Abstract
Proteomic diversity is frequently achieved by alternative RNA-splicing events that can be fine-tuned in tissue-specific and developmentally regulated ways. Understanding this type of genetic regulation is compelling because of the extensive complexity of alternative splicing found in the nervous system. quaking (qk), one of the classical mouse dysmyelination mutants, is defective for the expression of myelin-associated glycoprotein (MAG), and the misregulation of MAG pre-mRNA alternative splicing is implicated as a causal factor. The qk locus encodes several RNA-binding proteins with heterogeneous nuclear ribonucleoprotein K-type homology, a characteristic of several known alternative splicing regulators. Here we test the nuclear-localized qk isoform (QKI-5) for its ability to regulate alternative splicing of MAG pre-mRNA in transient coexpression assays. QKI-5 exhibits properties of a negative regulator of MAG exon 12 alternative splicing. An intronic sequence element required for the repressive function and binding of QKI-5 is also identified. Direct evidence for irregularities in alternative splicing of MAG and other myelin protein transcripts in the qk mouse is demonstrated.
Alternative splicing is a powerful way to regulate gene expression at the posttranscriptional level and to generate protein diversity (1, 2). Understanding the molecular basis of such mechanisms is an urgent challenge, because the number of protein-coding genes in humans has been estimated at barely double the number in Drosophila and Caenorhabditis elegans. Between 35% and 59% of human genes are predicted to generate alternatively spliced mRNA isoforms (3).
Mouse quaking (qk), also known as quaking viable (qkv), is a classical recessive dysmyelination mutation (4). The qk locus produces a diverse set of proteins by alternative splicing (5, 6). They all contain a single heterogeneous nuclear ribonucleoprotein K homology (KH) RNA-binding domain and belong to the evolutionarily conserved signal transduction and activator of RNA (STAR) family (7, 8). The first three studied in detail (QKI-5, -6, and -7) are constructed with the same 311-aa body, but have different carboxyl tails. QKI-5 is the only nuclear isoform and shuttles between the nucleus and cytoplasm (9, 10). The expression of QKI isoforms is developmentally regulated, with QKI-5 being highly expressed throughout the embryogenesis and neonatal stages and decreasing gradually thereafter (7, 9). In postnatal day 14 (P14) mutant mice QKI proteins are decreased exclusively in myelin-forming cells. In addition, the QKI-5 expression level in qkv brain correlates with the severity of dysmyelinating phenotype, suggesting a function of QKI-5 in myelination (9).
The relationship between the decreased QKI protein in affected mice and their myelination defects is not understood. It has been shown that several myelin-specific genes are alternatively spliced, including myelin basic protein (MBP), proteolipid protein (PLP), and MAG (11–13). Some splicing events seem to be abnormal in qkv mice. The best-documented candidate target of QKI regulation is the MAG transcript. MAG protein is a myelin-specific transmembrane protein believed to be important for the initiation and maintenance of the myelin sheath (14). MAG pre-mRNA contains 13 exons, and exon 12 is alternatively spliced by inclusion or skipping (see Fig. 1). This alternative splicing is responsible for generating two MAG protein isoforms with distinct carboxyl termini because of the presence of an in-frame stop codon within exon 12. The long and short protein isoforms are designated L- and S-MAG, respectively (11, 15). At the RNA level, exon 12 is skipped in L-MAG and included in S-MAG message. L-MAG and S-MAG coexist in myelin-forming cells, but their ratio is developmentally regulated (11). L-MAG is the major isoform in young mice, whereas S-MAG is more abundant in adults. However, in qkv mice, L-MAG is scarcely expressed, but S-MAG is overexpressed (16). This alteration is thought to be one of the causes of dysmyelination in qkv. Because exon 12 inclusion is increased when a relatively low level of QKI exists, we hypothesized that the nuclear isoform QKI-5 regulates the alternative splicing of MAG by repressing the inclusion of exon 12.
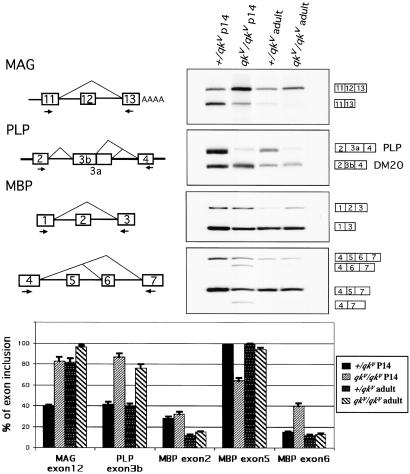
Figure 1.
Abnormal myelin gene alternative splicing in the qkv mutant. (Upper Right) RT-PCR of MAG, PLP, and MBP messages from normal (+/qkv) and mutant (qkv/qkv) brain RNA from P14 and adult mice. (Upper Left) Diagram of MAG, PLP, and MBP alternative splicing pattern. The arrows represent the primers. The content of each PCR product is indicated next to the band. (Lower) Graphical comparison of the percentage of exon selection.
