Abstract
Transcription-coupled repair (TCR) is essential for the rapid, preferential removal of DNA damage in active genes. The large subunit of RNA polymerase (Pol) II is ubiquitinated in cells after UV-irradiation or cisplatin treatment, which induces DNA damage preferentially repaired by TCR. Several human mutations, such as Cockayne syndrome complementation groups A and B, are defective in TCR and incapable of Pol II ubiquitination upon DNA damage. Here we demonstrate a correlation between ubiquitination of RNA Pol II and arrest of transcription in vitro. Ubiquitination of Pol II is significantly induced by α-amanitin, an amatoxin that blocks Pol II elongation and causes its degradation in cells. Pol II undergoes similar ubiquitination on DNA containing cisplatin adducts that arrest transcription. Stimulation of ubiquitination requires the addition of template DNA and does not occur in the presence of an antibody to the general transcription factor TFIIB, indicating the transcription dependence of the reaction. We propose that components of the reaction recognize elongating Pol II–DNA complexes arrested by α-amanitin or cisplatin lesions, triggering ubiquitination.
Transcription-coupled repair (TCR) is a cellular pathway for the removal of the many lesions that block and arrest transcription. TCR is responsible for the rapid and preferential repair of damage in the transcribed strand of active genes (1–3). Specific mutations in genes are known to cause defects in TCR, including Cockayne syndrome (CS) complementation groups A and B which debilitate TCR of UV- and oxidative-damage- induced lesions (4, 5), and the breast cancer susceptibility gene BRCA1, which is defective in TCR of oxidative DNA damage (6, 7). Lesions induced by chemotherapeutic agents such as cisplatin are also removed by TCR (8, 9), implicating TCR in the differential response of cells to cancer treatment.
The recent discovery that UV-irradiation or cisplatin treatment of cells induces ubiquitination of the large subunit of RNA polymerase II (Pol II LS) and its degradation (10, 11) suggested a biochemical link between ubiquitination and DNA repair. The ubiquitinated Pol II is hyperphosphorylated on the carboxyl-terminal domain (CTD) of Pol II LS (10). Ubiquitination was not observed in TCR-deficient CSA and CSB cells but could be restored upon transfection of their respective cDNAs (10). These results further indicate a relevance of Pol II ubiquitination to the mechanism of action of cisplatin. This chemotherapeutic agent is routinely used in cancer treatment against otherwise resistant solid tumors, such as encountered in testicular, ovarian, and breast cancer (12).
Interestingly, Pol II undergoes degradation in cells treated with α-amanitin, an inhibitor of its transcription (13). This amatoxin binds specifically to RNA Pol II (14, 15), and it arrests the elongating polymerase in a conformation that inhibits incorporation of a subsequent nucleoside triphosphate to the nascent RNA transcript (16, 17). Pol II transcription inhibition by α-amanitin may resemble its transcription arrest at a DNA lesion. It is not known whether α-amanitin dependent degradation of Pol II is mediated by the ubiquitin pathway.
To understand the cellular response to DNA damage during transcription, we have investigated transcription- and DNA-damage-dependent ubiquitination of RNA Pol II in vitro. We significantly modified the original in vitro assay for ubiquitination of Pol II (18) to increase its sensitivity. In addition, we compared nuclear extracts from cells at different cell cycle stages to optimize ubiquitination activity.
Materials and Methods
Chemical Reagents.
α-Amanitin and His-tagged ubiquitin (His-Ub) were purchased from Calbiochem. Aphidicolin, hydroxyurea, and nocodazole (methyl [5-(2-thienylcarbonyl)-1H-benzimidazol-2-yl]-carbamate) were purchased from Sigma–Aldrich. N-Phospho-N-acetyl-l-aspartate (PALA) was obtained from the Drug Biosynthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment, National Cancer Institute (Bethesda, MD). Cisplatin was provided by Johnson Matthey (West Chester, PA).
Preparation of Nuclear Extracts.
Nuclear extracts were prepared from HeLa cells as described (19). Cells were maintained in MEM with 10% horse serum (GIBCO/BRL) at 37°C at a cell density of 0.5–1.0 × 106 cells per ml.
To arrest HeLa cells at different stages, cells were grown either in spinner flasks to 0.5 × 106 cells per ml as above, or on plates in DME containing 10% heat-inactivated FBS (GIBCO/BRL) to 30–50% confluency. The cells were then serum-starved for 16 h by replacing the serum-containing medium with serum-free medium. Serum was added back to the medium to a final concentration of 10% on the following day, and cells were incubated for a further 8 h before the addition of various drugs to a final concentration of 1 μg/ml aphidicolin, 750 μM hydroxyurea, 0.1 μg/ml nocodazole, and 500 μM PALA, respectively. Cells were incubated with the drugs for 16 h before harvesting.
For analysis of drug-treated cells, 1 × 106 cells in PBS were permeabilized with 95% ethanol and stained by incubating with RNase A (200 μg/ml, Calbiochem) and propidium iodide (4 μg/ml, Sigma–Aldrich). The stained cells were analyzed in a fluorescence-activated cell sorter (FACSCalibur, Becton Dickinson) and cell cycle stages were determined with the modfit lt program.
Platination of Plasmid DNA.
Plasmids pUC19 (New England Biolabs), pG5MLP-G380 (obtained from D. Tantin, Massachusetts Institute of Technology), pUC118-296 (obtained from C. Kneip, Massachusetts Institute of Technology), pHIV+TAR-G400 and pHIVΔTAR-G100 (20) were prepared from overnight cultures of plasmid-harboring JM109 (Promega) in LB medium by using a Promega Wizard Midi-prep kit. pG5MLP-G380 contained the major late promoter (MLP) with a 380-bp G-less cassette inserted 10 bp downstream of the initiation site. pUC118-296 contained the MLP and 5S rDNA nucleosome positioning sequences (21).
