Abstract
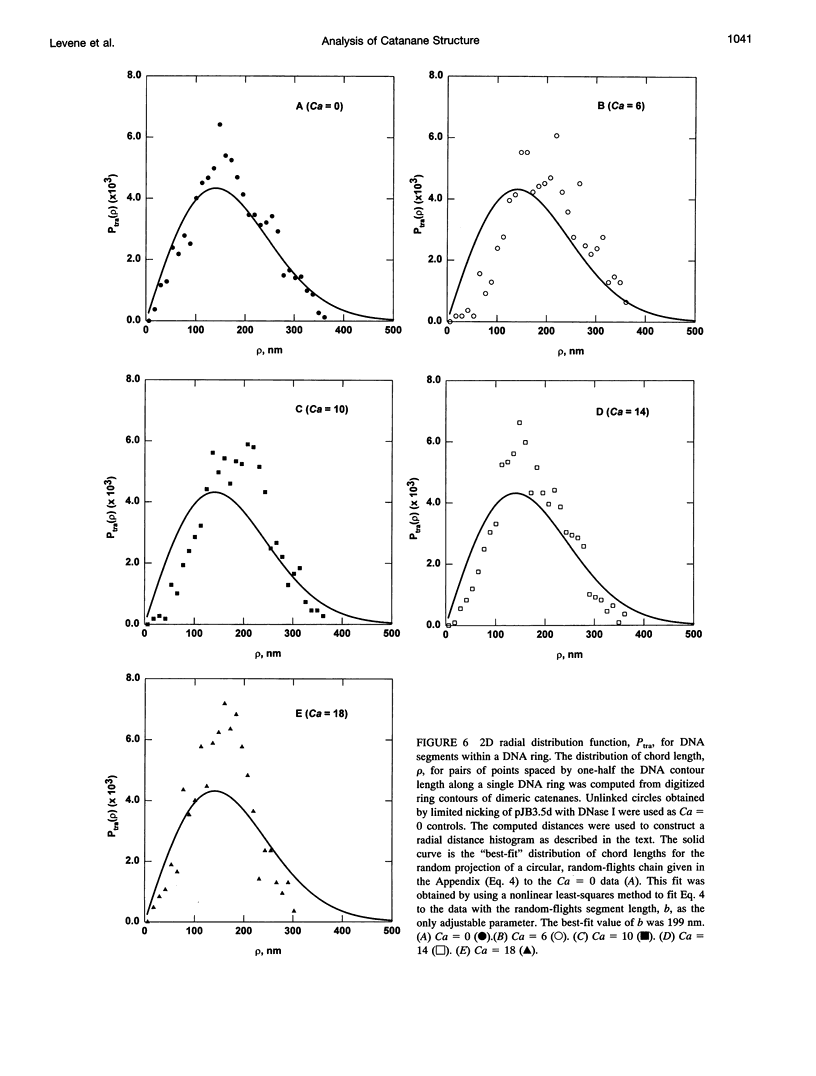
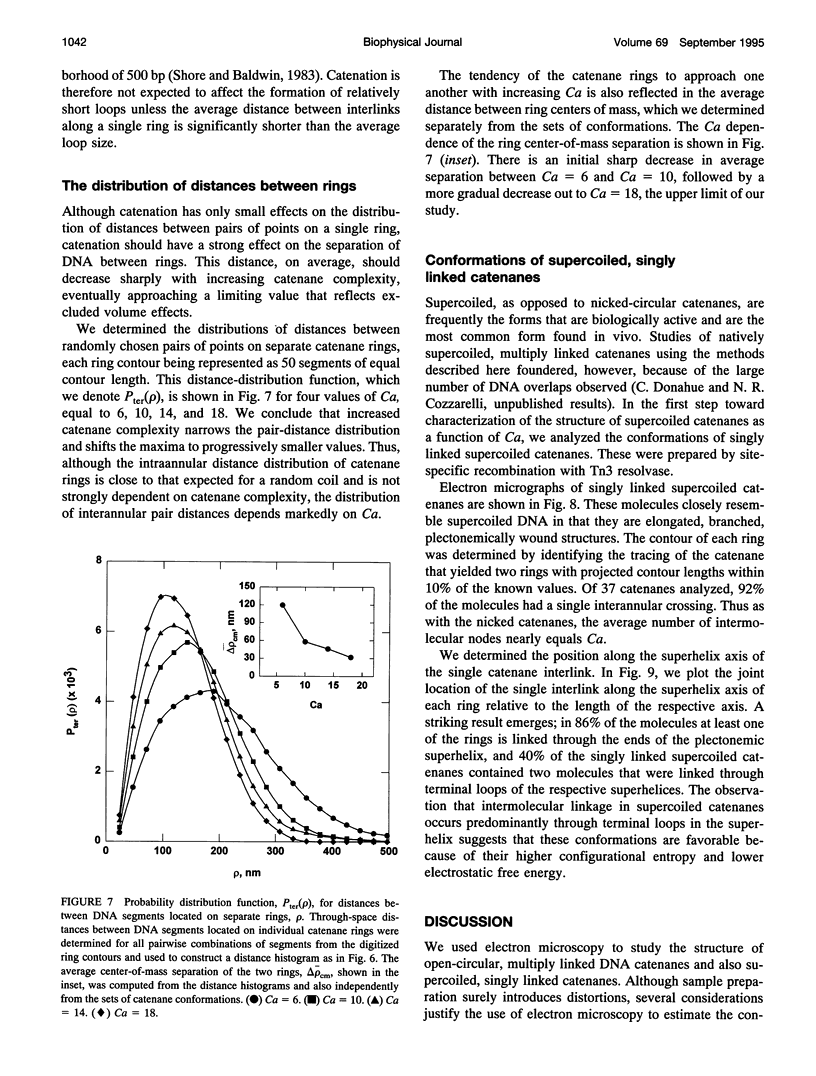
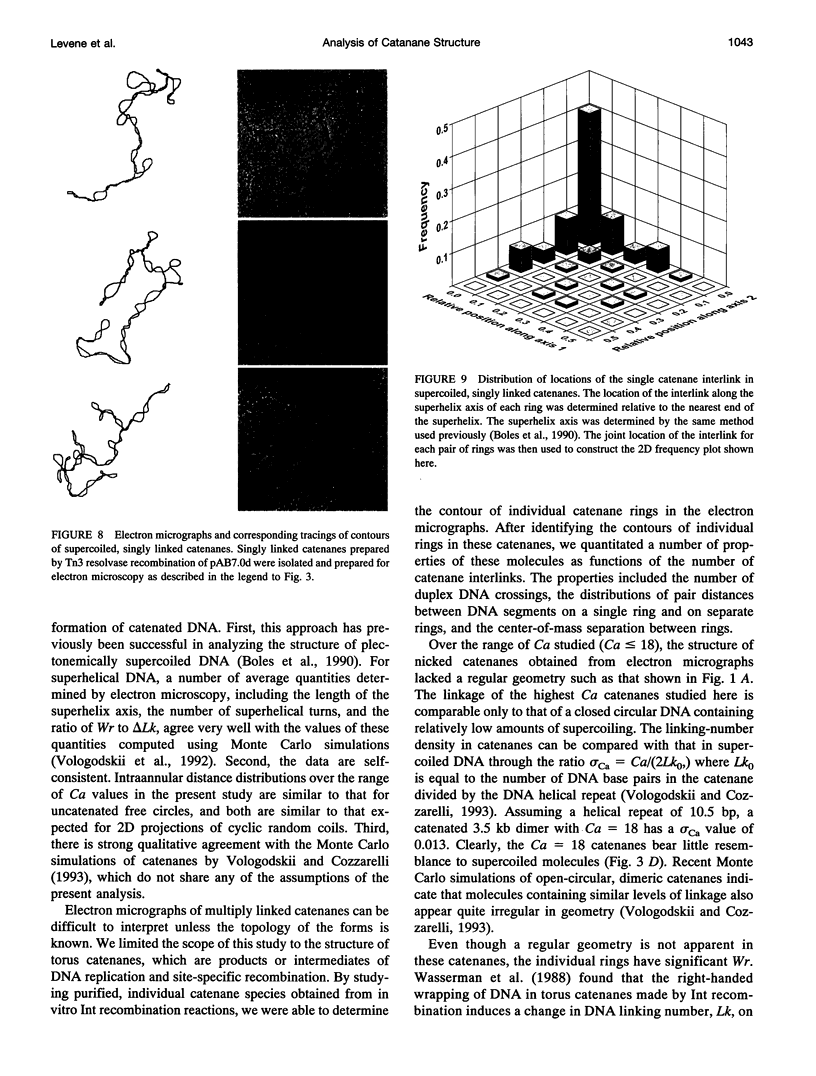
We analyzed the structure of open-circular and supercoiled dimeric DNA catenanes generated by site-specific recombination in vitro. Electron microscopy of open-circular catenanes shows that the number of duplex crossings in a plane is a linear function of the number of catenane interlinks (Ca/2), and that the length of the catenane axis is constant, independent of Ca. These relationships are similar to those observed with supercoiled DNA. Statistical analyses reveal, however, that the conformations of the individual rings of the catenanes are similar to those of unlinked circles. The distribution of distances between randomly chosen points on separate rings depends strongly on Ca and is consistent with a sharp decrease in the center-of-mass separation between rings with increasing Ca. Singly linked supercoiled catenanes are seen by microscopy to be linked predominantly through terminal loops in the respective superhelices. The observations suggest that chain entropy is a major factor determining the conformation of DNA catenanes.
