Abstract
The transcription factor NtcA is a global regulator of nitrogen homeostasis in cyanobacteria. It thus positively regulates the expression of genes related to nitrogen assimilation such as glnA (which encodes glutamine synthetase) and ntcA itself in response to nitrogen shortage or depletion. The binding of NtcA to the glnA and ntcA promoters of Synechococcus sp. PCC 7942 in vitro now has been shown to be enhanced by 2-oxoglutarate. In vitro analysis of gene transcription also revealed that the interaction of NtcA with its promoter element was not sufficient for activation of transcription, and 2-oxoglutarate was required for transcriptional initiation by NtcA. Given that the intracellular concentration of 2-oxoglutarate is inversely related to nitrogen availability, it is proposed that this metabolite functions as a signaling molecule that transmits information on cellular nitrogen status to NtcA and thereby regulates the transcription of genes related to nitrogen assimilation in cyanobacteria.
Keywords: regulatory factor|cyanobacteria|RNA polymerase|nitrogen assimilation
Cyanobacteria constitutes a diverse group of eubacterial strains that are characterized by the ability to perform oxygen-evolving photosynthesis. Although the morphology and the ability to develop into differentiated cells such as N2-fixing heterocysts vary greatly among cyanobacteria, most of these organisms share many metabolic characteristics (1, 2).
The predominant nitrogen compounds assimilated by cyanobacteria are ammonium and nitrate ions. Nitrate is reduced by the successive actions of nitrate reductase and nitrite reductase, and the resulting ammonium usually is incorporated by the glutamine synthetase (GS; ref. 1) and glutamate synthase (GOGAT) cycle (1–3). Diazotrophic strains of cyanobacteria are able to use gaseous nitrogen, but the fixed nitrogen compound, ammonium, is also assimilated by the GS-GOGAT cycle. The carbon skeleton for nitrogen assimilation is 2-oxoglutarate (2-OG), which is synthesized from isocitrate by isocitrate dehydrogenase. Because of the lack of 2-OG dehydrogenase, isocitrate is used solely for nitrogen assimilation in cyanobacteria. Also as a result of the lack of 2-OG dehydrogenase, the tricarboxylic acid cycle usually is incomplete in this phylum (1).
The regulation of nitrogen assimilation is relatively well characterized in enteric bacteria (4, 5). Expression of the glnA gene, which encodes GS, and other nitrogen assimilation-related genes requires an alternative RNA polymerase σ factor known as σN (5). The RNA polymerase that contains σN is controlled by a two-component regulatory system comprising NtrB and NtrC, the activity of which in turn is determined by the uridylylation status of the trimeric protein PII (the glnB product; refs. 6 and 7). The uridylylation status of PII is determined by the activity of the dual-function uridylyl-removing enzyme uridylyltransferase, which is regulated by the ratio of the intracellular concentrations of 2-OG and glutamine (8). PII itself also binds 2-OG and thereby senses the nitrogen status of the cell. Furthermore, the uridylylation status of PII is a determinant of the adenylylation of GS, which regulates the activity of this enzyme (9). Thus, nitrogen status is assessed on the basis of the concentrations of 2-OG and glutamine and modulates the expression and activity of the key assimilatory enzyme GS.
The regulation of nitrogen assimilation in cyanobacteria seems to differ markedly from that in enteric bacteria. Although a conserved PII protein has been identified in cyanobacteria (10), it is unlikely to participate in the transcriptional control of nitrogen assimilation-related genes (11). Rather, a unique transcription factor, NtcA, acts as a global regulator of nitrogen homeostasis in cyanobacteria (12, 13) by promoting the expression of various genes important in nitrogen assimilation. The ntcA gene also is required for the initiation of heterocyst differentiation in filamentous species (14), indicating that it contributes generally to nitrogen-related signal transduction in cyanobacteria. Moreover, NtcA is known to control the expression of genes implicated in carbon assimilation (3). Despite the importance of the NtcA signaling pathway, the molecular basis of the regulation of transcription by NtcA remains unclear. NtcA is a member of the family of transcription factors defined by CRP of Escherichia coli (13) and is thought to activate transcription by binding to a conserved sequence motif centered ≈40 bp upstream from the transcription start site (3, 15). However, the mechanism by which NtcA senses changes in nitrogen status of the cyanobacterial cell is unknown.