Besides MAG, MBP and PLP are also possible targets of QKI. MBP, a major myelin component, produces at least seven isoforms by including or skipping exons 2, 5, or 6. Point mutations and misregulation of the plp gene cause dysmyelination in both mice and humans (17, 18). In the plp gene, two splicing products named PLP and DM20 are produced by choosing different 5′ splice sites of exon 3 (12). Although several studies of MBP and PLP expression in qkv mice have been reported, these experiments characterized the total gene expression, but not that of individual isoforms (19, 20). Thus, it is still not clear whether their alternative splicing is disturbed in the mutant.
Here we investigate the role of the nuclear isoform of QKI, QKI-5, in alternative splicing regulation of MAG pre-mRNA. Using a MAG minigene in transfected cells, we show that overexpression of QKI-5 represses the inclusion of exon 12 in a dosage-dependent manner. We identify the QKI-5 alternative splicing element (QASE) as a 53-nt region in the downstream intron, which is necessary for QKI-5 interaction and regulation. We also address the possibility that QKI-5 regulates alternative splicing of other myelin targets.
Materials and Methods
Plasmid Constructions.
The mammalian expression constructs pcDNA3-QKI-5 and QKI-5ΔKH were generated by inserting the coding region of QKI-5 and a mutation in vector pcDNA3.1/hygro (Invitrogen). To generate MAG minigene constructs, an exon-trapping vector pSPL3 from Life Technologies (Grand Island, NY) was used. A piece of MAG genomic DNA including exon 12 and part of exon 13 was obtained by PCR with primers me11 and me13. The 1.1-kb EcoRI fragment was cloned into pSPL3 (pMAG). pMAGPY was generated from pMAG with its polypyrimidine tract changed from CGTTTCCTTCTTCAAT to ATGTTTCATGCCTCCCTGC by using the QuickChange site-directed mutagenesis kit (Stratagene). pMAG and pMAGPY were then used as templates for other deletions and PCR-based mutagenesis. pMAG-5′SS was made by mutating the 5′ splice site of exon 12 from GTAGGT to GTAAGT. The gel-shift probe templates E12QASE, ΔQASE, and EP160 were obtained by PCR and cloned in PCR4-TOPO vector (Invitrogen) and pBluescript KS(+) separately. The wild-type N-methyl-D-aspartate (NMDA) exon 21 splicing reporter (NMDA E21) contains the complete genomic sequence of the NMDA R1 receptor from exon 20 to exon 22. The DNA fragment was generated from rat genomic DNA by PCR with primers 3021-RI and 3255A-RI and inserted into the EcoRI site of the mammalian expression vector pBPSVPA+. Plasmid rGγ25 was generated as described (21). The sequences of all primers are available upon request.
Mice and RNA Preparation.
Age-matched qkv/qkv and +/qkv mice from our colony were maintained as described (7). Total RNA from P14 mice and the brains of 2-month-old littermates were isolated by the LiCl/urea method (22).
Cell Culture and Transfection.
COS7 or C2C12 cells were grown in DMEM supplemented with 10% FBS. Cells grown in 12-well plates (Corning) were transfected with 1 μg of plasmid DNA total with Fugene6 reagent (Roche Molecular Biochemicals) and harvested 24 h after transfection. The RNA was extracted with Trizol reagent (Life Technologies). Western blotting was performed as described (10).
Detection of RNA.
MAG isoforms in brain and transfected cells were analyzed by reverse transcription (RT)-PCR. RNA (2 μg) was reverse transcribed with Superscript II (Lifetech). me13 or SA2 was used as RT primer. The RT reaction (20%) was used for a 25-μl PCR. The 5′ primer me11 or SD6 was end-labeled with [γ-32P]ATP and a 0.5 μM concentration of each primer was used. PCR conditions were: 94°C for 1 min 30 s; followed by 18 cycles at 94°C for 30 s, at 55°C for 45 s, and at 72°C for 1 min; finally, at 72°C for 5 min. The PCR products (10 μl) were run on a 5% polyacrylamide gel and analyzed with a PhosphorImager. The percentage of exon 12 inclusion is the average value of at least three assays. The primers used for PLP/DM20 RT-PCR are PLP2 and PLP4. MBPe1 and MBPe3 were used for MBP exon 2 RT-PCR, whereas MBPe4 and MBPe7 were used to examine exon 5 and 6 splicing. Oligo(dT) was used as an RT primer for MBP and PLP. Transfected γ-aminobutyric acid (GABA) receptor mRNA products were detected by RT-PCR with primers rP1 and rP2. NMDA receptor mRNAs were detected with primers 3021 and 3255A.
Gel Mobility-Shift and UV Crosslinking Assays.
32P-labeled RNA probes E12QASE and ΔQASE were in vitro transcribed in the presence of [α-32P]UTP. HeLa cell nuclear extract was prepared as described (23). RNA (105 cpm) was incubated with 1 μl of HeLa nuclear extract in 25 μl of 1/2 D buffer (20 mM Hepes, pH 7.6/20% glycerol/100 mM KCl/2 mM EDTA/0.5 mM DTT), 0.5 mg/ml tRNA, 0.025% Nonidet P-40, 0.8 unit of RNase inhibitor/μl (Promega), 1 mM DTT at 30°C for 30 min. Heparin (1 μl of 25 mg/ml) was added to the mixture followed by an additional 5-min incubation. After 3 μl of glycerol had been added, 10 μl of this final mixture was loaded onto a 3.75% prechilled polyacrylamide gel and electrophoresed at 200 V for 2 h at 4°C. Complexes were visualized by autoradiography. In the competition assay, RNA competitors were preincubated with proteins for 5 min before the addition of labeled RNA probe. In the antibody supershift assay, the indicated amount of anti-QKI-5 antiserum or preimmune serum was added after heparin treatment, followed by another 5-min incubation before adding the glycerol.
