Abstract
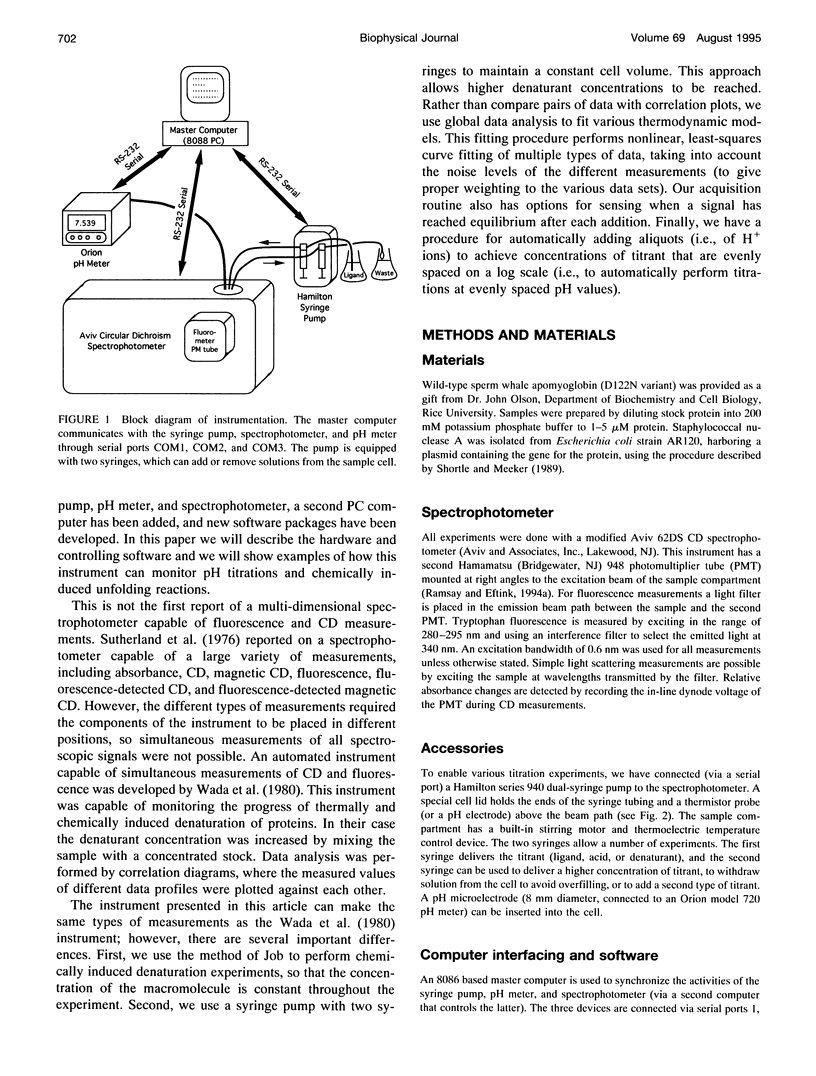
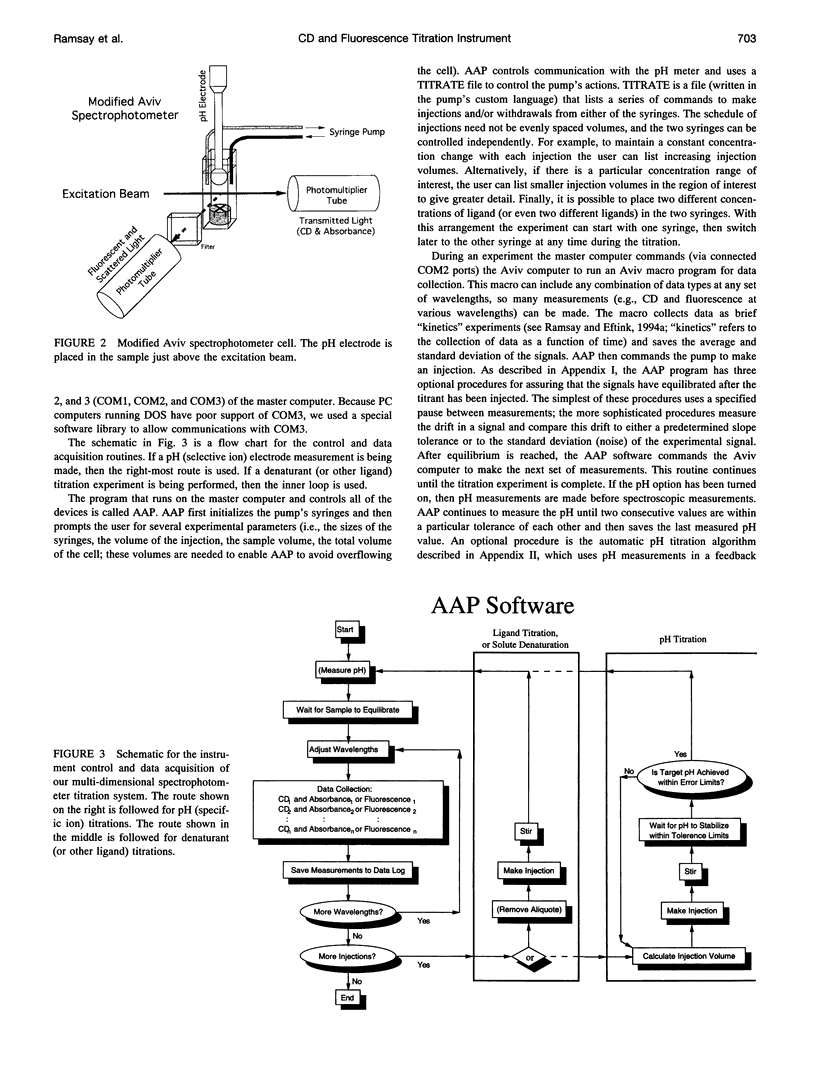
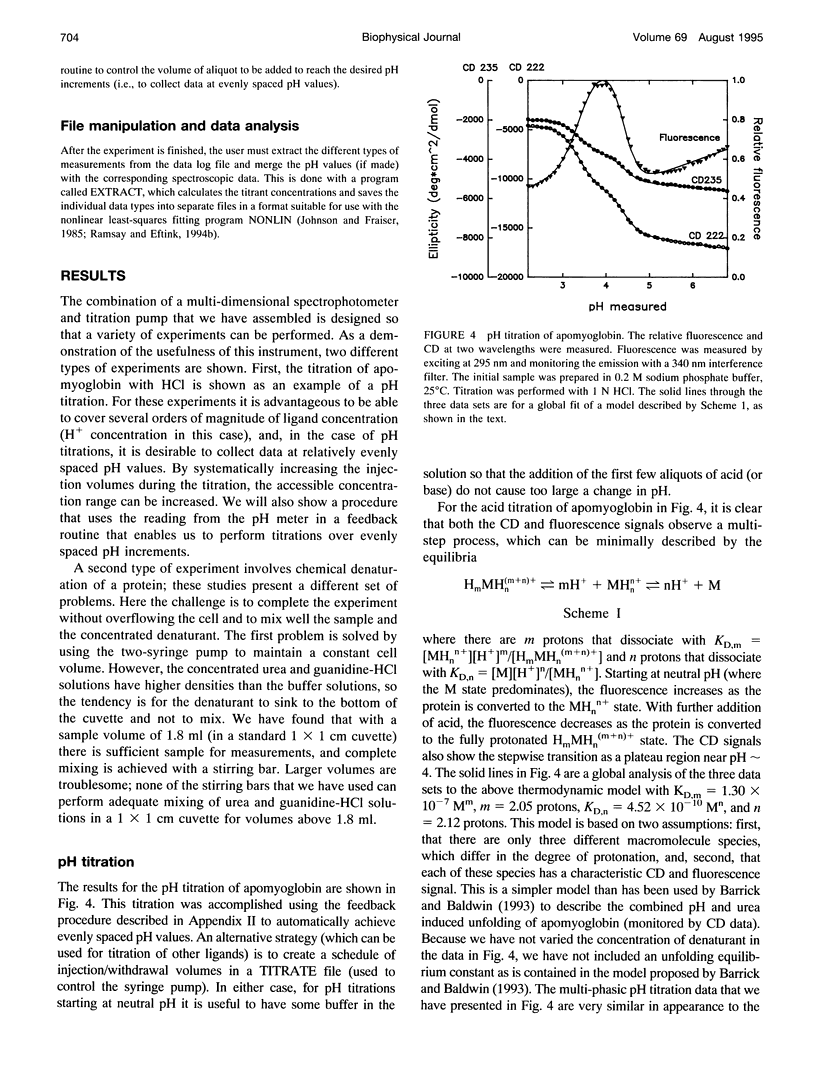
In a previous paper (Ramsay and Eftink, Biophys. J. 66:516-523) we reported the development of a modified spectrophotometer that can make nearly simultaneous circular dichroism (CD) and fluorescence measurements. This arrangement allows multiple data sets to be collected during a single experiment, resulting in a saving of time and material, and improved correlation between the different types of measurements. The usefulness of the instrument was shown by thermal melting experiments on several different protein systems. This CD/fluorometer spectrophotometer has been further modified by interfacing with a syringe pump and a pH meter. This arrangement allows ligand, pH, and chemical denaturation titration experiments to be performed while monitoring changes in the sample's CD, absorbance, fluorescence, and light scattering properties. Our data acquisition program also has an ability to check whether the signals have approached equilibrium before the data is recorded. For performing pH titrations we have developed a procedure which uses the signal from a pH meter in a feedback circuit in order to collect data at evenly spaced pH intervals. We demonstrate the use of this instrument with studies of the unfolding of sperm whale apomyoglobin, as induced by acid pH and by the addition of guanidine-HCI.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balestrieri C., Colonna G., Giovane A., Irace G., Servillo L. Equilibrium evidence of non-single step transition during guanidine unfolding of apomyoglobins. FEBS Lett. 1976 Jul 1;66(1):60–64. doi: 10.1016/0014-5793(76)80585-6. [DOI] [PubMed] [Google Scholar]