Full text
PDF

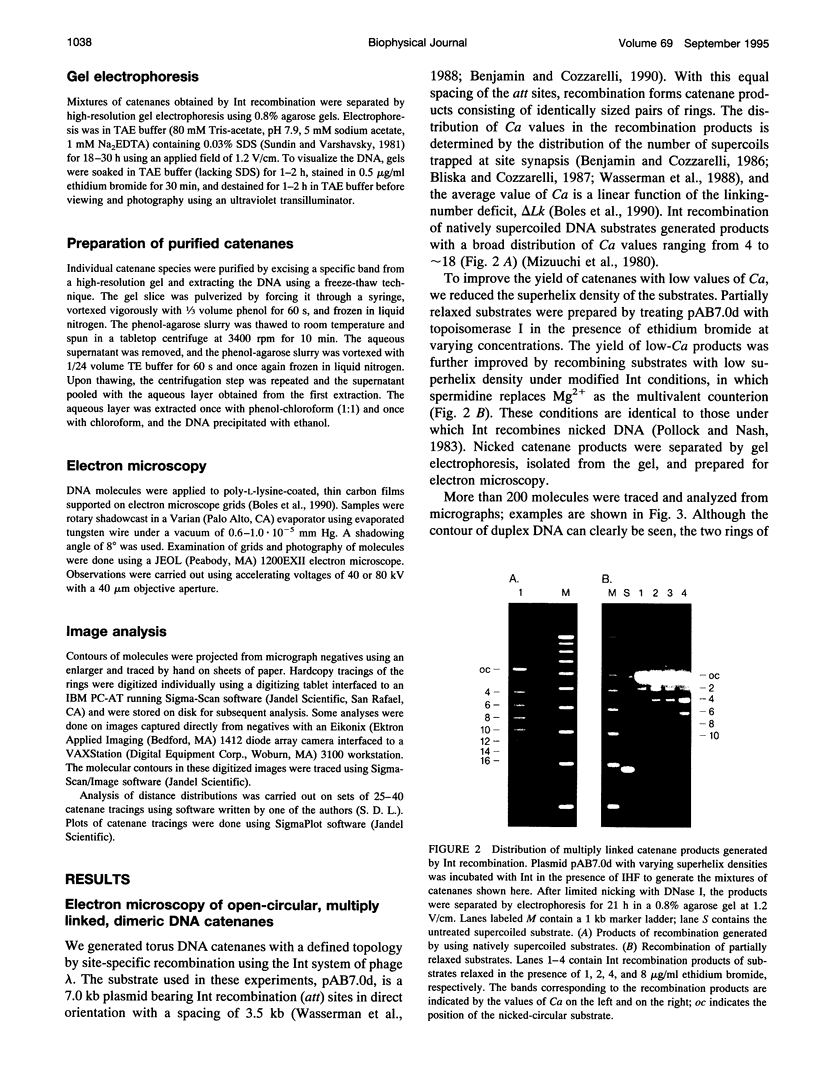
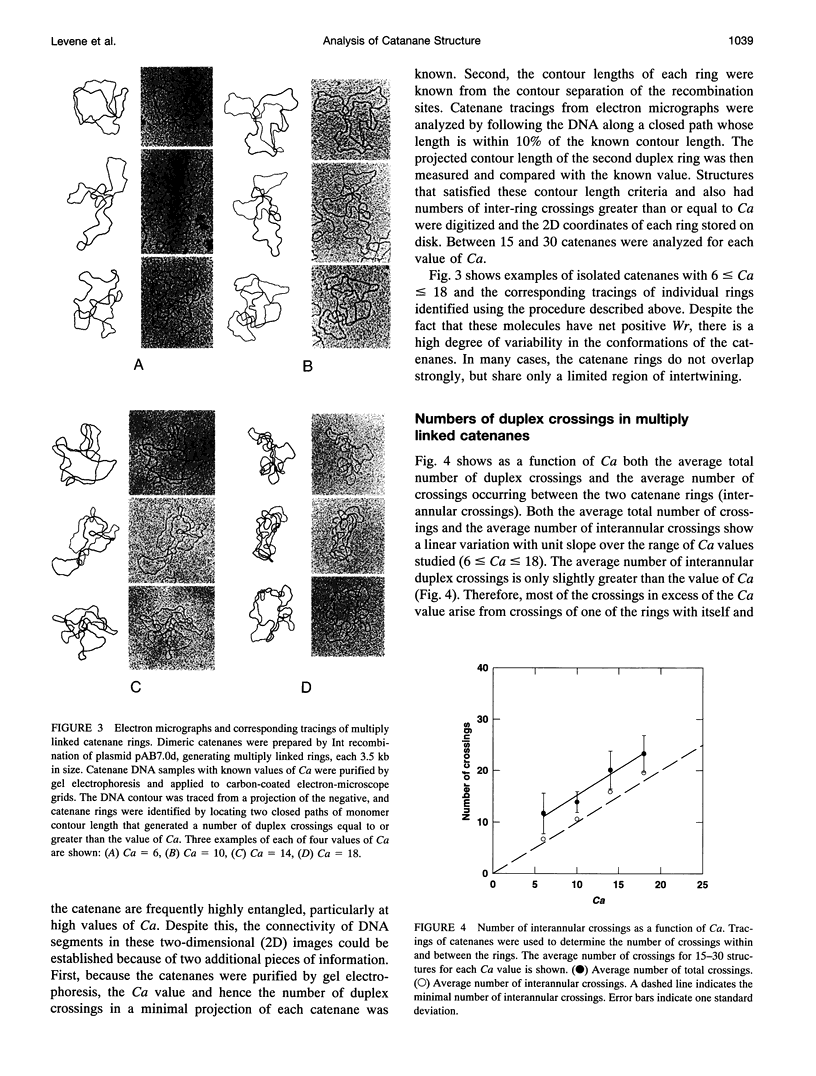
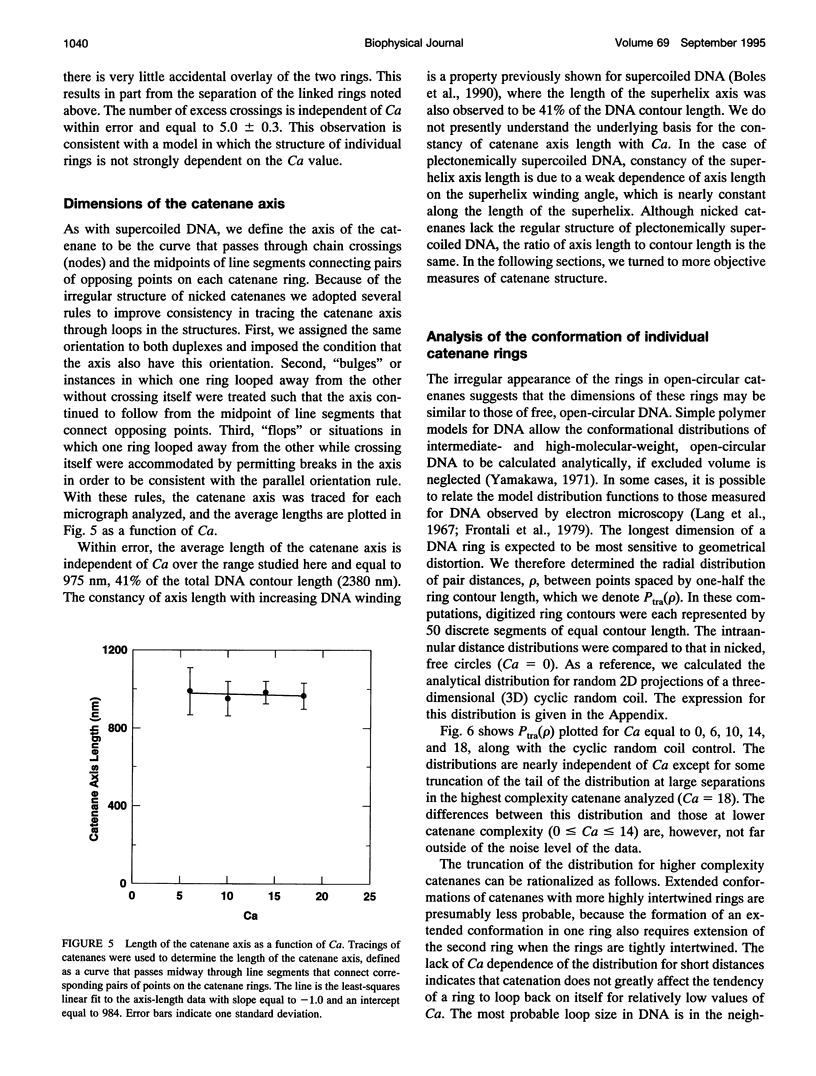


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams D. E., Shekhtman E. M., Zechiedrich E. L., Schmid M. B., Cozzarelli N. R. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992 Oct 16;71(2):277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- Barzilai R. SV40 DNA: quantitative conversion of closed circular to open circular form by an ethidium bromide-restricted endonuclease. J Mol Biol. 1973 Mar 15;74(4):739–742. doi: 10.1016/0022-2836(73)90062-4. [DOI] [PubMed] [Google Scholar]
- Benjamin H. W., Cozzarelli N. R. Geometric arrangements of Tn3 resolvase sites. J Biol Chem. 1990 Apr 15;265(11):6441–6447. [PubMed] [Google Scholar]
- Bliska J. B., Cozzarelli N. R. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987 Mar 20;194(2):205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Boles T. C., White J. H., Cozzarelli N. R. Structure of plectonemically supercoiled DNA. J Mol Biol. 1990 Jun 20;213(4):931–951. doi: 10.1016/S0022-2836(05)80272-4. