi: 10.1016/s0968-0004(97)01043-8. [DOI] [PubMed] [Google Scholar]
- 22.Qin H, Srinivasula S M, Wu G, Fernandes-Alnemri T, Alnemri E S, Shi Y. Nature (London) 1999;399:547–555. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- 23.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Alnemri E S. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 24.Hu Y, Ding L, Spencer D M, Nunez G. J Biol Chem. 1998;273:33489–33494. doi: 10.1074/jbc.273.50.33489. [DOI] [PubMed] [Google Scholar]
- 25.Hu Y, Benedict M A, Ding L, Nunez G. EMBO J. 1999;18:3586–3595. doi: 10.1093/emboj/18.13.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chai J, Shiozaki E, Srinivasula S M, Wu Q, Datta P, Alnemri E S, Shi Y. Cell. 2001;104:769–780. doi: 10.1016/s0092-8674(01)00272-0. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 29.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Brunger A T, Adams P D, Clore G M, Delano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Renatus M, Stennicke H R, Scott F L, Liddington R C, Salvesen G S. Proc Natl Acad Sci USA. 2001;98:14250–14255. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purring C, Zou H, Wang X, McLendon G L. J Am Chem Soc. 1999;121:7435–7436. [Google Scholar]
- 33.Cain K, Bratton S B, Langlais C, Walker G, Brown D G, Sun X-M, Cohen G M. J Biol Chem. 2000;275:6067–6070. doi: 10.1074/jbc.275.9.6067. [DOI] [PubMed] [Google Scholar]
- 34.Cain K, Brown D G, Langlais C, Cohen G M. J Biol Chem. 1999;274:22686–22692. doi: 10.1074/jbc.274.32.22686. [DOI] [PubMed] [Google Scholar]
- 35.Bratton S B, Walker G, Roberts D L, Cain K, Cohen G M. Cell Death Differ. 2001;8:425–433. doi: 10.1038/sj.cdd.4400834. [DOI] [PubMed] [Google Scholar]
- 36.Lauber K, Appel H A E, Schlosser S F, Gregor M, Schulze-Osthoff K, Wesselborg S. J Biol Chem. 2001;276:29772–29781. doi: 10.1074/jbc.M101524200. [DOI] [PubMed] [Google Scholar]