We have now examined the effects of 2-OG on the binding of NtcA to, and on the transcriptional activity of, two NtcA-dependent promoters, those of glnA and ntcA, of Synechococcus sp. PCC 7942. Our data indicate that 2-OG not only promotes the binding of NtcA to these promoters but also is essential for initiation of transcription. On the basis of our in vitro results and previous in vivo observations (16, 17), we propose a molecular mechanism for the sensing of cellular nitrogen status in cyanobacteria.
Materials and Methods
Bacterial Strains and Growth Conditions: Synechococcus sp.
PCC 7942 was grown photoautotrophically on BG-11 plates or in BG-11 liquid medium (18) at 30°C under continuous illumination from fluorescent lamps. Liquid cultures were bubbled with 1% CO2 in air. E. coli DH5α, used as the host for plasmid propagation and recombinant protein expression, was cultivated in LB medium at 37°C.
Enzymes and Reagents.
Enzymes were obtained from Takara Shuzo (Kyoto), and 2-OG and glutamine were from Sigma and Kanto Chemicals (Tokyo), respectively. Other reagents were obtained from Sigma or as indicated.
Proteins.
Expression and purification of recombinant NtcA were performed essentially as described (19) with some modifications. In brief, E. coli DH5α harboring pMNTCA, which encodes a fusion of maltose-binding protein and NtcA, was cultivated at 37°C in LB medium supplemented with 1 mM isopropyl-β-d-thiogalactopyranoside, and the chimeric protein was purified with the use of an amylose resin (New England Biolabs). The purified fusion protein was cleaved with factor Xa and applied to a Poros HQ anion-exchange column (5 × 100 mm, PerSeptive Biosystems, Framingham, MA), which then was washed with HGED buffer (10 mM Hepes-KOH, pH 8.0/5% glycerol/0.1 mM EDTA/0.1 mM DTT) containing 0.1 M NaCl. Elution was performed with a linear gradient of 0.1–1 M NaCl in the same buffer. Fractions containing NtcA were concentrated with an Ultrafree-15 centrifugal concentrator (Millipore), dialyzed against HGED buffer containing 50% glycerol, and stored at −20°C. RNA polymerase from Synechococcus sp. PCC 7942 and the principal σ factor (RpoD1) were purified as described (20).
DNA Fragments.
A 309-bp DNA fragment corresponding to nucleotides −172 to +137 (relative to the NtcA-dependent transcription start site) of glnA (15, 21) was amplified by PCR from total Synechococcus DNA with the primers glnA-i (5′-ACCATGCAGACTAGTCCTGCC-3′) and glnA-ii (5′-CTCCTTGTGGGATCCTGGTGG-3′). The PCR product was inserted into the polylinker site of pBluescript KS(+) (Toyobo, Osaka) with the use of the attached SpeI and BamHI restriction sites. The resulting plasmid, pKS-glnA, then was used as a template for PCR with the same primer set (glnA-i and gln-ii) or the primers glnA-i and glnA-iii (5′-AAACAACGCTGCTGCGATCGCAGCTGGATC-3′) to generate DNA fragments termed templates I and II, respectively. The 235-bp template II corresponds to nucleotides −172 to +63 of glnA.
Similarly, with the primers ntcA-i (5′-GCCTGTGACTAGTGCTTTCTT-3′) and ntcA-ii (5′-TGCCTTATGGATCCGCGCAAG-3′), a 262-bp DNA fragment corresponding to nucleotides −161 to +101 (relative to the NtcA-dependent transcription start site) of ntcA (13, 15) was amplified from total Synechococcus DNA and then inserted into the polylinker site of pBluescript KS with the use of the attached SpeI and BamHI restriction sites. The resulting plasmid, pKS-ntcA, was used as a template for PCR with the same primer set (ntcA-i and ntcA-ii) to generate template III.
