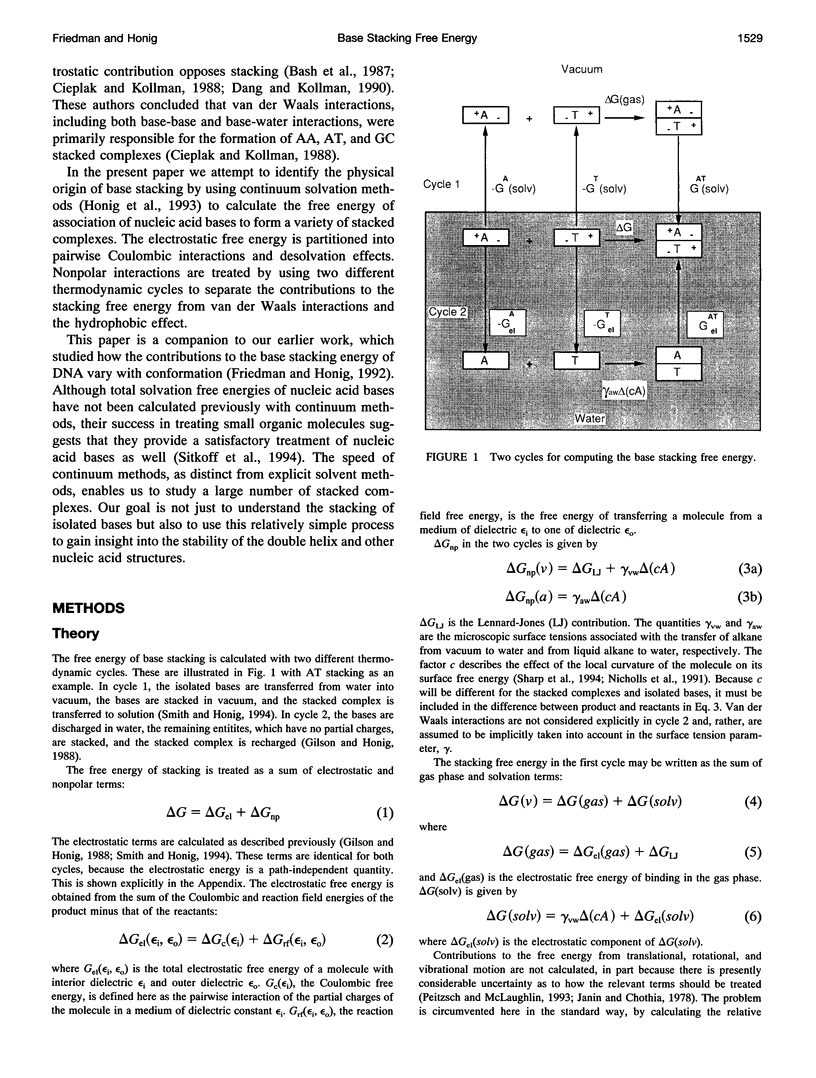
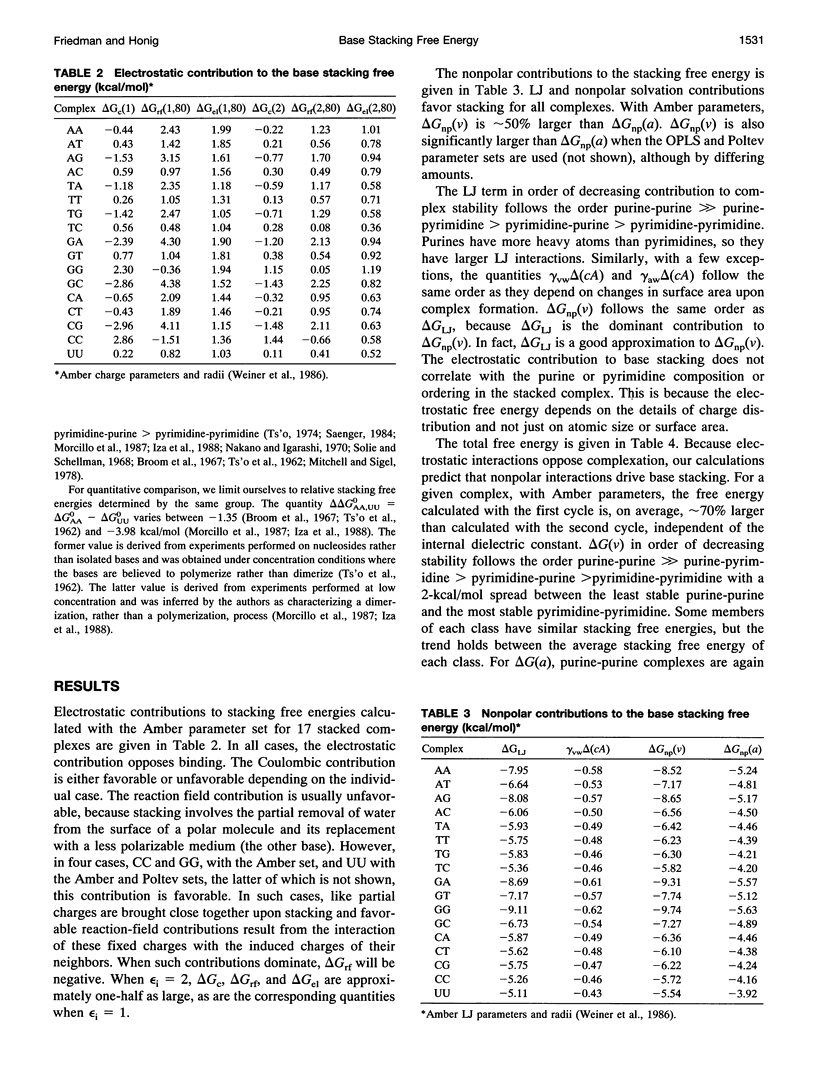
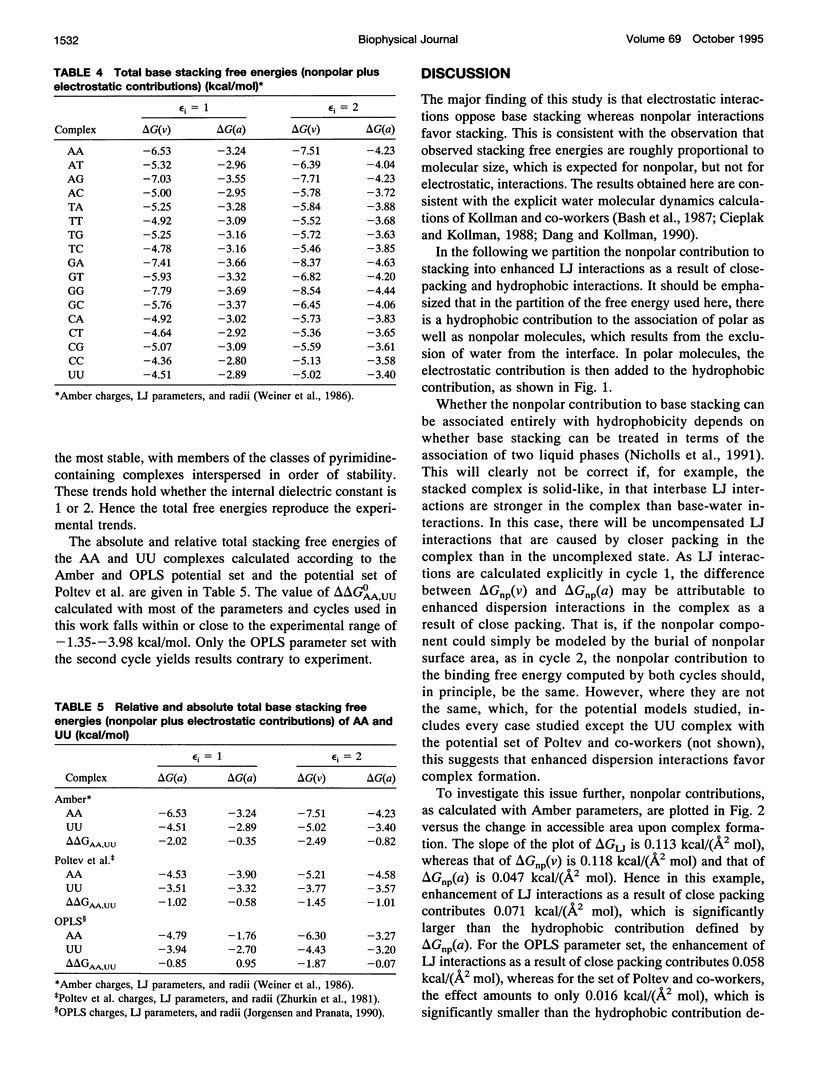
Abstract
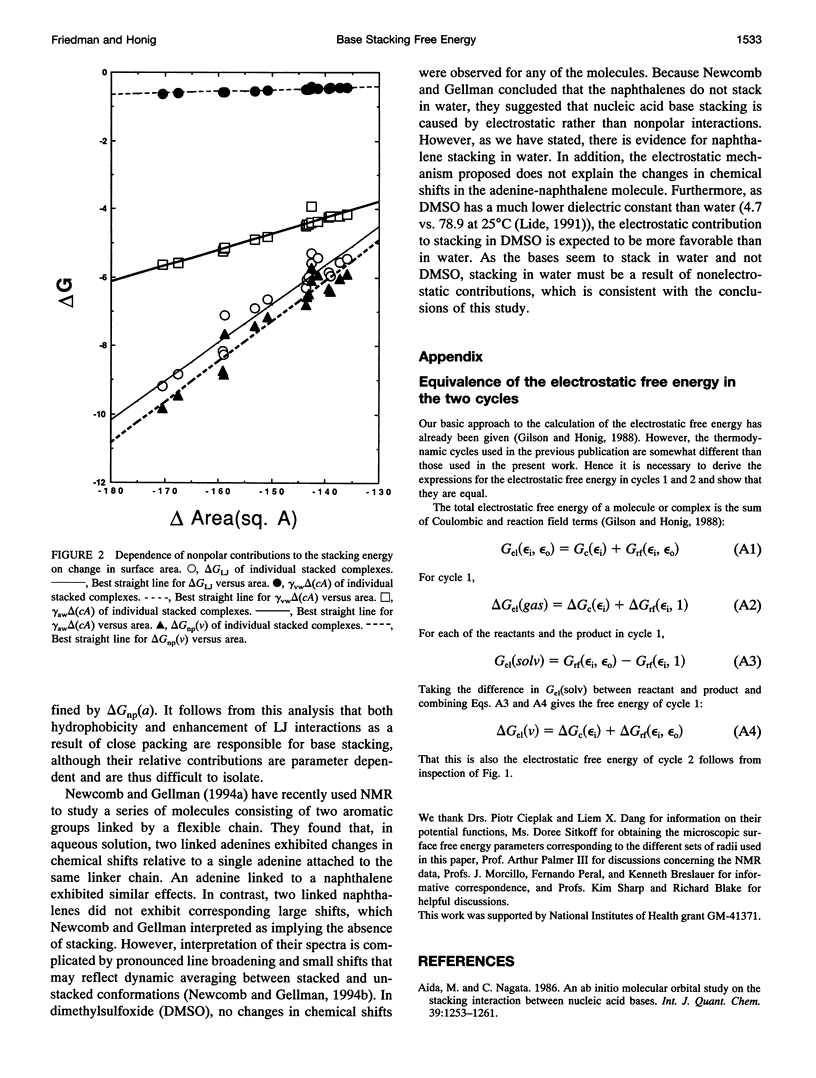
This paper reports a theoretical study of the free energy contributions to nucleic acid base stacking in aqueous solution. Electrostatic interactions are treated by using the finite difference Poisson-Boltzmann method and nonpolar effects are treated with explicit calculation of van der Waals interactions and/or free energy-surface area relationships. Although for some pairs of bases there is a favorable Coulombic interaction in the stacked conformation, generally the net effect of electrostatic interactions is to oppose stacking. This result is caused by the loss of favorable base-solvent electrostatic interactions, that accompany the partial removal of polar atoms from water in the stacked conformation. Nonpolar interactions, involving the hydrophobic effect and enhancement of van der Waals interactions caused by close-packing, drive stacking. The calculations qualitatively reproduce the experimental dependence of stacking free energy on purine-pyrimidine composition.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bash P. A., Singh U. C., Langridge R., Kollman P. A. Free energy calculations by computer simulation. Science. 1987 May 1;236(4801):564–568. doi: 10.1126/science.3576184. [DOI] [PubMed] [Google Scholar]