UV crosslinking experiments were performed under the same conditions as the gel mobility-shift assay but without RNase inhibitor. E12QASE probe (105 cpm) and 2 μl of HeLa nuclear extract were used. After the 30°C incubation, the reactions were irradiated with 254-nm UV light for 30 min. RNase A (0.1 unit) and RNase T1 (5 units) (Ambion, Austin, TX) were added to the reactions, followed by a 30-min incubation at 37°C. The digested mixtures were resolved on an SDS/10% polyacrylamide gel. In vitro translation of QKI-5 was accomplished with TNT coupled reticulocyte lysate (Promega).
Results
Alternative splicing of several myelin genes is misregulated in qkv mice. In qkv mice, the ratio of the MAG isoforms seems to be abnormal, as judged by Northern blot analysis (16). The normal pattern of MAG alternative splicing involves exon 12 inclusion or skipping (Fig. 1). To test for splicing abnormalities of exon 12 in qkv mice, an RT-PCR assay was performed with primers specific for MAG exons 11 and 13 (Fig. 1). In this experiment, the 5′ primer was end-labeled with 32P, and 18 cycles of PCR amplification were used. We have observed that the ratio of the spliced mRNA isoforms with and without exon 12 is stable when the PCR cycles are varied between 16 and 20 (data not shown). These results confirm that the exon 12-skipped mRNA isoform of MAG predominates in normal P14 brain when QKI-5 expression is at a high level, whereas the exon 12-included mRNA predominates in qkv brain, when QKI-5 is much reduced (Fig. 1, MAG inset, left lanes). In adult mice, the abnormal pattern in exon 12 alternative splicing becomes less pronounced, as expected from a reduction in QKI-5 protein (right lanes). Thus, these results show a correlation between reduced QKI-5 levels and an increase in MAG exon 12 inclusion, implicating QKI-5 as a negative regulator of exon 12 during development.
The abnormality of MAG alternative splicing is not likely the sole cause of the dysmyelination phenotype, because MAG knockout mice have a much less striking phenotype than qkv (24, 25). Notably, several other myelin genes are also alternatively spliced (12, 13). Among them, PLP and MBP represent possible targets of QKI-5. In PLP, exon 3 has two possible 5′ splice sites. In MBP, exons 2, 5, and 6 are either included or skipped. To investigate the splicing patterns of PLP and MBP in qkv mice, RT-PCR experiments were performed (Fig. 1). By using primers within the flanking exons, all of the possible splicing isoforms were obtained. A comparison of exon inclusion between normal and qkv homozygous brain RNAs indicates that at P14, the alternative splicing patterns of some exons are aberrant in qkv. In PLP, the usage of 5′ splice site of exon 3b is substantially increased. In MBP, relatively more exon 6 and less exon 5 are included. However, little or no effect on MBP exon 2 is seen. In adults, the effect is generally weaker. Thus, PLP and MBP may also be targets of QKI.
QKI-5 Regulates MAG Exon 12 Splicing in a Dosage-Dependent Manner.
To investigate the functional effect of QKI-5 on MAG alternative splicing, a splicing reporter was generated with exon 12 and flanking sequences (pMAG; Fig. 2A). The mRNA products generated from pMAG were detected by RT-PCR after transfection into COS7 cells (Fig. 2B). Two resulting PCR fragments of 241 bp and 286 bp were observed. Sequencing confirmed that they represent the alternatively spliced MAG mRNAs, including or skipping exon 12. Although exon 12 was alternatively spliced in this system, its inclusion is weak, and negative regulation might prove difficult to detect (Fig. 2B, lanes 1 and 2). Thus, we exchanged its polypyrimidine tract for the one upstream of exon 11, generating pMAGPY plasmid (Fig. 2A). With this construct, the percentage of exon 12 inclusion increased to 63% (Fig. 2B, lane 3). This system was then amenable to study the sequence requirement for exon 12 inclusion and the effect of QKI-5 on the splicing.
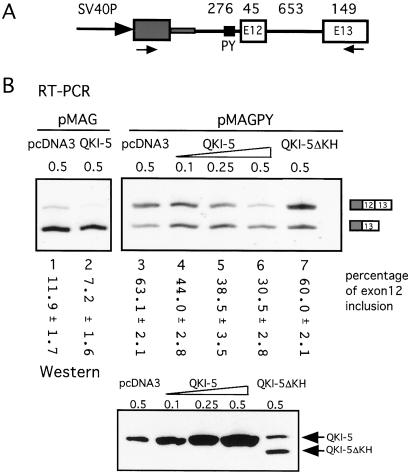
Figure 2.