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Jendrisak J. J., Hager D. A., Burgess R. R. Purification and characterization of wheat germ DNA topoisomerase I (nicking-closing enzyme). J Biol Chem. 1981 Jun 10;256(11):5860–5865. [PubMed] [Google Scholar]
- Frontali C., Dore E., Ferrauto A., Gratton E., Bettini A., Pozzan M. R., Valdevit E. An absolute method for the determination of the persistence length of native DNA from electron micrographs. Biopolymers. 1979 Jun;18(6):1353–1373. doi: 10.1002/bip.1979.360180604. [DOI] [PubMed] [Google Scholar]
- Krasnow M. A., Cozzarelli N. R. Site-specific relaxation and recombination by the Tn3 resolvase: recognition of the DNA path between oriented res sites. Cell. 1983 Apr;32(4):1313–1324. doi: 10.1016/0092-8674(83)90312-4. [DOI] [PubMed] [Google Scholar]
- Lang D., Bujard H., Wolff B., Russell D. Electron microscopy of size and shape of viral DNA in solutions of different ionic strengths. J Mol Biol. 1967 Jan 28;23(2):163–181. doi: 10.1016/s0022-2836(67)80024-x. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Gellert M., Weisberg R. A., Nash H. A. Catenation and supercoiling in the products of bacteriophage lambda integrative recombination in vitro. J Mol Biol. 1980 Aug 25;141(4):485–494. doi: 10.1016/0022-2836(80)90256-9. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A., Flamm E., Weisberg R. A., Miller H. I. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J Bacteriol. 1987 Sep;169(9):4124–4127. doi: 10.1128/jb.169.9.4124-4127.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Purification and properties of the Escherichia coli protein factor required for lambda integrative recombination. J Biol Chem. 1981 Sep 10;256(17):9246–9253. [PubMed] [Google Scholar]
- Pollock T. J., Nash H. A. Knotting of DNA caused by a genetic rearrangement. Evidence for a nucleosome-like structure in site-specific recombination of bacteriophage lambda. J Mol Biol. 1983 Oct 15;170(1):1–18. doi: 10.1016/s0022-2836(83)80224-1. [DOI] [PubMed] [Google Scholar]
- Ryan K. A., Shapiro T. A., Rauch C. A., Englund P. T. Replication of kinetoplast DNA in trypanosomes. Annu Rev Microbiol. 1988;42:339–358. doi: 10.1146/annurev.mi.42.100188.002011. [DOI] [PubMed] [Google Scholar]