- Barrick D., Baldwin R. L. Three-state analysis of sperm whale apomyoglobin folding. Biochemistry. 1993 Apr 13;32(14):3790–3796. doi: 10.1021/bi00065a035. [DOI] [PubMed] [Google Scholar]
- Barrick D., Hughson F. M., Baldwin R. L. Molecular mechanisms of acid denaturation. The role of histidine residues in the partial unfolding of apomyoglobin. J Mol Biol. 1994 Apr 15;237(5):588–601. doi: 10.1006/jmbi.1994.1257. [DOI] [PubMed] [Google Scholar]
- De Young L. R., Dill K. A., Fink A. L. Aggregation and denaturation of apomyoglobin in aqueous urea solutions. Biochemistry. 1993 Apr 20;32(15):3877–3886. doi: 10.1021/bi00066a006. [DOI] [PubMed] [Google Scholar]
- Eftink M. R., Helton K. J., Beavers A., Ramsay G. D. The unfolding of trp aporepressor as a function of pH: evidence for an unfolding intermediate. Biochemistry. 1994 Aug 30;33(34):10220–10228. doi: 10.1021/bi00200a002. [DOI] [PubMed] [Google Scholar]
- Hargrove M. S., Krzywda S., Wilkinson A. J., Dou Y., Ikeda-Saito M., Olson J. S. Stability of myoglobin: a model for the folding of heme proteins. Biochemistry. 1994 Oct 4;33(39):11767–11775. doi: 10.1021/bi00205a012. [DOI] [PubMed] [Google Scholar]
- Irace G., Balestrieri C., Parlato G., Servillo L., Colonna G. Tryptophanyl fluorescence heterogeneity of apomyoglobins. Correlation with the presence of two distinct structural domains. Biochemistry. 1981 Feb 17;20(4):792–799. doi: 10.1021/bi00507a022. [DOI] [PubMed] [Google Scholar]