The cis-[Pt(NH3)2(H2O)2]2+ cation (activated form of cisplatin) was prepared by stirring a solution of cis-Pt(NH3)2Cl2 (6.4 mg, 0.021 mmol) with 1.98 molar equivalents of AgNO3 overnight in the dark. Any precipitated AgCl was removed by centrifugation, and the supernatant was diluted to a final concentration of 1 mM cis-[Pt(NH3)2(H2O)2]2+ in water. Platination of plasmids was achieved by incubating 0.043 nmol of DNA with cis-[Pt(NH3)2(H2O)2]2+ at rf = 0.1 (where rf equals the number of moles of drug added per mole of DNA nucleotide) in 100 μl of 10 mM sodium phosphate (pH 6.8) for 16 h at 37°C. The unbound platinum salt was removed by dialysis. The ratio of platinum bound per nucleotide—i.e., the rb value—was determined by flameless atomic absorption spectroscopy (Perkin–Elmer HGA-800 Analyst 300) and UV absorption spectroscopy (Hewlett Packard 8453).
In Vitro Transcription Assay.
Transcription reactions (25 μl) were performed essentially as described (20). Nuclear extracts (30 μg) were incubated at 30°C for 30 min in buffer 1 [10 mM Hepes–KOH (pH 7.9)/10% (vol/vol) glycerol/60 mM KCl/7 mM MgCl2/0.1 mM EDTA/1 mM ATP/10 mM creatine phosphate/12 μg/ml poly(I⋅C)/2 μg/ml poly(dG⋅dC)] containing 7 mM DTT, C/G/UTP mix (25:200:200 μM), 10 μCi of [α-32P]CTP [800 Ci (29,600 GBq)/mmol], and 200 ng of DNA template. To study the effect of α-amanitin, the reactions were preincubated at 30°C for 15 min in the absence or presence of 5 μM α-amanitin under the above conditions, but with the NTPs and creatine phosphate added subsequently. The reactions were then incubated for a further 30 min. To observe a complete inhibition of transcription by α-amanitin, the preincubation step was included to allow for sufficient drug binding to Pol II. For the in vitro ubiquitination assay (see below), the results obtained with or without preincubation with α-amanitin were similar. All reactions were subjected to RNase T1 digestion at 37°C for 5 min, and G-less RNA transcripts were isolated and analyzed by urea/PAGE as described (22). The gels were vacuum-dried and autoradiographed with Kodak X-Omat film at −80°C for 24–40 h.
In Vitro Ubiquitination Assay.
Reactions were set up as in in vitro transcription assays with 60–70 μg nuclear extract in buffer 1 (which contains 1 mM ATP) with 1 μg of DNA template, 200 μM each of CTP, UTP, and GTP, and 1.25 μg of His-Ub. Saturating levels of α-amanitin (5 μM) were added to the mix where indicated. Reactions were incubated at 30°C for 45 min, and His-ubiquitinated proteins were isolated by incubating at 4°C for 1 h with 20 μl of Ni-nitrilotriacetate (NTA) agarose (Qiagen) in a final volume of 200 μl in buffer 2 [50 mM sodium phosphate (pH 7.9)/0.3 M NaCl/0.05% Tween 20] containing 10 mM imidazole. The Ni-NTA agarose was preblocked with 1 mg/ml BSA before use. After low-speed centrifugation (735 × g), the Ni-agarose beads containing His-ubiquitinated proteins were washed twice with 1 ml of buffer 2 containing 50 mM imidazole. The Ni-bound proteins were eluted with SDS-loading buffer [20 mM Tris⋅HCl (pH 6.8)/10% (vol/vol) glycerol/100 mM 2-mercaptoethanol/1% (wt/vol) SDS/0.02% bromophenol blue] containing 0.1 M EDTA (pH 7.0). To analyze the protein content, samples were boiled in SDS-loading buffer and electrophoresed in SDS/7.5% polyacrylamide gels, followed by electrotransfer to Immobilon-P membranes (Millipore).
Pol II was detected with N20 (Santa Cruz), a non-phospho-specific rabbit polyclonal IgG. The phosphorylated form of Pol II was detected with H5 or H14 (Covance), a mouse monoclonal IgM that recognizes phosphoserine-2 or -5 of the CTD heptapeptide repeat.
Results
α-Amanitin Stimulates Ubiquitination of RNA Pol II.
To investigate a possible correlation between ubiquitination of RNA Pol II and the arrest of transcription, we examined the effect of α-amanitin, a Pol II inhibitor, on polymerase ubiquitination in vitro. Previous results suggested little or no effect of α-amanitin on in vitro ubiquitination of polymerase in nuclear extracts from unsynchronized cells (18). We prepared nuclear extracts from cells arrested in G1/S phase by aphidicolin, a DNA α-Pol inhibitor (23), and tested for α-amanitin-dependent ubiquitination of Pol II LS (see below for drug and cell cycle dependence). Cell cycle stages were verified by fluorescence-activated cell sorter analysis (not shown). To increase the sensitivity of the assay, His-Ub was added to the reaction, and ubiquitinated proteins were selected on Ni-NTA agarose and analyzed by Western blotting. The reaction conditions were those used for transcription (see Material and Methods).