All PCRs were performed for 24 cycles with 1 μg of Synechococcus DNA or 0.1 μg of plasmid DNA as template. Each cycle comprised incubation at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, and the products were purified by PAGE. Cyanobacterial DNA was prepared as described (22).
A 24-bp double-stranded DNA fragment containing the NtcA-binding sequence of the glnA promoter region (15, 21), used as a specific competitor for electrophoretic mobility-shift assay (EMSA) analysis, was prepared by annealing the oligonucleotides 5′-TTTATGTATCAGCTGTTACAAAAG-3′ and 5′-CTTTTGTAACAGCTGATACATAAA-3′. A nonspecific competitor was prepared similarly by annealing the oligonucleotides 5′-TTTATACGTCAGCTGTCGTAAAAG-3′ and 5′-CTTTTACGACAGCTGACGTATAAA-3′, in which conserved nucleotides of the NtcA binding site were specifically substituted.
EMSA Analysis.
The 5′ ends of DNA fragments were labeled by incubation with polynucleotide kinase and [γ-32P]ATP (Amersham Pharmacia). 32P-Labeled DNA fragments (1 nM) corresponding to the glnA promoter (template I) or the ntcA promoter (template III) were mixed with purified NtcA and various concentrations of additives as indicated in 20 μl of T buffer [50 mM Hepes-KOH, pH 8.0/3 mM MgCl2/20% glycerol/1 mM DTT/50 mM potassium glutamate/BSA (25 μg/ml)]. After incubation for 20 min at 30°C, the mixtures were subjected to PAGE on a native 4% gel. Gels were analyzed with a BAS1000 image analyzer (Fuji).
In Vitro Transcription.
Transcription reactions were performed under standard conditions for E. coli RNA polymerase (23) with modifications. The Synechococcus core enzyme was mixed with a 3-fold molar excess of the purified σ protein (RpoD1), and the mixture was incubated for 20 min at 30°C to allow formation of the holoenzyme. A transcription reaction mixture (35 μl) comprising 5.7 nM template DNA, 86 nM RNA polymerase, and 229 nM NtcA in T buffer was incubated for 20 min at 30°C, after which RNA synthesis was initiated by the addition of 15 μl of a prewarmed substrate mixture containing 160 μM each of ATP, GTP, and CTP as well as 50 μM UTP and 2 μCi (1 Ci = 37 GBq) of [α-32P]UTP (Amersham Pharmacia) in T buffer. When added, 2-OG was included in both the reaction mixture and substrate mixture. After incubation for 5 min at 30°C, the reaction was terminated by the addition of 50 μl of ice-cold stop solution containing 40 mM EDTA and E. coli tRNA (300 μg/ml), and the nucleic acids then were precipitated with ethanol. Transcripts were fractionated by PAGE on a 7% gel containing 8 M urea, and they were analyzed with a BAS1000 instrument.
Results
Sequence-Specific Binding of NtcA to DNA.
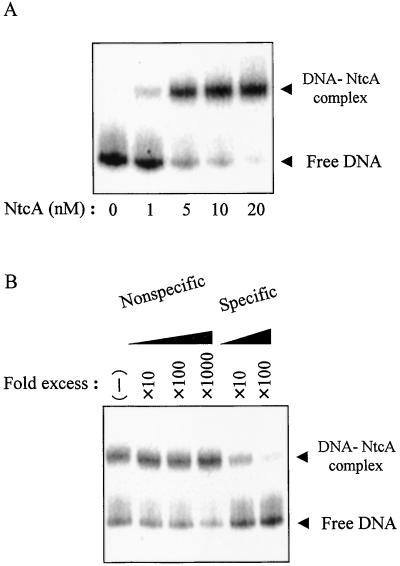
Previous studies have demonstrated that NtcA binds to a 14-bp conserved DNA sequence and thereby activates, or in a few instances represses, the initiation of transcription (3, 15, 19, 24, 25). We monitored the binding of recombinant NtcA to a 32P end-labeled glnA promoter fragment containing the NtcA binding site by EMSA analysis. NtcA reduced the electrophoretic mobility of the DNA probe, and the amount of the DNA–NtcA complex increased in proportion to the concentration of NtcA (Fig. 1A). To examine the sequence specificity of the binding of NtcA to the DNA probe, we performed the binding reaction in the presence of either a synthetic double-stranded oligonucleotide containing an NtcA binding site or a control oligonucleotide that does not contain the binding sequence as competitors for the 32P-labeled probe. Whereas the nonspecific competitor had virtually no effect on the binding of NtcA to the 32P-labeled probe even at a 1,000-fold molar excess, the specific competitor inhibited the interaction of NtcA with the probe almost completely at a 100-fold molar excess (Fig. 1B). We thus were able to observe the sequence-specific binding of NtcA to DNA in our assay system.