- Claverie P., Pullman B., Caillet J. Van der Waals-London interactions between stacked purines and pyrimidines. J Theor Biol. 1966 Dec;12(3):419–434. doi: 10.1016/0022-5193(66)90154-8. [DOI] [PubMed] [Google Scholar]
- DEVOE H., TINOCO I., Jr The stability of helical polynucleotides: base contributions. J Mol Biol. 1962 Jun;4:500–517. doi: 10.1016/s0022-2836(62)80105-3. [DOI] [PubMed] [Google Scholar]
- Danilov V. I., Tolokh I. S. Nature of the stacking of nucleic acid bases in water: a Monte Carlo simulation. J Biomol Struct Dyn. 1984 Aug;2(1):119–130. doi: 10.1080/07391102.1984.10507551. [DOI] [PubMed] [Google Scholar]
- Friedman R. A., Honig B. The electrostatic contribution to DNA base-stacking interactions. Biopolymers. 1992 Feb;32(2):145–159. doi: 10.1002/bip.360320205. [DOI] [PubMed] [Google Scholar]
- Gilson M. K., Honig B. Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins. 1988;4(1):7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- HERSKOVITS T. T., SINGER S. J., GEIDUSCHEK E. P. Nonaqueous solutions of DNA. Denaturation in methanol and ethanol. Arch Biochem Biophys. 1961 Jul;94:99–114. doi: 10.1016/0003-9861(61)90016-9. [DOI] [PubMed] [Google Scholar]
- Hanlon S. The importance of London dispersion forces in the maintenance of the deoxyribonucleic acid helix. Biochem Biophys Res Commun. 1966 Jun 21;23(6):861–867. doi: 10.1016/0006-291x(66)90567-5. [DOI] [PubMed] [Google Scholar]