The repression of MAG exon 12 inclusion by QKI-5. (A) Schematic drawing of the MAG minigene construct. A 1.1-kb genomic DNA fragment around MAG exon 12 is cloned downstream of an HIV tat exon (gray box) in the vector pSPL3. SV40P, simian virus 40 promoter. The numbers above refer to the size (bp) of corresponding exon or intron. The arrows represent the primers used in the RT-PCR in B. The polypyrimidine (PY) tract of exon 12 is indicated by the small black box. In the pMAGPY construct, it was replaced by the PY of exon 11 (from CGTTTCCTTCTTCAAT to ATGTTTCATGCCTCCCTGC). (B) Representative RT-PCR results of RNA from COS7 cells cotransfected with 0.5 μg of MAG minigene and indicated amounts of QKI expression plasmid. The vector pcDNA3 was used to maintain the total level of transfected DNA and as a negative control. The intensity of the bands was measured with a PhosphorImager, and the percentage of exon 12 inclusion is marked below each lane with standard deviations (n = 3). A Western blot with anti-QKI-5 antibody indicates the expression of QKI-5 and QKI-5ΔKH proteins in cells cotransfected with pMAGPY and indicated amounts of QKI-5 expression constructs. The endogenous QKI-5 protein was indicated by the band in the pcDNA3 only lane.
Next, we asked whether overexpression of QKI-5 would change exon 12 splicing. An expression construct, pcDNA3-QKI-5, was generated for this purpose. When QKI-5 is overexpressed in COS7 cells in the presence of pMAG, exon 12 inclusion is reduced from 12% to 7% (Fig. 2B, lanes 1 and 2), whereas in the presence of pMAGPY exon 12 inclusion is decreased from 63% to 30% (Fig. 2B, lanes 3–6). These results show that QKI-5 represses the inclusion of exon 12 in a dose-dependent manner. We also tested the effect of a shortened form of the protein, QKI-5ΔKH, which is missing part of the RNA-binding domain (amino acids 96–136), and which accumulates in the nucleus (10). As expected, without an intact RNA-binding domain QKI-5ΔKH has little or no effect on MAG alternative splicing (Fig. 2B, lane 7). QKI-5 and QKI-5ΔKH protein expression in the transfected cells was confirmed by Western blotting. Thus, the regulation of MAG alternative splicing by QKI-5 depends on the presence of the RNA-binding domain.
These experiments establish the correlation between MAG exon 12 skipping and QKI-5 coexpression in mammalian cells. To assess whether the effect of QKI-5 on exon 12 inclusion is general or substrate-specific, additional splicing reporters containing alternative exons were assayed for their sensitivity to QKI-5 regulation. These results show that the neural exon of the GABA receptor γ2 subunit is positively regulated (Fig. 3, lanes 10–12), whereas exon 21 of the NMDA receptor is essentially unaffected by QKI-5 coexpression (Fig. 3, lanes 16–18). Thus, the effects of QKI-5 on the MAG exon 12 splicing event cannot be explained by a general effect on alternative splicing. It is curious that QKI-5 enhances exon inclusion for the GABA receptor.
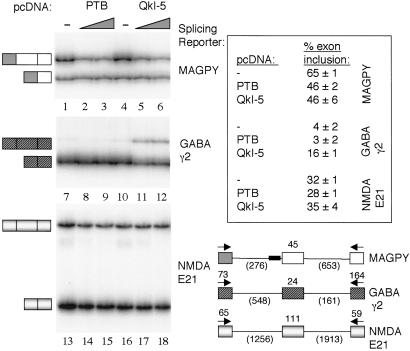
Figure 3.
Comparison of QKI-5 effects on alternative splicing in vivo. Splicing reporters: MAGPY (lanes 1–6), GABA γ2 (lanes 7–12), NMDA E21 (lanes 13–18); and protein expression plasmids, pcDNA QKI-5 and pcDNA PTB, were transfected into the nonneural cell line C2C12, and alternative splicing products were assayed by RT-PCR with primers (arrows) specific for sequences in the first and last exons. Exon-included and exon-skipped products are shown at left of each panel. Wedge indicates a 1:4 or 1:6 ratio of splicing reporter substrate to protein expression plasmid, left and right lanes, respectively. Control lanes (−) were transfected with empty pcDNA vector backbone. Structures of splicing reporter substrates are indicated schematically with exon and intron lengths in nucleotides (bottom right).
To investigate further the functional properties of QKI-5 in vivo, we compared the magnitude of its effect on exon 12 with the effects of the polypyrimidine tract-binding protein (PTB), which is a well-characterized negative regulator of alternative splicing events in the nervous system and muscle (26). When PTB is coexpressed with the pMAGPY splicing reporter, exon 12 inclusion is negatively regulated (Fig. 3, lanes 1–3), and this effect is similar to that observed for QKI-5 (Fig. 3, lanes 4–6). That is, both PTB and QKI-5 reduce exon 12 inclusion by approximately 19%.
A 53-nt Sequence Is Required for QKI-5 Regulation of MAG Exon 12 Inclusion.