Figure 1.
Specific binding of NtcA to a glnA promoter fragment. (A) The binding of NtcA at the indicated concentrations to a 32P-labeled glnA promoter fragment (template I) was examined by EMSA analysis. (B) The binding of 5 nM NtcA to the 32P-labeled glnA promoter fragment was assayed in the absence or presence of unlabeled specific or nonspecific competitor oligonucleotides at the indicated molar excesses. The positions of the free probe and the DNA–NtcA complex are indicated.
Promotion of the Specific Binding of NtcA to DNA by 2-OG.
Our data indicated that NtcA does not require a specific effector for its interaction with DNA (Fig. 1A) if the protein concentration is sufficiently high. Physiologically, NtcA-dependent promoters are repressed in the presence of ammonium and are activated by either nitrogen starvation or growth on poor nitrogen sources. Such effects might result from either activation or inactivation of the NtcA transcription factor. In the case of positive regulation, the default status of NtcA-dependent promoters would be inactive, and a specific molecule or signaling mechanism might enhance either the binding of NtcA to DNA or a later step of transcriptional initiation. In the case of negative regulation, the default status of NtcA-dependent promoters would be active, and an effector might inhibit the binding of NtcA to DNA or a subsequent event. To distinguish between these possibilities, we first examined the effects of 2-OG and glutamine on the NtcA-DNA interaction, given that the concentrations of these metabolites are most likely to reflect the nitrogen status of the cell. The intracellular concentration of 2-OG increases, that of glutamine decreases during nitrogen depletion, and vice versa during nitrogen repletion (17).
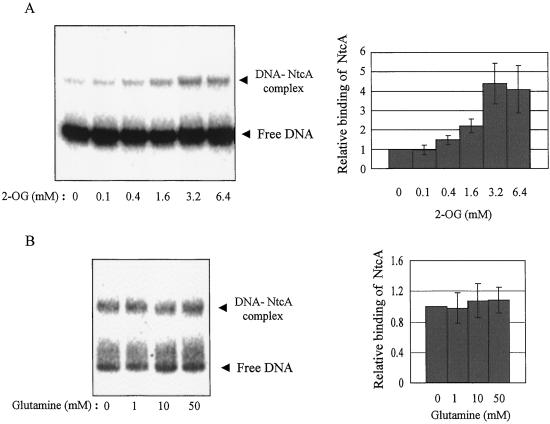
If 2-OG is an effector for positive transcriptional regulation, then the specific binding of NtcA to DNA might be promoted in the presence of this metabolite. To examine this possibility, we performed EMSA analysis in the presence of various concentrations of 2-OG. The assays were performed with a relatively low concentration (1 nM) of NtcA (Fig. 1A) to increase the sensitivity for detection of a positive effect of 2-OG. The specific binding of NtcA to DNA indeed was increased by 2-OG in a concentration-dependent manner, with the maximal effect being apparent at a 2-OG concentration of 3.2 mM (Fig. 2A). Similar experiments performed with a relatively high concentration (3 nM) of NtcA to detect a possible negative effect of glutamine on the NtcA-DNA interaction failed to reveal any such effect of glutamine at concentrations of up to 50 mM (Fig. 2B). EMSA analysis with an ntcA promoter fragment containing the NtcA binding site yielded similar results with regard to the effects of 2-OG and glutamine on the binding of NtcA (data not shown), indicating that the stimulatory action of 2-OG is not restricted to the glnA promoter.