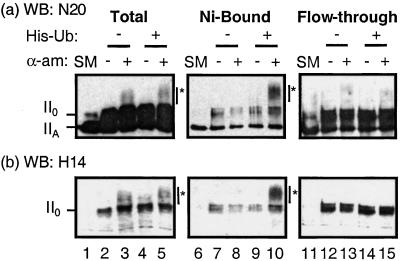
SDS/PAGE of total unselected extracts and Western blot analysis with a non-phospho-specific antibody to the N terminus of Pol II LS (N20) revealed that hypophosphorylated Pol IIA (Fig. 1a, lane 1) was converted to hyperphosphorylated Pol II0 during the reaction (Fig. 1a, lanes 2 and 4). Upon addition of α-amanitin, a new slow-migrating band appeared (marked with an asterisk in Fig. 1a, lanes 3 and 5). This slow-migrating band was observed in the Ni-bound fraction only when His-Ub was present in the reaction (Fig. 1a, lane 10), indicative of ubiquitination of RNA Pol II. The Pol IIA and II0 bands recovered in the Ni-NTA fractions in the absence of His-Ub are due to nonspecific binding to Ni-agarose (Fig. 1a, lanes 7–8). When the Western blot was stripped and reprobed with an antibody to the phosphorylated CTD (H14), ubiquitinated Pol II was also detected (marked with an asterisk in Fig. 1b, lanes 3, 5, and 10). H14 recognizes phosphoserine-5 of the CTD heptapeptide repeat. Similar results were obtained by using H5, another antibody that specifically recognizes phosphoserine-2 of the CTD repeat (not shown). Thus α-amanitin stimulates ubiquitination of Pol II LS, and hyperphosphorylated Pol II0 constitutes the ubiquitinated species, consistent with previous results (10). Only a small percentage of Pol II (≈5%) is ubiquitinated since, relative to the input material (Fig. 1 a and b, lanes 1–5), most of the Pol II was recovered in the fraction not bound to the column (flow-through) (Fig. 1 a and b, lanes 11–15).
Figure 1.
Ubiquitination of Pol II is induced by α-amanitin in an in vitro assay. Nuclear extracts from cells treated with aphidicolin were incubated in an in vitro ubiquitination reaction with pUC118-296 in the absence or presence of His-Ub and α-amanitin (α-am). His-ubiquitinated proteins were selected on Ni-NTA-agarose. Total (lanes 1–5), Ni-bound (lanes 6–10), and flow-through fractions from the Ni-NTA-agarose (lanes 11–15) were analyzed by SDS/7.5% PAGE and transferred to a poly(vinylidene difluoride) (PVDF) membrane. The immunoblot was probed consecutively with antibodies to the N terminus of RNA Pol II (a; antibody N20), and the phosphorylated CTD of Pol II (b; antibody H14). Lanes 1, 6, and 11 show untreated nuclear extracts (SM, starting material).
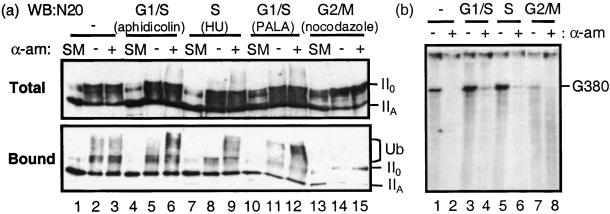
To determine whether ubiquitination activity varied at different stages of the cell cycle when the cells were arrested by drug treatment, we compared the activities of extracts from cells treated with different cell-cycle inhibitors. The fraction of cells at each stage was determined by fluorescence-activated cell sorter analysis (not shown). Extracts from cells treated with aphidicolin, hydroxyurea (a ribonucleotide reductase inhibitor; ref. 24), or PALA (an inhibitor of de novo pyrimidine biosynthesis; ref. 25), all showed similar levels of α-amanitin-induced ubiquitination (Fig. 2a, lanes 5 and 6, 8 and 9, and 11 and 12, respectively). Consistent with previous findings (18), extracts from unsynchronized cells did not show an appreciable αamanitin induction of ubiquitination (Fig. 2a, lanes 2 and 3). G2/M extracts from cells treated with the microtubule inhibitor nocodazole (26) gave a higher proportion of Pol II0 to Pol IIA under the reaction conditions (Fig. 2a, total, lanes 13–15), but showed little or no ubiquitination (Fig. 2a, bound, lanes 14 and 15). In parallel work, we compared in vitro transcription activities of the different extracts by using pG5MLP-G380, which contains an MLP and a 380-bp G-less cassette (G380). The amount of G380 transcript produced by G1/S and S extracts after RNase T1 digestion was approximately 2-fold higher (Fig. 2b, lanes 3 and 5) than extracts from unsynchronized cells (Fig. 2b, lane 1), whereas that of G2/M extracts was considerably lower (Fig. 2b, lane 7). In all cases, α-amanitin inhibited transcription as judged by disappearance of the transcript (Fig. 2b, lanes 2, 4, 6, and 8).
Figure 2.