- Ramsay G. D., Eftink M. R. Analysis of multidimensional spectroscopic data to monitor unfolding of proteins. Methods Enzymol. 1994;240:615–645. doi: 10.1016/s0076-6879(94)40066-0. [DOI] [PubMed] [Google Scholar]
- Ramsay G., Eftink M. R. A multidimensional spectrophotometer for monitoring thermal unfolding transitions of macromolecules. Biophys J. 1994 Feb;66(2 Pt 1):516–523. doi: 10.1016/s0006-3495(94)80803-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Meeker A. K. Residual structure in large fragments of staphylococcal nuclease: effects of amino acid substitutions. Biochemistry. 1989 Feb 7;28(3):936–944. doi: 10.1021/bi00429a003. [DOI] [PubMed] [Google Scholar]
- Sugawara T., Kuwajima K., Sugai S. Folding of staphylococcal nuclease A studied by equilibrium and kinetic circular dichroism spectra. Biochemistry. 1991 Mar 12;30(10):2698–2706. doi: 10.1021/bi00224a018. [DOI] [PubMed] [Google Scholar]
- Sutherland J. C., Cimino G. D., Lowe J. T. Emission and polarization spectrometer for biophysical spectroscopy. Rev Sci Instrum. 1976 Mar;47(3):358–360. doi: 10.1063/1.1134624. [DOI] [PubMed] [Google Scholar]
- Wada A., Tachibana H., Hayashi H., Saito Y. Multidimensional spectroscopic data correlation in the conformation transition of biological macromolecules. J Biochem Biophys Methods. 1980 May;2(5):257–269. doi: 10.1016/0165-022x(80)90050-0. [DOI] [PubMed] [Google Scholar]