Figure 2.
Effects of 2-OG and glutamine on the specific binding of NtcA to DNA. (A) The binding of 1 nM NtcA to a 32P-labeled glnA promoter fragment (template I) was examined by EMSA analysis in the presence of the indicated concentrations of 2-OG (Left). The relative band intensities for each DNA–NtcA complex were quantified, and data are expressed as means ± SD of values from three independent experiments (Right). (B) The binding of 3 nM NtcA to DNA was analyzed as described for A with the exception that 2-OG was replaced by glutamine.
Transcriptional Activation of glnA and ntcA Promoters by 2-OG in Vitro.
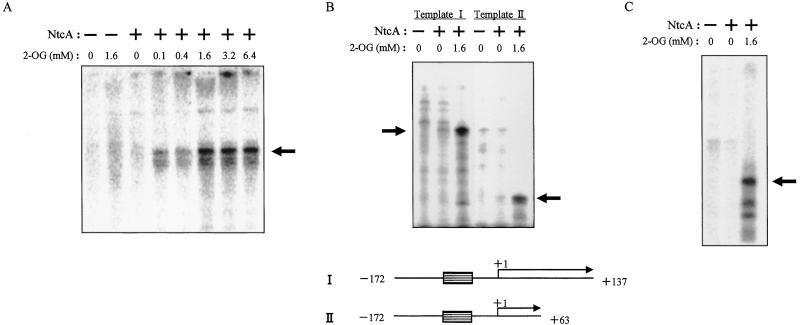
To examine whether the 2-OG-induced enhancement of the binding of NtcA to DNA actually results in transcriptional activation, we performed in vitro transcription experiments with the NtcA-dependent promoters of glnA and ntcA. An RNA polymerase holoenzyme was reconstituted from the purified core enzyme and recombinant principal σ factor (RpoD1) and then was used for run-off transcription assays. Transcriptional activation was not observed with both promoters in the absence of 2-OG even when the concentration of NtcA was sufficiently high to saturate its binding sites (data not shown). Under conditions in which the binding sites were fully occupied by NtcA even in the absence of 2-OG, this metabolite induced a marked increase in NtcA-dependent transcription from the glnA promoter (Fig. 3A). The effect of 2-OG was concentration-dependent, being maximal (5.8 ± 0.7-fold increase; mean ± SD of three independent experiments) at 1.6 mM. The identity of the glnA transcripts was confirmed with the use of two template DNA fragments that were expected to produce transcripts of different lengths (Fig. 3B). Activation of NtcA-dependent transcription from the ntcA promoter by 2-OG was demonstrated also (Fig. 3C). Thus, 2-OG appears to activate NtcA-dependent transcription by promoting both the interaction of NtcA with DNA and transcriptional initiation.
Figure 3.
Effects of 2-OG on NtcA-dependent transcription in vitro. (A) Run-off transcription reactions were performed with the glnA promoter (template I) in the absence (−) or presence (+) of NtcA and in the presence of the indicated concentrations of 2-OG. The position of the 137-nt transcript is indicated by the arrow. (B) Run-off transcription reactions were performed as described for A but with two different glnA promoter fragments (templates I and II) that are expected to yield transcripts of different sizes [137 and 63 nt (indicated by arrows on the autoradiogram), respectively]. The structures of the templates are represented schematically below the autoradiogram with the NtcA binding sites indicated as hatched boxes and the direction of transcription indicated by arrows. (C) Run-off transcription assays were performed as described for A but with an ntcA promoter fragment (template III). The position of the 101-nt transcript is indicated by the arrow.