Comparison of ubiquitination and transcription activities of various drug-induced cell cycle stage nuclear extracts. (a) The in vitro ubiquitination assay was performed as described in the legend of Fig. 1 with nuclear extracts from unsynchronized cells (lanes 1–3) as well as with cells treated, respectively, with aphidicolin (lanes 4–6), hydroxyurea (HU) (lanes 7–9), PALA (lanes 10–12), and nocodazole (lanes 13–15). Reactions (containing His-Ub) were analyzed by SDS/7.5% PAGE, and the immunoblots for the total (Upper) and Ni-bound (Lower) proteins are shown. The immunoblot of total extracts is shown at a lower exposure time than that for the bound proteins to allow better resolution of protein bands. Lanes 1, 4, 7, 10, and 13 show untreated nuclear extracts. (b) Nuclear extracts from unsynchronized cells (lanes 1 and 2), and cells arrested at the cell cycle stages of G1/S (lanes 3 and 4), S (lanes 5 and 6), and G2/M (lanes 7 and 8), were incubated in an in vitro transcription reaction with pG5MLP-G380 containing a 380-bp G-less cassette (G380), in the absence and presence of α-amanitin, respectively. Reactions were subsequently treated with RNase T1, and G-less transcripts were isolated and analyzed by 8 M urea/6% PAGE. An autoradiograph of the gel is shown.
Dependence of Ubiquitination on DNA.
Because α-amanitin binds to Pol II itself (15, 27), we studied whether ubiquitination of polymerase caused by α-amanitin depends on the presence of DNA. We titrated the ubiquitination reaction against increasing concentrations of template DNA (Fig. 3). In the absence of DNA, only low-level or background ubiquitination of Pol was observed (Fig. 3, lanes 3 and 4). When a promoter-containing DNA (pUC118-296) was present, the extent of ubiquitination with α-amanitin increased with increasing amounts of DNA until 1.2 μg of template was added to the reaction (Fig. 3, lanes 5–10). The level of ubiquitination, however, declined upon further addition of DNA (Fig. 3, lane 12). Therefore, α-amanitin induces ubiquitination dependent upon the presence of DNA, probably by interacting with Pol II–DNA complexes during transcription. This stimulation could not be recapitulated by nucleotide depletion or termination of transcription by addition of 3′-O-methyl-GTP (not shown). At high DNA concentrations in vitro, total transcription by Pol II is typically not promoter-dependent and occurs in the absence of a promoter (not shown). Consistent with this behavior, duplex plasmid DNA stimulated α-amanitin-dependent ubiquitination independent of the promoter sequence (not shown). Similarly, supercoiled and linearized plasmid were equally as effective in stimulating ubiquitination (not shown).
Figure 3.
α-Amanitin induction of Pol II ubiquitination is DNA-dependent. Ubiquitination reactions with nuclear extracts from aphidicolin-treated cells and His-Ub were titrated against increasing amounts of pUC118-296 (0–2.4 μg, lanes 3–12), and the effect of α-amanitin (α-am) was assayed. Reactions were analyzed as described in the legend of Fig. 1, and immunoblots of total (Upper) and Ni-NTA-bound proteins (Lower) against N20 are shown. Lanes 1, untreated extracts; lanes 2, ubiquitination reaction containing 1 μg of DNA, incubated in the absence of both His-Ub and α-amanitin.
Ubiquitination Is Induced by Arrest of Transcription on Cisplatin-Damaged DNA.
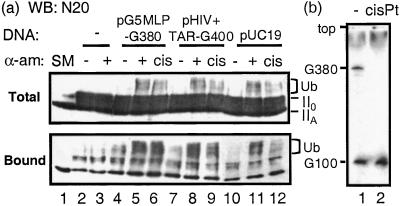
The above data strongly suggest that Pol II–DNA complexes stalled during transcription are targeted for ubiquitination. To investigate a role for DNA damage in stimulating Pol II ubiquitination, DNA templates containing lesions that arrest transcription by Pol II were added to the reaction. Plasmid DNAs modified by cisplatin having an rb value (bound platinum to DNA nucleotide ratio) of 0.055, or an average of one platinum adduct per 18 DNA bases, were tested for induction of Pol II ubiquitination. Reactions containing α-amanitin were also performed in parallel for comparison of activities.
As observed earlier, ubiquitination of Pol II LS was negligible in a reaction lacking DNA or containing unmodified DNA (Fig. 4a, lanes 2–4). In contrast, ubiquitination was induced significantly by each cisplatin-modified template (Fig. 4a, lanes 6, 9, and 12), to levels similar to that induced by α-amanitin with unmodified DNA (Fig. 4a, lanes 5, 8, and 11). This result indicates that both cisplatin and α-amanitin induce ubiquitination in a similar fashion, probably by inhibiting elongation of Pol II transcription on DNA. At this level of cisplatin modification, transcription from cisplatinated DNA containing a 380-bp G-less cassette (G380) was abolished, whereas transcription from an unmodified DNA template with a 100-bp G-less cassette (G100) was unaffected in the same reaction (Fig. 4b, lanes 1 and 2). Thus, the absence of transcript from the G380 cassette is specific to cisplatin modification of the DNA, suggesting that ubiquitination is induced by arrest of transcription attributable to the lesions in the DNA. As with α-amanitin, ubiquitination caused by cisplatin-modified DNA is independent of the presence of a specific promoter on the template DNA. Ubiquitination was observed with cisplatinated pUC19 DNA, albeit at a lower level (Fig. 4a, lanes 11 and 12).
Figure 4.
Ubiquitination of Pol II is induced to a similar extent by cisplatin-modified DNA and α-amanitin. (a) The effects of α-amanitin and various cisplatin-modified DNAs on Pol II ubiquitination were examined in the in vitro ubiquitination assay. Immunoblots of total (Upper) and bound (Lower) proteins are shown. Lanes 1, untreated nuclear extracts. Lanes 2 and 3, ubiquitination reactions performed in the absence of DNA, with α-amanitin (α-am) added where indicated. Lanes 4 and 5, 7 and 8, and 10 and 11, reactions containing unmodified DNA with the addition of α-amanitin where indicated. Lanes 6, 9, and 12, reactions containing cisplatinated DNA (cis). (b) In vitro transcription reactions were performed simultaneously with pG5MLP-G380 containing a G380 cassette and pHIVΔTAR-G100 containing a G100 cassette. Lane 1, transcription with unmodified DNA. Lane 2, transcription with cisplatinated pG5MLP-G380 but unmodified pHIVΔTAR-G100. Autoradiograph of gel is shown.