- Rybenkov V. V., Cozzarelli N. R., Vologodskii A. V. Probability of DNA knotting and the effective diameter of the DNA double helix. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5307–5311. doi: 10.1073/pnas.90.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S. Y., Wang J. C. Knotting of a DNA chain during ring closure. Science. 1993 Apr 23;260(5107):533–536. doi: 10.1126/science.8475384. [DOI] [PubMed] [Google Scholar]
- Shore D., Baldwin R. L. Energetics of DNA twisting. I. Relation between twist and cyclization probability. J Mol Biol. 1983 Nov 15;170(4):957–981. doi: 10.1016/s0022-2836(83)80198-3. [DOI] [PubMed] [Google Scholar]
- Simpson L. The mitochondrial genome of kinetoplastid protozoa: genomic organization, transcription, replication, and evolution. Annu Rev Microbiol. 1987;41:363–382. doi: 10.1146/annurev.mi.41.100187.002051. [DOI] [PubMed] [Google Scholar]
- Spengler S. J., Stasiak A., Cozzarelli N. R. The stereostructure of knots and catenanes produced by phage lambda integrative recombination: implications for mechanism and DNA structure. Cell. 1985 Aug;42(1):325–334. doi: 10.1016/s0092-8674(85)80128-8. [DOI] [PubMed] [Google Scholar]
- Sundin O., Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981 Sep;25(3):659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. V., Cozzarelli N. R. Monte Carlo analysis of the conformation of DNA catenanes. J Mol Biol. 1993 Aug 20;232(4):1130–1140. doi: 10.1006/jmbi.1993.1465. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. V., Levene S. D., Klenin K. V., Frank-Kamenetskii M., Cozzarelli N. R. Conformational and thermodynamic properties of supercoiled DNA. J Mol Biol. 1992 Oct 20;227(4):1224–1243. doi: 10.1016/0022-2836(92)90533-p. [DOI] [PubMed] [Google Scholar]
- Wasserman S. A., Cozzarelli N. R. Biochemical topology: applications to DNA recombination and replication. Science. 1986 May 23;232(4753):951–960. doi: 10.1126/science.3010458. [DOI] [PubMed] [Google Scholar]
- Wasserman S. A., White J. H., Cozzarelli N. R. The helical repeat of double-stranded DNA varies as a function of catenation and supercoiling. Nature. 1988 Aug 4;334(6181):448–450. doi: 10.1038/334448a0. [DOI] [PubMed] [Google Scholar]