Abstract
The structure of Escherichia coli core RNA polymerase (RNAP) was determined by cryo-electron microscopy and image processing of helical crystals to a nominal resolution of 15 Å. Because of the high sequence conservation between the core RNAP subunits, we were able to interpret the E. coli structure in relation to the high-resolution x-ray structure of Thermus aquaticus core RNAP. A very large conformational change of the T. aquaticus RNAP x-ray structure, corresponding to opening of the main DNA/RNA channel by nearly 25 Å, was required to fit the E. coli map. This finding reveals, at least partially, the range of conformational flexibility of the RNAP, which is likely to have functional implications for the initiation of transcription, where the DNA template must be loaded into the channel.
RNA in all cellular organisms is synthesized by a complex molecular machine, the DNA-dependent RNA polymerase (RNAP). In its simplest bacterial form, the enzyme comprises at least four subunits with a total molecular mass of around 400 kDa. The eukaryotic enzymes comprise upward of a dozen subunits with a total molecular mass of around 500 kDa. The core component of the bacterial RNAP (subunit composition α2ββ′ω) is evolutionarily conserved from bacteria to human (1–3).
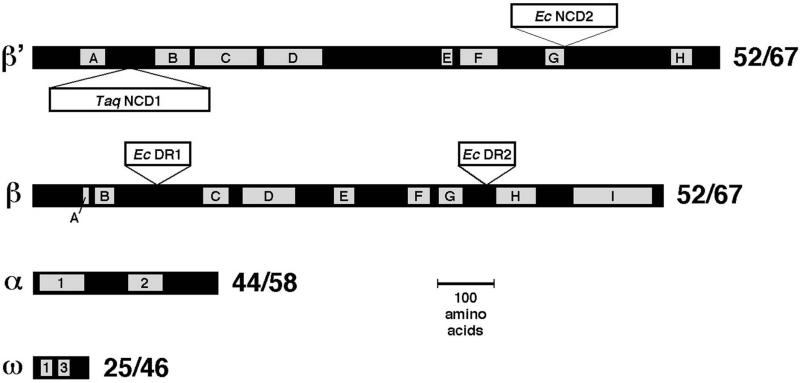
From a functional point of view, by far the best-characterized cellular RNAP is that from Escherichia coli. The structure of this enzyme has been determined to a resolution of 19 Å by cryo-electron microscopy (EM) and image processing of helical crystals (4, 5). The 3.3-Å resolution crystal structure of a bacterial RNAP has been determined from x-ray analysis of the core RNAP from Thermus aquaticus (Taq; refs. 6 and 7). The structures reveal a “crab claw”-shaped molecule with a 27-Å wide internal channel between the claws. Each subunit of the Taq core RNAP is highly conserved in sequence with E. coli (Fig. 1), and the copious literature obtained from biochemical, biophysical, and genetic studies of E. coli RNAP (8) is, for the most part, interpretable in terms of the Taq RNAP structure (9, 10). Nevertheless, the enzymes are not identical (Fig. 1).
Figure 1.
Sequence comparison between E. coli and Taq core RNAP subunits. The black bars represent the primary sequences of the core RNAP subunits (β′, β, α, and ω). The gray boxes indicate evolutionarily conserved regions among all prokaryotic, chloroplast, archaebacterial, and eukaryotic sequences, labeled A–H for β′ (7, 34) and A–I for β (7, 35). For α, motifs 1 and 2 are shown (2, 36). For ω, conserved regions 1 and 3 are shown (3). Insertions (>15 aa) in β′ and β are shown as white bars above (for E. coli insertions) or below (for Taq insertions). To the right of each subunit, the sequence identity (%)/sequence similarity (%) between the E. coli and Taq subunit is shown, calculated by ignoring the large, nonconserved inserts.
The large β and β′ subunits contain colinearly arranged regions of high sequence conservation among prokaryotes, chloroplasts, archaebacteria, and eukaryotes (Fig. 1; refs. 1 and 7). These highly conserved regions are separated by relatively nonconserved spacer regions. Large gaps or insertions can occur within these spacer regions. Comparing E. coli and Taq, each of the core subunits is highly conserved throughout (Fig. 1). Nevertheless, E. coli β′ lacks a 283-residue domain present in Taq β′ between conserved regions A and B (Taq β′ nonconserved domain 1, or Taq β′NCD1), and contains a 188-residue domain immediately C terminal of conserved region G (E. coli β′NCD2) that is missing in Taq. In β, the E. coli subunit harbors two large domains missing in the Taq subunit, a 115-residue domain inserted between conserved regions B and C, and a 99-residue domain between conserved regions G and H. Because large deletions in these two regions of E. coli β do not affect RNAP assembly and basic function in vitro, they have been termed dispensable regions I (DR1) and 2 (DR2), respectively (11, 12). DR2 of the E. coli β subunit has been localized on the low-resolution structure of core RNAP (13).
In this study, we present a 15-Å resolution structure of E. coli core RNAP, determined by cryo-EM and image processing of helical crystals. The high sequence conservation between each of the core RNAP subunits of E. coli and Taq allows us to interpret the cryo-EM map of E. coli core RNAP in relation to the x-ray structure of Taq core RNAP (6, 7). The comparison reveals the precise locations of DR1 and DR2 of E. coli β. A large conformational change of the Taq RNAP x-ray structure, corresponding to opening of the claws by nearly 25 Å, was required to fit the x-ray structure into the cryo-EM map, revealing, at least partially, the range of conformational flexibility available to the RNAP.