Dependence of Ubiquitination on Transcription.
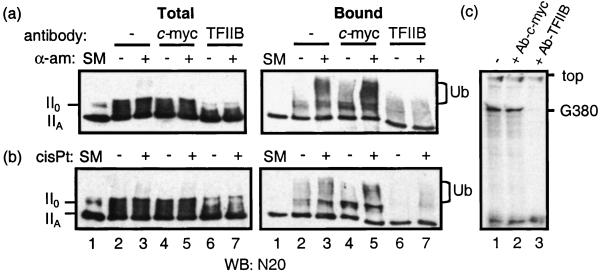
The transcription dependence of the ubiquitination reactions was examined further by using antibodies to a general transcription factor involved in preinitiation complex assembly, namely TFIIB (28). Addition of an antibody to TFIIB inhibited transcription (Fig. 5c, lane 3), as indicated by disappearance of the transcript present in a normal in vitro transcription reaction (Fig. 5c, lane 1). As expected, addition of an antibody to c-myc had no effect on transcription (Fig. 5c, lane 2). When the ubiquitination reaction was tested in the presence of an anti-TFIIB antibody, the ability of either α-amanitin or cisplatin-modified DNA to stimulate ubiquitination was severely reduced (Fig. 5 a and b, bound fractions, lanes 6 and 7) relative to a control without antibody (Fig. 5 a and b, bound fractions, lanes 2 and 3). Ubiquitination overall was unaffected by an antibody to c-myc (Fig. 5 a and b, bound fractions, lanes 4 and 5). Analysis of the total extracts revealed that phosphorylation of CTD was largely inhibited by addition of antibody to TFIIB (Fig. 5 a and b, total, lanes 6 and 7). This behavior is probably a consequence of inhibition of transcription initiation, which is accompanied by Pol II phosphorylation.
Figure 5.
Stimulation of Pol ubiquitination by α-amanitin or cisplatin-modified DNA is suppressed by inhibiting transcription. (a) A typical ubiquitination reaction was carried out in the absence (lane 2) or presence (lane 3) of α-amanitin (α-am). Corresponding reactions were performed with the addition of antibodies to c-myc (lanes 4 and 5) or TFIIB (lanes 6 and 7). Figure shows immunoblots of total extracts (Left) and Ni-bound fractions (Right), which were developed with antibodies against Pol II LS (N20). The immunoblot of total extracts is shown at a lower exposure time than that for the bound proteins. Lanes 1 show untreated nuclear extracts. (b) Ubiquitination reactions were performed in the presence of unmodified DNA (lane 2) or cisplatin-modified DNA (cisPt) (lane 3). Corresponding reactions were conducted with the addition of the different antibodies, followed by immunoblotting as described for a. Lanes 1 show untreated nuclear extracts. (c) In vitro transcription reactions using pG5MLP-G380 were performed in the absence (lane 1) and in the presence of an antibody to c-myc (lane 2) or TFIIB (lane 3). Reactions were analyzed as in Fig. 2b. Autoradiograph of gel is shown.
Discussion
We report evidence for transcription-coupled and DNA-damage-dependent ubiquitination of RNA Pol II in vitro. Addition of α-amanitin, an inhibitor of Pol II elongation, stimulates ubiquitination of Pol II LS. Hyperphosphorylated Pol II is ubiquitinated, consistent with an engagement in elongation. This stimulation requires the addition of template DNA, also indicating a dependence upon Pol II being engaged in transcription. Moreover, addition of DNA templates containing cisplatin adducts, which arrest transcription, stimulates ubiquitination of Pol II LS as compared with reactions containing unmodified templates. The transcription dependence of both α-amanitin and cisplatin-induced ubiquitination was further demonstrated by the abrogation of ubiquitination after addition of antisera to a component of the transcription preinitiation complex, TFIIB.
Implications for Transcription-Coupled DNA Repair.
Previous in vivo experiments pointed to a possible role for ubiquitination of Pol II in transcription-dependent DNA repair. Bregman et al. (10) observed that Pol II was ubiquitinated upon DNA damage from UV irradiation or cisplatin treatment of repair-competent cells and that this ubiquitination was absent in TCR-deficient CS cells. Our finding that ubiquitination is triggered by α-amanitin as well as cisplatin-DNA adducts in vitro, both of which arrest transcription, is consistent with a role for ubiquitination in the cellular response to DNA damage. The transcription dependence of ubiquitination in vitro suggests that components of the reaction specifically recognize a form of Pol II generated by its arrest at lesions or by α-amanitin. This same arrested complex may be a key structure signaling TCR.