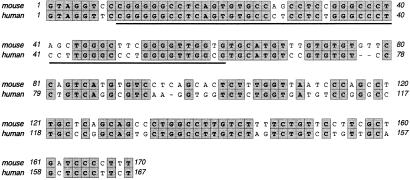
To define the RNA element required for QKI-5 regulation, modifications were made around exon 12. One construct (pMAG-B) that contains exon 12, all of the upstream sequence, and 265 nt from the downstream intron is sufficient for exon 12 inclusion and QKI-5 repression (Fig. 4, construct B). The importance of the upstream intron was also tested by exchanging it for MAG intron 10. The splicing products from this pMAG-B-I10 construct contained a significant amount of exon 12 with QKI-5 still repressing its inclusion (Fig. 4, construct C). Thus, the RNA elements responsible for QKI-5 regulation are in the region encompassing exon 12 and 265 nt of downstream intron. To examine whether this effect of QKI-5 is exon-specific, we exchanged exon 12 for the 100-nt exon 11. All the splicing products contain exon 11 (Fig. 4, construct D). Because an increase in exon size can improve its inclusion, we deleted part of exon 11 to reduce its size to 45 nt. Downstream of this modified exon is 170 nt from intron 12. Now this 45-nt exon is alternatively spliced, and QKI-5 can repress the inclusion of the exon (from 48% to 30%) (Fig. 4, construct E). When the 170-nt fragment was deleted from the construct, overexpression of QKI-5 did not repress the inclusion of this modified exon 11 (Fig. 4, construct F). It is slightly elevated. Thus the 170-nt-downstream intron fragment is sufficient to introduce QKI-5 repression function to an unrelated pre-mRNA, whereas the sequence of exon 12 is not essential. In addition, the comparison of the 170-nt region between mouse and human (Fig. 5) revealed high sequence conservation, suggesting a regulatory function for it.
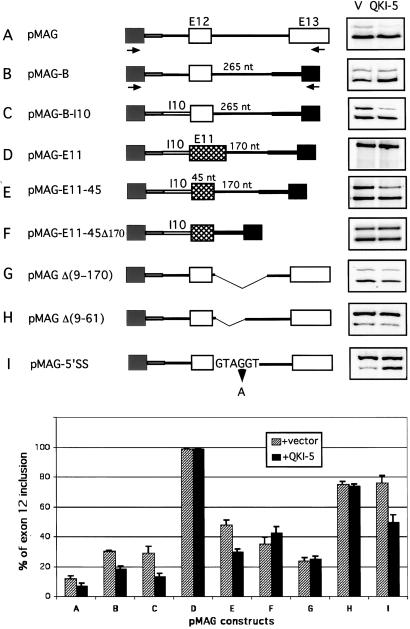
Figure 4.
Sequence requirement for exon 12 inclusion and QKI-5 regulation. Mutations and deletions of MAG minigene are drawn schematically. MAG exons 12 and 13 are shown as white boxes and exon 11 as a cross-hatched box, and tat exons in the vector are shown in gray. Construct A, the wild-type pMAG. In constructs B–F, exon 12 or 11 and their flanking region were cloned between two tat exons. I10 refers to MAG intron 10. In constructs D–F, exon 12 is replaced with exon 11 (100 nt) or a modified exon 11 (45 nt). Constructs G and H represent two internal deletions downstream of exon 12. In construct I, the 5′ splice site of exon 12 was changed to the consensus GTAAGT. The arrows indicate primers. The right side represents the RT-PCR results of MAG minigene splicing products from COS7 cells cotransfected with these constructs and QKI-5 expression plasmid or the vector (V) alone. At the bottom is shown graphical comparison of the percentage of exon 12 inclusion from these mutations with SD (n = 3).
Figure 5.
Sequence alignment between mouse and human MAG intron 12. The 170 nt immediately downstream of mouse MAG exon 12 is compared with the corespondent sequence in human MAG. The identical nucleotides are marked in gray boxes. The 53-nt QASE element is underlined.
To identify the QKI-5 regulatory elements further, internal deletions in the downstream intron were generated while conserving the 5′ splice site. The mutations are illustrated in Fig. 4. When tested, deletions of nucleotides 9–170 and 9–61 were totally unresponsive to QKI-5 (Fig. 4, constructs G and H). Deletions of other regions or smaller deletions in region 9–61 caused partial loss of QKI-5 function (data not shown), suggesting that the entire region of 9–61 is necessary for QKI-5 function. Thus, we named this 53-nt element QASE for the QKI-5 alternative splicing element. From the same data, it was evident that deletion of QASE increases exon 12 inclusion from 12% to 73% (compare Fig. 4, constructs A and H). Therefore, QASE behaves like an intronic splicing silencer. The comparison of Δ(9–170) and Δ(9–61) suggests that, in addition, a splicing enhancer exists between nt 61 and 170 (Fig. 4, constructs G and H).
The 5′ splice site of exon 12 deviates from the consensus sequence (27). Because of its close proximity to the QKI-5 regulatory region, we were curious to see the effect of QKI-5 on a canonical 5′ splice site. Thus this site was changed to match the consensus sequence at position 4 (GUAGGU to GUAAGU) (Fig. 4, construct I). In the absence of QKI-5, exon 12 inclusion increases from 12% to 76%, whereas in the presence of QKI-5, exon 12 selection is substantially down-regulated. Thus, the effect of QKI-5 on MAG pre-mRNA is robust in the presence of a weak or strong 5′ splice site.
Specific Interaction Between QKI-5 and QASE.