- Helmkamp G. K., Kondo N. S. Preferred orientations in purine stacking. Biochim Biophys Acta. 1967 Aug 22;145(1):27–30. doi: 10.1016/0005-2787(67)90650-8. [DOI] [PubMed] [Google Scholar]
- Holtzer A. The use of Flory-Huggins theory in interpreting partitioning of solutes between organic liquids and water. Biopolymers. 1992 Jun;32(6):711–715. doi: 10.1002/bip.360320611. [DOI] [PubMed] [Google Scholar]
- Hunter C. A. Sequence-dependent DNA structure. The role of base stacking interactions. J Mol Biol. 1993 Apr 5;230(3):1025–1054. doi: 10.1006/jmbi.1993.1217. [DOI] [PubMed] [Google Scholar]
- Janin J., Chothia C. Role of hydrophobicity in the binding of coenzymes. Appendix. Translational and rotational contribution to the free energy of dissociation. Biochemistry. 1978 Jul 25;17(15):2943–2948. doi: 10.1021/bi00608a001. [DOI] [PubMed] [Google Scholar]
- Martel P. Base crystallization and base stacking in water. Eur J Biochem. 1979 May 15;96(2):213–219. doi: 10.1111/j.1432-1033.1979.tb13031.x. [DOI] [PubMed] [Google Scholar]
- Mitchell P. R., Sigel H. A proton nuclear-magnetic-resonance study of self-stacking in purine and pyrimidine nucleosides and nucleotides. Eur J Biochem. 1978 Jul 17;88(1):149–154. doi: 10.1111/j.1432-1033.1978.tb12432.x. [DOI] [PubMed] [Google Scholar]