Discussion
NtcA is a global regulator of nitrogen homeostasis in cyanobacteria, being required for the activation of genes related to nitrogen assimilation under conditions of nitrogen limitation. Given that transcription from all known NtcA-dependent promoters is repressed under conditions of nitrogen repletion (that is, in the presence of ammonium in the medium), NtcA has been thought to sense cellular nitrogen status and regulate transcription accordingly. The promoters of NtcA-dependent genes contain the consensus sequence GTAN8TACN22TAN3T, in which GTAN8TAC is the NtcA binding motif and TAN3T is the −10 promoter element (3, 15). In vitro analysis has established that NtcA recognizes and binds to the sequence GTAN8TAC (26), but the molecular basis for the nitrogen responsiveness of transcription has remained unknown. As shown by previous studies (15, 19, 24, 25) and confirmed by our present results, the binding of NtcA to target promoters does not require the presence of any specific compound. However, we have now shown that 2-OG enhances the binding of NtcA to DNA at low protein concentrations. Our in vitro transcription analysis also revealed that DNA binding per se is insufficient for promotion of transcription by NtcA, and 2-OG is required for transcription initiation. Transcriptional activity thus was increased by 2-OG in a concentration-dependent manner with the maximal effect apparent at a concentration of 1.6 mM, suggesting that 2-OG might act as a metabolic signal in the regulation of NtcA-dependent gene expression. Given that 2-OG is synthesized from isocitrate and serves as the substrate for the GS-GOGAT pathway of ammonium assimilation in cyanobacteria, the intracellular concentration of 2-OG likely reflects both the nitrogen status and the carbon status of the cell. The intracellular concentration of 2-OG was shown recently to change according to cellular nitrogen status in cyanobacteria (17); it thus increased from 0.06 mM in ammonium-grown cells to 0.44 mM in nitrogen-depleted cells, as calculated from the data provided. Together, the results of in vitro and in vivo experiments therefore suggest that the nitrogen responsiveness of NtcA-dependent promoters is attributable primarily to the dependence of transcriptional initiation on 2-OG.
In addition to its role in transcription initiation, 2-OG was shown to promote the binding of NtcA (at low concentrations) to DNA in a concentration-dependent manner. In Synechococcus sp. PCC 7942, expression of ntcA is activated by NtcA itself in response to nitrogen shortage (15). Hence, the concentration of NtcA fluctuates depending on cellular nitrogen status, being low in nitrogen-replete cells and high in nitrogen-limited cells (15). During adaptation from nitrogen-replete conditions to nitrogen-limited conditions, the enhancement by 2-OG of the binding of NtcA to DNA would be expected to play an important role in transcriptional activation of ntcA and of other NtcA-dependent genes until the concentration of NtcA becomes sufficiently high to allow 2-OG-independent binding.
The addition of ammonium to nitrogen-limited cells, which express NtcA-dependent genes at a high level, results in the immediate repression of transcription of these genes (17, 27). Given that the intracellular abundance of NtcA in such cells presumably is also high, NtcA would be expected to occupy its binding sites irrespective of the 2-OG concentration. The observed ammonium-induced repression of gene transcription therefore is likely attributable not to the dissociation of NtcA from DNA but to the rapid decrease in the concentration of 2-OG that occurs after the addition of ammonium (17). Thus, the dual role of 2-OG in the regulation of NtcA-dependent genes (enhancement of both NtcA binding to DNA and initiation of transcription) ensures rapid activation and repression, respectively, of transcription in response to changes in cellular nitrogen status.
Transcription from NtcA-dependent promoters is induced not only when cells are deprived of a nitrogen source but also when ammonium assimilation by the GS-GOGAT cycle is inhibited by l-methionine-D,L-sulfoxime (MSX, an inhibitor of GS) or by 5-diazo-6-oxo-l-norleucine (DON; an inhibitor of glutamine amidotransferases including GOGAT; ref. 27). Given that preincubation of cells with glutamine prevented MSX-induced transcription but not DON-induced transcription, we previously hypothesized that a nitrogenous compound produced by transfer of the amide nitrogen of glutamine or a metabolite thereof acts as a negative regulator of transcription (27, 28). However, inhibition of the GS-GOGAT cycle would be expected to result in accumulation of 2-OG, which should be prevented by exogenous glutamine in MSX-treated cells but not in DON-treated cells, because DON inhibits consumption of 2-OG by the GOGAT reaction. These previous observations thus are accounted for mostly by the effects of 2-OG on both NtcA binding to DNA and transcriptional initiation. We therefore propose that 2-OG is the major regulator of NtcA-dependent transcription, although an additional role for a negative regulator cannot be excluded.