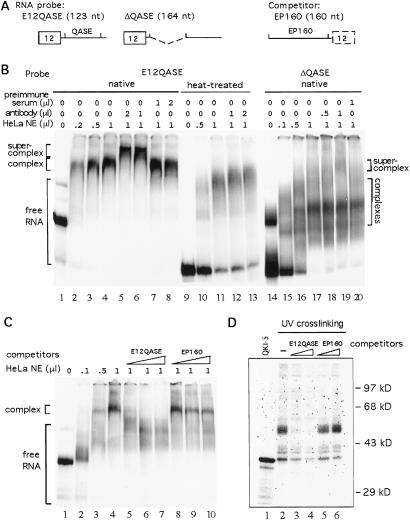
The experiments above suggested that the QASE is responsible for the repression of exon 12 inclusion by QKI-5. The next question was whether QKI-5 binds directly to the QASE. In the cotransfection assay, the disruption of QKI-5 KH domain eliminates its effect on exon 12 splicing, suggesting the importance of RNA binding in this regulation. Because RNA secondary structure can play a role in splicing regulation, gel mobility-shift assays were performed to compare QKI-5 binding with native or heat-denatured RNA probes. E12QASE, a probe containing exon 12 and QASE, was used in a QKI-5 antibody supershift assay. ΔQASE, with its QASE element deleted, was used as a control (Fig. 6 A and B). Previous studies have shown that HeLa cell nuclei contain a significant amount of QKI-5 protein, and the QKI-5 antibody recognizes QKI-5 protein with high specificity (10). Thus, HeLa nuclear extract was used in the gel mobility-shift assays. Increasing amounts of HeLa nuclear extract were mixed with heat-treated or native E12QASE probes. Complexes were formed with both probes (Fig. 6B, lanes 1–4, 9–11). However, anti-QKI-5 antibody supershifted only the complexes formed with the native probe (Fig. 6B, lanes 5, 6, 12, 13). Preimmune serum did not affect the migration of the complexes (Fig. 6B, lanes 7 and 8). In addition, the apparent mobility difference between native and denatured E12QASE clearly indicates the existence of a high-order structure and its importance for QKI-5 association (Fig. 6B, lanes 1 and 9). When native ΔQASE was used as probe, several complexes could be seen (Fig. 6B, lanes 15–17). However, on adding the QKI-5 antibody, only a small portion of the complexes were supershifted. The major complex remained unchanged (Fig. 6B, lanes 18–20). This result suggests that QKI-5 does not bind to ΔQASE efficiently, which is consistent with the cotransfection assay results.
Figure 6.
Specific association of QKI-5 and QASE. (A) Diagrams of the RNA probe, E12QASE, ΔQASE, and competitor, EP160. (B) QKI-5 supershift assay. 32P-labeled E12QASE (native or heat-denatured) and ΔQASE (native) RNA probes were mixed with increasing amounts of HeLa nuclear extract (lanes 1–4, 9–11, 15–17). The amounts of anti-QKI-5 antibody or preimmune serum added to supershift reactions (lanes 5–8, 12, 13, 18–20) are indicated above. (C) Competition assay. Indicated amounts of HeLa nuclear extract were used in the gel mobility-shift assay. Used was 100-, 200-, or 400-fold of unlabeled RNA probe or competitor (lanes 5–10) in the competition reactions. (D) UV crosslinking assay of HeLa nuclear extract to native E12QASE probe (lanes 2–6). In lanes 3–6, 100- or 200-fold of competitors were added to the reactions before adding the probes. 35S-labeled in vitro translated QKI-5 protein was analyzed on the same gel (lane 1).
Competition experiments were used to examine further whether the interaction of QKI-5 with E12QASE is specific. E12QASE or the control RNA, EP160, was used as competitor in these assays. EP160 contains 160 nt located immediately upstream of exon 12 (Fig. 6A). At 200-fold excess, unlabeled E12QASE completely competed away QKI-5 binding, whereas a significant amount of the complex remained when EP160 was used as competitor (Fig. 6C, compare lanes 5–7 with 8–10). Thus we conclude that the association between QKI-5 and QASE is specific and that MAG is a direct target of QKI regulation.
In HeLa nuclear extract, QKI-5 exists in a protein complex formed on E12QASE. However, purified recombinant His-QKI-5 alone does not bind to E12QASE probe in the gel mobility-shift assay (data not shown). Thus, the interaction between QKI-5 and E12QASE may require other protein factors in the nuclear extract. To test this interaction, a UV crosslinking assay was performed (Fig. 6D). Four prominent bands from HeLa nuclear extract were detected after UV crosslinking. These included proteins with molecular mass of 50, 42, 40, and 38 kDa. The 38-kDa band comigrated with in vitro translated QKI-5 (Fig. 6D, lane 1). When 200-fold unlabeled E12QASE was used as competitor, all protein bands, especially the 50-kDa band, decreased. However, the same amount of EP160 did not compete away any of the proteins. This experiment suggests that a protein complex in the nuclear extract interacts with the E12QASE region.
Discussion
QKI-5 Is a Negative Regulator of MAG Alternative Pre-mRNA Splicing.
QKI-5 was previously postulated to play a role in splicing regulation because of its predominantly nuclear localization, its putative function as a sequence-specific RNA-binding protein, and the close correlation between its diminished expression in the developing brain of the qkv mouse and abnormal myelin gene mRNA transcripts. Here we provide direct evidence for the involvement of QKI-5 in the regulation of MAG alternative splicing. These results demonstrate a switch in the ratio of MAG mRNA isoforms from predominant exon 12 skipping in wild type to predominant exon 12 selection in qkv mice. MAG exon 12 inclusion is repressed in a substrate-specific manner by the coexpression of QKI-5. A 53-nt intronic RNA element termed QASE is required for the negative effect of QKI-5, and this RNA element binds specifically to a complex containing QKI-5 protein in HeLa splicing extracts.