- Nakano N. I., Igarashi S. J. Molecular interactions of pyrimidines, purines, and some other heteroaromatic compounds in aqueous media. Biochemistry. 1970 Feb 3;9(3):577–583. doi: 10.1021/bi00805a019. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp K. A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Peitzsch R. M., McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry. 1993 Oct 5;32(39):10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- Pohorille A., Pratt L. R., Burt S. K., MacElroy R. D. Solution influence on biomolecular equilibria: nucleic acid base associations. J Biomol Struct Dyn. 1984 Mar;1(5):1257–1280. doi: 10.1080/07391102.1984.10507516. [DOI] [PubMed] [Google Scholar]
- Poltev V. I., Shulyupina N. V. Simulation of interactions between nucleic acid bases by refined atom-atom potential functions. J Biomol Struct Dyn. 1986 Feb;3(4):739–765. doi: 10.1080/07391102.1986.10508459. [DOI] [PubMed] [Google Scholar]
- Privalov P. L., Gill S. J. Stability of protein structure and hydrophobic interaction. Adv Protein Chem. 1988;39:191–234. doi: 10.1016/s0065-3233(08)60377-0. [DOI] [PubMed] [Google Scholar]
- Rymdén R., Stilbs P. Nucleotide aggregation in aqueous solution. A multicomponent self-diffusion study. Biophys Chem. 1985 Feb;21(2):145–156. doi: 10.1016/0301-4622(85)85016-x. [DOI] [PubMed] [Google Scholar]
- Sharp K. A., Nicholls A., Fine R. F., Honig B. Reconciling the magnitude of the microscopic and macroscopic hydrophobic effects. Science. 1991 Apr 5;252(5002):106–109. doi: 10.1126/science.2011744. [DOI] [PubMed] [Google Scholar]