In enteric bacteria, nitrogen status is sensed by uridylyl-removing enzyme uridylyltransferase as the ratio of the concentrations of 2-OG and glutamine. The uridylyltransferase-dependent uridylylation status of PII determines the activity of GS as well as that of the RNA polymerase containing σN through the two-component regulatory system comprising NtrB and NtrC (5–9). PII is present also in cyanobacteria, and its functions related to regulation of the nitrate/nitrite uptake have been well characterized (11). However, the significance of this sensory protein for transcriptional regulation in these organisms remains obscure (11). Rather, NtcA seems to regulate the expression of nitrogen assimilation-related genes in cyanobacteria through direct sensing of the intracellular concentration of 2-OG. This difference between enteric bacteria and cyanobacteria may be caused by the metabolic divergence of these two groups; 2-OG is used only for nitrogen assimilation in cyanobacteria, whereas it is both a substrate for nitrogen assimilation and an intermediate in the tricarboxylic acid cycle in enteric bacteria. Thus the 2-OG concentration alone may be sufficient to represent cellular nitrogen status in cyanobacteria.
We propose that a change in the conformation of NtcA induced by 2-OG both promotes the binding of this transcription factor to DNA and facilitates a later step of transcriptional initiation. In E. coli, the galactose operon promoter (Pgal) is activated by CRP, which binds to the conserved sequence motif centered 41.5 bp upstream of the transcription start site (29–31). CRP interacts simultaneously with multiple regions of RNA polymerase, promoting recruitment of and conformational changes in this enzyme that result in the initiation of transcription from Pgal (30, 31). Given that NtcA is a homolog of CRP and NtcA binds its target promoters at a position similar to that targeted by CRP in Pgal, it is possible that transcriptional activation by NtcA occurs in a manner similar to that proposed for CRP. Further in vitro analysis should clarify the kinetics of transcriptional activation by NtcA and provide insight into the regulation of nitrogen homeostasis in cyanobacteria at the molecular level.
Acknowledgments
We thank laboratory members for technical assistance. This work was supported by Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants 09460046, 11480197, and 11694196 for scientific research (to K.T. and H.T.).
Abbreviations
- GS
glutamine synthetase
- GOGAT
glutamate synthase
- 2-OG
2-oxoglutarate
- CRP
cAMP receptor protein
- EMSA
electrophoretic mobility-shift assay
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Stanier R Y, Cohen-Bazire G. Annu Rev Microbiol. 1977;31:225–274. doi: 10.1146/annurev.mi.31.100177.001301. [DOI] [PubMed] [Google Scholar]
- 2.Meeks J C, Wolk C P, Lockau W, Schilling N, Shaffer P W, Chien W S. J Bacteriol. 1978;134:125–130. doi: 10.1128/jb.134.1.125-130.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herrero A, Muro-Pastor A M, Flores E. J Bacteriol. 2001;183:411–425. doi: 10.1128/JB.183.2.411-425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senior P J. J Bacteriol. 1975;123:407–418. doi: 10.1128/jb.123.2.407-418.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merrick M J, Edwards R A. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang P, Peliska J A, Ninfa A J. Biochemistry. 1998;37:12795–12801. doi: 10.1021/bi9802420. [DOI] [PubMed] [Google Scholar]