Several KH-type RNA-binding proteins function in splicing regulation. Studies of neuron-specific splicing lead to the identification of two mammalian KH proteins, KSRP and Nova-1, as splicing activators (28, 29). In the STAR family, SF1/BBP1 is a constitutive splicing factor. The cooperative binding of SF1 to branch point and U2AF to polypyrimidine tract facilitates the recognition of branch point by U2 small nuclear ribonucleoprotein (30). Recently, it has been reported that rat SLM-2, a SAM68-related STAR protein, regulates selection of alternative splice sites (31). QKI, the only STAR protein with a developmentally defined phenotype, is now added to this interesting group.
Because QASE is immediately downstream of the 5′ splice site of exon 12, it is possible that QKI-5 binding could interfere with 5′ splice site recognition, exon definition, or both. Furthermore, the close proximity of QASE to the intronic enhancer (nucleotides 62–170) might interfere with enhancement. Deletion of the enhancer region alone results in the complete exclusion of exon 12 (data not shown), making it difficult to examine the effect of QKI-5 on the enhancer. However, the deletion of the QASE repressor along with the enhancer (nucleotides 9–170) results in an increase in exon 12 inclusion (compare Fig. 4 constructs A and G), suggesting that the effect of the QASE is dominant.
QKI-5 specifically binds to QASE and regulates MAG splicing. However, MAG is not likely the only target of QKI-5. Indeed, splicing of PLP and MBP is also misregulated in qkv mutants at the peak of the myelination process. It is highly possible that the low level of QKI-5 in mutants is responsible for some or all of the abnormal splicing. Besides the nervous system, QKI-5 is also highly expressed in the early embryo, heart, lung, and testis (6, 7). Therefore, it is appealing to think that many more “non-myelin” targets exist for QKI-5.
What Is the Cause of the Dysmyelination Phenotype of qkv Mice?
In this study, we have shown that QKI-5, the nuclear isoform of QKI, is likely to be involved in regulating MAG alternative splicing in vivo. Cell culture experiments imply that MAG protein plays an important role in initiating and maintaining myelin and in axon–glial cell interaction (14). The analysis of an engineered null mutation confirmed the role of MAG in myelination, although the phenotype is surprisingly subtle (24, 25). In qkv mice, MAG is expressed at a normal level, but exhibits an abnormal splicing profile (16). L-MAG, the dominant isoform in young mice, is greatly reduced. L-MAG and S-MAG have different cytoplasmic domains and L-MAG can be phosphorylated by tyrosine kinase Fyn. It is noteworthy that fyn knockout mice have problems initiating myelination (32). Recently, mutant mice were generated with only the L-MAG cytoplasmic domain being truncated (33). Their phenotype in the central nervous system is very similar to MAG null mutants, whereas peripheral nervous system myelin remains intact. However, in these mice, a 50% reduction of total MAG expression occurred. Thus, this particular mutant is not strictly comparable with the situation in qkv, because, although qkv mice have little L-MAG, they have a compensatory increase in S-MAG. In any case, it would be hard to compare the knockout of a single myelin gene to qkv because QKI seems to affect multiple target RNAs.
In the qkv mouse brain, MBP and PLP, two major myelin components, have aberrant splicing patterns as well. It has also been reported that MBP RNA is destabilized and mislocalized in qkv mice (34). Therefore, our data imply that QKI regulates RNA metabolism of a subset of myelin structural genes and the combination of the abnormalities leads to the ultimate phenotype.
Acknowledgments
This work was supported by National Institutes of Health Grant HD10668 (to K.A.). P.J.G. is an Associate Investigator of the Howard Hughes Medical Institute.