Only a small proportion of Pol is ubiquitinated (≈5%) in a reaction containing saturating levels of α-amanitin. Strikingly, the levels of ubiquitination induced by α-amanitin and by DNA with an average of one platinum adduct per 18 DNA bases (≈1 platinum adduct per helical turn) are qualitatively similar. α-Amanitin binds with high affinity (Kd = 108 to 1010 M−1) (15) to a conserved region of Pol II LS (27). It is a very specific and potent inhibitor of the elongating Pol II (14, 29). Biochemical studies reveal that α-amanitin neither dissociates the Pol II–DNA–RNA complex nor competes with nucleotide incorporation (14, 15). Rather, under conditions of elongation, it blocks Pol II after formation of a phosphodiester bond, and probably inhibits a transition in the conformation of Pol II required for its movement along the DNA template (16, 17). Thus the binding of α-amanitin may arrest the elongating Pol II in a transitional conformation that is recognized by components in the reaction specifying ubiquitination. Based on the similarities in ubiquitination of Pol II because of α-amanitin and cisplatin–DNA damage, the platinum lesion most likely also blocks translocation of Pol II along the DNA, stabilizing a similar transitional conformation. The major cisplatin–DNA adduct is the intrastrand crosslink between adjacent purine residues, with the 1,2-d(GpG) and 1,2-d(ApG) intrastrand crosslinks being the predominant lesions (30). Both the 1,2-d(GpG) and 1,3-d(GpTpG) adducts are efficient blocks to transcript elongation in vitro (31, 32) and in vivo (33).
Evidence suggests that the inability to remove an RNA Pol II stalled at a DNA lesion prevents access to repair enzymes, thus enhancing the mutation rate. The same Pol II would also block elongation by other Pol molecules and hence interfere with gene expression. An RNA Pol II stalled at a UV-induced cyclobutane pyrimidine dimer prevented access of a small bacterial repair protein in vitro (34). Human mutant cells impaired in TCR of oxidative lesions, namely CSA and CSB, as well as subgroups of xeroderma pigmentosum XPB, XPD, and XPG that show CS, exhibit a high mutation frequency of 30–40% at an 8-oxoguanine lesion compared with a normal level of 1–4% (35). TCR of oxidative lesions, including thymine glycols, is also defective in cells derived from brca1−/− mice (6) and the human breast carcinoma cell line HCC1937 (7, 36), which harbors a mutation of the BRCA1 gene inherent in many incidences of breast and ovarian cancer (37, 38). Transcription was not observed beyond the 8-oxoguanine lesion on plasmid DNA transfected into the XPG/CS and HCC1937 cells (35, 36). Further, TCR-deficient CSA and CSB cells are more susceptible to apoptosis upon UV-irradiation, perhaps because of the inefficient removal of RNA Pol II at UV-induced lesions and subsequent blockage of transcription (39).
The Cellular Signals Triggering Ubiquitination.
Ubiquitination occurs by covalent attachment of multiple ubiquitin molecules to the protein substrate. This process is typically mediated by three enzymes, the ubiquitin-activating enzyme E1, the ubiquitin-conjugating enzyme E2, and the substrate-specific ubiquitin ligase complex E3 (40, 41). Ubiquitination by E3 ligase is highly regulated and, in many cases, substrate recognition requires covalent modification—e.g., in the phosphorylation of IκB (42) and β-catenin (43). Immunoblot analyses of whole cell extracts from UV-irradiated/cisplatin-treated cells (10) and of Ni-selected nuclear extracts from the in vitro ubiquitination assay (this work) demonstrate that ubiquitinated RNA Pol II is recognized by antibodies specific to phosphorylation of either serine-2 or serine-5 of the CTD. Previous results using nuclear extracts from growing cells indicated that a glutathione S-transferase-CTD fusion protein was ubiquitinated in a kinase-dependent manner (18), suggesting some modification of this repetitive domain may be important for ubiquitination.
An interesting finding of the present work is that nuclear extracts from cells arrested with drugs that affect DNA synthesis in the G1/S phase exhibit significantly enhanced ubiquitination activity relative to unsynchronized cells. The reason for this behavior is unclear because ubiquitination of Pol II after DNA damage is observed in vivo in unsynchronized cells (10), which are primarily in the G1 phase of the cell cycle. Of possible relevance is that all three drug inhibitors activate or induce proteins involved in repair of DNA damage. Included are the DNA damage cell cycle checkpoint protein p53 (44–47), and the breast cancer susceptibility protein BRCA1 (48), which in addition to transcription-coupled repair of oxidative damage (6, 7, 36) is involved in double-strand break DNA repair (49, 50). p53 interacts with proteins involved in TCR, namely CSB, as well as the XPB and XPD subunits of TFIIH (51), with the latter interactions being important for induction of apoptosis (52). BRCA1, on the other hand, undergoes phosphorylation and is relocalized into replicating DNA structures upon treatment of S-phase cells with hydroxyurea or DNA damaging agents (48). BRCA1 also interacts with RNA Pol II holoenzyme (53) and contains a ring finger domain that interacts with the ring finger domain of another protein, BARD1 (54). Ring finger domains are generally involved in the ubiquitination pathway (55), and BRCA1 can facilitate E2-dependent ubiquitination in vitro (56). BARD1, which colocalized with BRCA1 upon DNA damage (48), enhances ubiquitination activity; a breast cancer-derived cysteine mutation in the ring finger of BRCA1, which is deficient in its interaction with BARD1, abolishes this activity (57). The same mutant is defective as a tumor suppressor. The substrate for ubiquitination by BRCA1 is currently not known but may be RNA Pol II (also see ref. 58).
Implications for the Cisplatin Mechanism of Action.
Previous studies revealed that specific architectural changes induced upon formation of the major cisplatin 1,2-intrastrand DNA crosslinks lead to binding of minor groove intercalating proteins including HMGB1 (59, 60) and TBP (61). Such processes inhibit nucleotide excision repair (NER) in vitro (62) and retard NER in vivo (63, 64), leaving the adducts intact for subsequent recognition through arrest of Pol II elongation during TCR. The present results suggest that ubiquitination of RNA Pol II may be an important event in the repair of cisplatin damage. Because removal of the stalled Pol II from the site of damage is critical for repair, inhibition of ubiquitination of Pol II could well enhance cell killing by cisplatin.