- Sharp K. A., Nicholls A., Friedman R., Honig B. Extracting hydrophobic free energies from experimental data: relationship to protein folding and theoretical models. Biochemistry. 1991 Oct 8;30(40):9686–9697. doi: 10.1021/bi00104a017. [DOI] [PubMed] [Google Scholar]
- Sitkoff D., Sharp K. A., Honig B. Correlating solvation free energies and surface tensions of hydrocarbon solutes. Biophys Chem. 1994 Aug;51(2-3):397–409. doi: 10.1016/0301-4622(94)00062-x. [DOI] [PubMed] [Google Scholar]
- Smith K. C., Honig B. Evaluation of the conformational free energies of loops in proteins. Proteins. 1994 Feb;18(2):119–132. doi: 10.1002/prot.340180205. [DOI] [PubMed] [Google Scholar]
- Solie T. N., Schellman J. A. The interaction of nucleosides in aqueous solution. J Mol Biol. 1968 Apr 14;33(1):61–77. doi: 10.1016/0022-2836(68)90281-7. [DOI] [PubMed] [Google Scholar]
- Sowers L. C., Shaw B. R., Sedwick W. D. Base stacking and molecular polarizability: effect of a methyl group in the 5-position of pyrimidines. Biochem Biophys Res Commun. 1987 Oct 29;148(2):790–794. doi: 10.1016/0006-291x(87)90945-4. [DOI] [PubMed] [Google Scholar]
- Stokkeland I., Stilbs P. A multicomponent self-diffusion NMR study of aggregation of nucleotides, nucleosides, nucleic acid bases and some derivatives in aqueous solution with divalent metal ions added. Biophys Chem. 1985 Jun;22(1-2):65–75. doi: 10.1016/0301-4622(85)80026-0. [DOI] [PubMed] [Google Scholar]