Abbreviations
- MAG
myelin-associated glycoprotein
- MBP
myelin basic protein
- PLP
proteolipid protein
- KH
heterogeneous nuclear ribonucleoprotein K homology
- QASE
quaking alternative splicing element
- P14
postnatal day 14
- NMDA
N-methyl-d-aspartate
- GABA
γ-aminobutyric acid
- STAR
signal transduction and activator of RNA
- PTB
polypyrimidine tract-binding protein
References
- 1.Grabowski P J, Black D L. Prog Neurobiol. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 2.Lopez A J. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 3.Modrek B, Lee C. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 4.Sidman R L, Dickie M M, Appel S H. Science. 1964;144:309–311. doi: 10.1126/science.144.3616.309. [DOI] [PubMed] [Google Scholar]
- 5.Cox R D, Hugill A, Shedlovsky A, Noveroske J K, Best S, Justice M J, Lehrach H, Dove W F. Genomics. 1999;57:333–341. doi: 10.1006/geno.1999.5804. [DOI] [PubMed] [Google Scholar]
- 6.Kondo T, Furuta T, Mitsunaga K, Ebersole T A, Shichiri M, Wu J, Artzt K, Yamamura K, Abe K. Mamm Genome. 1999;10:662–669. doi: 10.1007/s003359901068. [DOI] [PubMed] [Google Scholar]
- 7.Ebersole T, Chen Q, Justice M J, Artzt K. Nat Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- 8.Vernet C, Artzt K. Trends Genet. 1997;13:479–484. doi: 10.1016/s0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]
- 9.Hardy R, Loushin C, Friedrich V, Chen Q, Ebersole T, Lazzarini R, Artzt K. J Neurosci. 1996;16:7941–7949. doi: 10.1523/JNEUROSCI.16-24-07941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J, Zhou L, Tonissen K, Tee R, Artzt K. J Biol Chem. 1999;274:29202–29210. doi: 10.1074/jbc.274.41.29202. [DOI] [PubMed] [Google Scholar]
- 11.Tropak M B, Johnson P W, Dunn R J, Roder J C. Brain Res. 1988;464:143–155. doi: 10.1016/0169-328x(88)90006-x. [DOI] [PubMed] [Google Scholar]
- 12.Nave K A, Lai C, Bloom F E, Milner R J. Proc Natl Acad Sci USA. 1987;84:5665–5669. doi: 10.1073/pnas.84.16.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Ferra F, Engh H, Hudson L, Kamholz J, Puckett C, Molineaux S, Lazzarini R A. Cell. 1985;43:721–727. doi: 10.1016/0092-8674(85)90245-4. [DOI] [PubMed] [Google Scholar]
- 14.Schachner M, Bartsch U. Glia. 2000;29:154–165. doi: 10.1002/(sici)1098-1136(20000115)29:2<154::aid-glia9>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 15.Lai C, Watson J B, Bloom F E, Sutcliffe J G, Milner R J. Immunol Rev. 1987;100:129–151. doi: 10.1111/j.1600-065x.1987.tb00530.x. [DOI] [PubMed] [Google Scholar]
- 16.Fujita N, Sato S, Ishiguro H, Inuzuka T, Baba H, Kurihara T, Takahashi Y, Miyatake T. J Neurochem. 1990;55:1056–1059. doi: 10.1111/j.1471-4159.1990.tb04596.x. [DOI] [PubMed] [Google Scholar]
- 17.Yool D A, Edgar J M, Montague P, Malcolm S. Hum Mol Genet. 2000;9:987–992. doi: 10.1093/hmg/9.6.987. [DOI] [PubMed] [Google Scholar]
- 18.Griffiths I, Klugmann M, Anderson T, Thomson C, Vouyiouklis D, Nave K A. Microsc Res Tech. 1998;41:344–358. doi: 10.1002/(SICI)1097-0029(19980601)41:5<344::AID-JEMT2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 19.Sorg B A, Smith M M, Campagnoni A T. J Neurochem. 1987;49:1146–1154. doi: 10.1111/j.1471-4159.1987.tb10005.x. [DOI] [PubMed] [Google Scholar]
- 20.Konat G, Trojanowska M, Gantt G, Hogan E. J Neurosci Res. 1988;20:19–22. doi: 10.1002/jnr.490200104. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Liu W, Grabowski P J. RNA. 1999;5:117–130. doi: 10.1017/s1355838299981530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geliebter J. Focus (Rochester, NY) 1987;9:5–6. [Google Scholar]
- 23.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montag D, Giese K P, Bartsch U, Martini R, Lang Y, Bluthmann H, Karthigasan J, Kirschner D A, Wintergerst E S, Nave K A, et al. Neuron. 1994;13:229–246. doi: 10.1016/0896-6273(94)90472-3. [DOI] [PubMed] [Google Scholar]
- 25.Li C, Tropak M B, Gerlai R, Clapoff S, Abramow-Newerly W, Trapp B, Peterson A, Roder J. Nature (London) 1994;369:747–750. doi: 10.1038/369747a0. [DOI] [PubMed] [Google Scholar]
- 26.Valcarcel J, Gebauer F. Curr Biol. 1997;7:705–708. doi: 10.1016/s0960-9822(06)00361-7. [DOI] [PubMed] [Google Scholar]
- 27.Burge C B, Tuschl T, Sharp P A. In: The RNA World. 2nd Ed. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 525–560. [Google Scholar]
- 28.Min H, Turck C W, Nikolic J M, Black D L. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 29.Jensen K B, Dredge B K, Stefani G, Zhong R, Buckanovich R J, Okano H J, Yang Y Y, Darnell R B. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 30.Berglund J A, Abovich N, Rosbash M. Genes Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoss O, Olbrich M, Hartmann A M, Konig H, Memmott J, Andreadis A, Stamm S. J Biol Chem. 2001;276:8665–8673. doi: 10.1074/jbc.M006851200. [DOI] [PubMed] [Google Scholar]
- 32.Umemori H, Sato S, Yagi T, Aizawa S, Yamamoto T. Nature (London) 1994;367:572–576. doi: 10.1038/367572a0. [DOI] [PubMed] [Google Scholar]
- 33.Fujita N, Kemper A, Dupree J, Nakayasu H, Bartsch U, Schachner M, Maeda N, Suzuki K, Popko B. J Neurosci. 1998;18:1970–1978. doi: 10.1523/JNEUROSCI.18-06-01970.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Zhang Y, Li D, Feng Y. J Neurosci. 2000;20:4944–4953. doi: 10.1523/JNEUROSCI.20-13-04944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]