Acknowledgments
We are grateful to C. Kneip and D. Tantin for pUC118-296 and pG5MLP-G380. We thank D. Tantin for critical review of the manuscript, and members of the Sharp laboratory for helpful discussions. This work was supported by the U.S. Public Health Service Grant PO1-CA42063 from the National Cancer Institute and R01-AI32486 from the National Institutes of Health to P.A.S.; and by National Institutes of Health Grant CA34992 from the National Cancer Institute to S.J.L. The work was also partially supported by the Cancer Center Support (Core) Grant P30-CA14051 from the National Cancer Institute.
Abbreviations
- α-am
α-amanitin
- CS
Cockayne syndrome
- CTD
carboxyl-terminal domain
- His-Ub
His-tagged ubiquitin
- LS
large subunit
- MLP
major late promoter
- NTA
nitrilotriacetate
- PALA
N-phospho-N-acetyl-l-aspartate
- Pol
polymerase
- TCR
transcription-coupled repair
- XP
xeroderma pigmentosum
References
- 1.Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 2.Mellon I, Spivak G, Hanawalt P C. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 3.Jones J C, Zhen W P, Reed E, Parker R J, Sancar A, Bohr V A. J Biol Chem. 1991;266:7101–7107. [PubMed] [Google Scholar]
- 4.Venema J, Mullenders L H, Natarajan A T, van Zeeland A A, Mayne L V. Proc Natl Acad Sci USA. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leadon S A, Cooper P K. Proc Natl Acad Sci USA. 1993;90:10499–10503. doi: 10.1073/pnas.90.22.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gowen L C, Avrutskaya A V, Latour A M, Koller B H, Leadon S A. Science. 1998;281:1009–1012. doi: 10.1126/science.281.5379.1009. [DOI] [PubMed] [Google Scholar]
- 7.Abbott D W, Thompson M E, Robinson-Benion C, Tomlinson G, Jensen R A, Holt J T. J Biol Chem. 1999;274:18808–18812. doi: 10.1074/jbc.274.26.18808. [DOI] [PubMed] [Google Scholar]
- 8.May A, Nairn R S, Okumoto D S, Wassermann K, Stevnsner T, Jones J C, Bohr V A. J Biol Chem. 1993;268:1650–1657. [PubMed] [Google Scholar]
- 9.Zamble D B, Mu D, Reardon J T, Sancar A, Lippard S J. Biochemistry. 1996;35:10004–10013. doi: 10.1021/bi960453+. [DOI] [PubMed] [Google Scholar]
- 10.Bregman D B, Halaban R, van Gool A J, Henning K A, Friedberg E C, Warren S L. Proc Natl Acad Sci USA. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratner J N, Balasubramanian B, Corden J, Warren S L, Bregman D B. J Biol Chem. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 12.Zamble D B, Lippard S J. Trends Biochem Sci. 1995;20:435–439. doi: 10.1016/s0968-0004(00)89095-7. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen V T, Giannoni F, Dubois M F, Seo S J, Vigneron M, Kedinger C, Bensaude O. Nucleic Acids Res. 1996;24:2924–2929. doi: 10.1093/nar/24.15.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kedinger C, Gniazdowski M, Mandel J L, Jr, Gissinger F, Chambon P. Biochem Biophys Res Commun. 1970;38:165–171. doi: 10.1016/0006-291x(70)91099-5. [DOI] [PubMed] [Google Scholar]
- 15.Cochet-Meilhac M, Chambon P. Biochim Biophys Acta. 1974;353:160–184. doi: 10.1016/0005-2787(74)90182-8. [DOI] [PubMed] [Google Scholar]
- 16.Vaisius A C, Wieland T. Biochemistry. 1982;21:3097–3101. doi: 10.1021/bi00256a010. [DOI] [PubMed] [Google Scholar]
- 17.de Mercoyrol L, Job C, Job D. Biochem J. 1989;258:165–169. doi: 10.1042/bj2580165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitsui A, Sharp P A. Proc Natl Acad Sci USA. 1999;96:6054–6059. doi: 10.1073/pnas.96.11.6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Q, Sharp P A. EMBO J. 1995;14:321–328. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simpson R T, Stafford D W. Proc Natl Acad Sci USA. 1983;80:51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parvin J D, Timmers H T, Sharp P A. Cell. 1992;68:1135–1144. doi: 10.1016/0092-8674(92)90084-p. [DOI] [PubMed] [Google Scholar]
- 23.Ikegami S, Taguchi T, Ohashi M, Oguro M, Nagano H, Mano Y. Nature (London) 1978;275:458–460. doi: 10.1038/275458a0. [DOI] [PubMed] [Google Scholar]
- 24.Timson J. Mutat Res. 1975;32:115–132. doi: 10.1016/0165-1110(75)90002-0. [DOI] [PubMed] [Google Scholar]
- 25.Collins K D, Stark G R. J Biol Chem. 1971;246:6599–6605. [PubMed] [Google Scholar]
- 26.De Brabander M J, Van de Veire R M, Aerts F E, Borgers M, Janssen P A. Cancer Res. 1976;36:905–916. [PubMed] [Google Scholar]
- 27.Bartolomei M S, Corden J L. Mol Gen Genet. 1995;246:778–782. doi: 10.1007/BF00290727. [DOI] [PubMed] [Google Scholar]
- 28.Lee T I, Young R A. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 29.Lindell T J, Weinberg F, Morris P W, Roeder R G, Rutter W J. Science. 1970;170:447–449. doi: 10.1126/science.170.3956.447. [DOI] [PubMed] [Google Scholar]