- 7.Jiang P, Ninfa A J. J Bacteriol. 1999;181:1906–1911. doi: 10.1128/jb.181.6.1906-1911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ninfa A J. Biochemistry. 1998;37:12782–12794. doi: 10.1021/bi980667m. [DOI] [PubMed] [Google Scholar]
- 9.Ninfa A J, Atkinson M R. Trends Microbiol. 2000;8:172–179. doi: 10.1016/s0966-842x(00)01709-1. [DOI] [PubMed] [Google Scholar]
- 10.Tsinoremas N F, Castets A M, Harrison M A, Allen J F, Tandeau de Marsac N. Proc Natl Acad Sci USA. 1991;88:4565–4569. doi: 10.1073/pnas.88.11.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H-M, Flores E, Herrero A, Houmard J, Tandeau de Marsac N. FEBS Lett. 1998;427:291–295. doi: 10.1016/s0014-5793(98)00451-7. [DOI] [PubMed] [Google Scholar]
- 12.Vega-Palas M A, Madueño F, Herrero A, Flores E. J Bacteriol. 1990;172:643–647. doi: 10.1128/jb.172.2.643-647.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vega-Palas M A, Flores E, Herrero A. Mol Microbiol. 1992;6:1853–1859. doi: 10.1111/j.1365-2958.1992.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 14.Ferias J E, Flores E, Herrero A. Mol Microbiol. 1994;14:823–832. doi: 10.1111/j.1365-2958.1994.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 15.Luque A, Flores E, Herrero A. EMBO J. 1994;13:2862–2869. doi: 10.1002/j.1460-2075.1994.tb06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merida A, Candau P, Florencio F J. J Bacteriol. 1991;173:4095–4100. doi: 10.1128/jb.173.13.4095-4100.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muro-Pastor M I, Reyes J C, Florencio F J. J Biol Chem. 2001;276:38320–38328. doi: 10.1074/jbc.M105297200. [DOI] [PubMed] [Google Scholar]
- 18.Rippka R. Methods Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- 19.Maeda S, Kawaguchi Y, Ohe T, Omata T. J Bacteriol. 1998;180:4080–4088. doi: 10.1128/jb.180.16.4080-4088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goto-Seki A, Shirokane M, Masuda S, Tanaka K, Takahashi H. Mol Microbiol. 1999;34:473–484. doi: 10.1046/j.1365-2958.1999.01608.x. [DOI] [PubMed] [Google Scholar]
- 21.Cohen-Kupiec R, Gurevitz M, Zilberstein A. J Bacteriol. 1993;175:7727–7731. doi: 10.1128/jb.175.23.7727-7731.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golden S S, Brusslan J, Haselkorn R. Methods Enzymol. 1987;153:215–231. doi: 10.1016/0076-6879(87)53055-5. [DOI] [PubMed] [Google Scholar]
- 23.Nomura T, Fujita N, Ishihama A. Nucleic Acids Res. 1986;14:6857–6870. doi: 10.1093/nar/14.17.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramasubramanian T S, Wei T-F, Golden J W. J Bacteriol. 1994;176:1214–1223. doi: 10.1128/jb.176.5.1214-1223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H-M, Vázquez-Bermúdez M F, Tandeau de Marsac N. J Bacteriol. 1999;181:2697–2702. doi: 10.1128/jb.181.9.2697-2702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang F, Wisen S, Widersten M, Bergman B, Mannervik B. J Mol Biol. 2000;301:783–793. doi: 10.1006/jmbi.2000.4000. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki I, Sugiyama T, Omata T. Plant Cell Physiol. 1993;34:1311–1320. [Google Scholar]
- 28.Suzuki I, Sugiyama T, Omata T. J Bacteriol. 1996;178:2688–2694. doi: 10.1128/jb.178.9.2688-2694.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan B, Busby S. Gene. 1989;84:227–236. doi: 10.1016/0378-1119(89)90496-4. [DOI] [PubMed] [Google Scholar]
- 30.Williams R M, Rhodius V A, Bell A I, Kolb A, Busby S. Nucleic Acids Res. 1996;24:1112–1118. doi: 10.1093/nar/24.6.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niu W, Kim Y, Tau G, Heyduk T, Ebright R H. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]