- 30.Eastman A. Biochemistry. 1986;25:3912–3915. doi: 10.1021/bi00361a026. [DOI] [PubMed] [Google Scholar]
- 31.Corda Y, Job C, Anin M F, Leng M, Job D. Biochemistry. 1993;32:8582–8588. doi: 10.1021/bi00084a027. [DOI] [PubMed] [Google Scholar]
- 32.Cullinane C, Mazur S J, Essigmann J M, Phillips D R, Bohr V A. Biochemistry. 1999;38:6204–6212. doi: 10.1021/bi982685+. [DOI] [PubMed] [Google Scholar]
- 33.Mello J A, Lippard S J, Essigmann J M. Biochemistry. 1995;34:14783–14791. doi: 10.1021/bi00045a020. [DOI] [PubMed] [Google Scholar]
- 34.Donahue B A, Yin S, Taylor J S, Reines D, Hanawalt P C. Proc Natl Acad Sci USA. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Page F, Kwoh E E, Avrutskaya A, Gentil A, Leadon S A, Sarasin A, Cooper P K. Cell. 2000;101:159–171. doi: 10.1016/s0092-8674(00)80827-2. [DOI] [PubMed] [Google Scholar]
- 36.Le Page F, Randrianarison V, Marot D, Cabannes J, Perricaudet M, Feunteun J, Sarasin A. Cancer Res. 2000;60:5548–5552. [PubMed] [Google Scholar]
- 37.Miki Y, Swensen J, Shattuck-Eidens D, Futreal P A, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett L M, Ding W, et al. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 38.Tomlinson G E, Chen T T, Stastny V A, Virmani A K, Spillman M A, Tonk V, Blum J L, Schneider N R, Wistuba I I, Shay J W, et al. Cancer Res. 1998;58:3237–3242. [PubMed] [Google Scholar]
- 39.Ljungman M, Zhang F. Oncogene. 1996;13:823–831. [PubMed] [Google Scholar]
- 40.Varshavsky A. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 41.Ciechanover A, Orian A, Schwartz A L. J Cell Biochem. 2000;77:40–51. doi: 10.1002/(sici)1097-4644(2000)77:34+<40::aid-jcb9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 42.Yaron A, Gonen H, Alkalay I, Hatzubai A, Jung S, Beyth S, Mercurio F, Manning A M, Ciechanover A, Ben-Neriah Y. EMBO J. 1997;16:6486–6494. doi: 10.1093/emboj/16.21.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linke S P, Clarkin K C, Di Leonardo A, Tsou A, Wahl G M. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 45.Zamble D B, Jacks T, Lippard S J. Proc Natl Acad Sci USA. 1998;95:6163–6168. doi: 10.1073/pnas.95.11.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor W R, Agarwal M L, Agarwal A, Stacey D W, Stark G R. Oncogene. 1999;18:283–295. doi: 10.1038/sj.onc.1202516. [DOI] [PubMed] [Google Scholar]
- 47.Gottifredi V, Shieh S, Taya Y, Prives C. Proc Natl Acad Sci USA. 2001;98:1036–1041. doi: 10.1073/pnas.021282898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scully R, Chen J, Ochs R L, Keegan K, Hoekstra M, Feunteun J, Livingston D M. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 49.Moynahan M E, Chiu J W, Koller B H, Jasin M. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 50.Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston D M. Mol Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 51.Wang X W, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J M, Wang Z, Freidberg E C, Evans M K, Taffe B G, et al. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 52.Wang X W, Vermeulen W, Coursen J D, Gibson M, Lupold S E, Forrester K, Xu G, Elmore L, Yeh H, Hoeijmakers J H, Harris C C. Genes Dev. 1996;10:1219–1232. doi: 10.1101/gad.10.10.1219. [DOI] [PubMed] [Google Scholar]
- 53.Scully R, Anderson S F, Chao D M, Wei W, Ye L, Young R A, Livingston D M, Parvin J D. Proc Natl Acad Sci USA. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu L C, Wang Z W, Tsan J T, Spillman M A, Phung A, Xu X L, Yang M C, Hwang L Y, Bowcock A M, Baer R. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 55.Freemont P S. Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 56.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T. J Biol Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- 58.Parvin J D. Proc Natl Acad Sci USA. 2001;98:5952–5954. doi: 10.1073/pnas.121184998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hughes E N, Engelsberg B N, Billings P C. J Biol Chem. 1992;267:13520–13527. [PubMed] [Google Scholar]
- 60.Pil P M, Lippard S J. Science. 1992;256:234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- 61.Vichi P, Coin F, Renaud J P, Vermeulen W, Hoeijmakers J H, Moras D, Egly J M. EMBO J. 1997;16:7444–7456. doi: 10.1093/emboj/16.24.7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang J C, Zamble D B, Reardon J T, Lippard S J, Sancar A. Proc Natl Acad Sci USA. 1994;91:10394–10398. doi: 10.1073/pnas.91.22.10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown S J, Kellett P J, Lippard S J. Science. 1993;261:603–605. doi: 10.1126/science.8342024. [DOI] [PubMed] [Google Scholar]
- 64.McA'Nulty M M, Lippard S J. Mutat Res. 1996;362:75–86. doi: 10.1016/0921-8777(95)00037-2. [DOI] [PubMed] [Google Scholar]