Materials and Methods
Purification and Crystallization.
Endogenous E. coli core RNAP was purified from MRE600 cells as described (14). A molar excess of rifampicin was added to the RNAP, and tubular, helical crystals were grown on positively charged liposomes as described (4, 5, 13, 14).
EM and Image Processing.
Specimen preparation, EM, and scanning were done as described (4, 5, 13). For three-dimensional reconstructions, selected tubes were analyzed by using helical processing programs and methods as described (4, 5, 15–17). The defocus and astigmatism, for calculating the contrast transfer function of each image, were determined by using a set of programs obtained from C. Toyoshima (University of Tokyo) (18). The final set of six processed tubes consisted of five different helical symmetries (Table 1). The two tubes with the same helical symmetry (−20 and 10), were averaged together in reciprocal space (4, 5). This reconstruction was used as a reference for alignment and averaging of the other helical symmetries by using the little-g averaging method (19). After reindexing and alignment of each of the images to the reference, the phases within each layer line were compared between images. Layer lines with total phase errors less than 60° were included in the final reconstruction, and these included layer lines out to 12.5-Å resolution. However, the layer lines included beyond 15-Å resolution did not comprise a complete set. We thus place the nominal resolution of the reconstruction at 15 Å.
Table 1.
Image statistics
| Tube symmetry (n1,0, n0,1) | Electron microscope | No. of repeats | No. of molecules | Defocus, nm | Unit cell
|
||
|---|---|---|---|---|---|---|---|
| a, Å | b, Å | γ, ° | |||||
| (−20, 10) | CM12 120 kV | 3 | 1,293 | −2,096 | 88.6 | 128.6 | 54.2 |
| (−20, 10) | CM200 200 kV | 3 | 841 | −1,968 | 90.3 | 133.8 | 52.7 |
| (−20, 11) | CM12 120 kV | 5 | 2,157 | −1,586 | 89.0 | 124.3 | 53.2 |
| (−24, 12) | CM12 120 kV | 2 | 1,028 | −1,718 | 89.4 | 128.4 | 55.0 |
| (−17, 10) | CM200 200 kV | 3 | 963 | −1,143 | 93.7 | 133.5 | 47.7 |
| Total | 6,282 | ||||||
Flexible Fitting.
We performed the flexing as described in detail (20), using connected skeletons of eight (coarse flexing) and 15 (fine flexing) positional markers. The flexible fitting requires that the atomic structure is completely accounted for by the EM density data. Therefore, we ignored the Taq ω subunit (which appeared to lack density) in the refinement. To remove extraneous density caused by neighboring RNAP subunits in the EM map or by E. coli insertions into the Taq sequence (the DRs), we devised the following iterative rigid-body and flexible refinement procedure to create a single molecule “Taq-like” map based on the E. coli density (see figure 2 of ref. 20). We used a three-dimensional cross-correlation-based template convolution tool (21) to scan the six translational and rotational degrees of freedom of the Taq structure relative to the (fixed) target map. The resolution of this docked template was lowered to that of the EM map, and the resulting density was subtracted from the EM map. Segmentation of the difference map allowed us to remove contiguous densities caused by neighboring molecules in the E. coli map (Fig. 2). The resulting single molecule map was then used as a template for coarse flexing. Subsequently, the procedure was repeated. This time, the E. coli insertions were also removed in preparation for the final fine-flexing refinement. In the flexing procedure, side chains were rearranged automatically to accommodate global conformational changes (20). Otherwise, the algorithm leaves the initial structure intact on the local level. All map manipulations were performed with situs, version 1.4.
Figure 2.
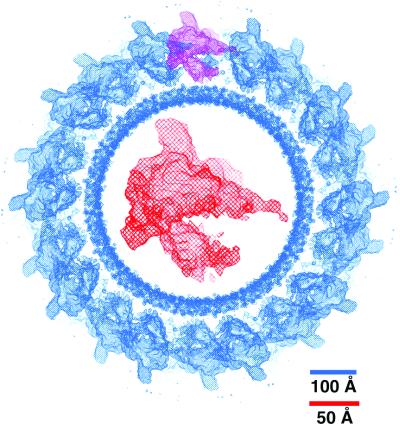
Cryo-EM structure of E. coli core RNAP. Shown in blue is a 200-Å thick cross section through the reconstructed tube, contoured at 1.1 σ. The noisy, inner circle of density is caused by the cylindrical lipid bilayer. Arranged around the outside of the lipid tube are the E. coli core RNAP molecules. The density caused by a single E. coli core RNAP molecule is highlighted in purple. The density for a single E. coli core RNAP molecule, extracted from the whole reconstruction and magnified, is shown in red in the middle of the cross section. Scale bars are for the whole reconstruction (blue) and magnified molecule (red).
Results
Comparison of E. coli Cryo-EM and Taq X-Ray Structures.
Previously, cryo-EM and image processing was used to determine the structure of E. coli core RNAP in helical crystals (4, 5, 13, 14), where the data from two tubular crystals were combined to achieve a nominal resolution estimated at around 19 Å but with significant information to 16-Å resolution (4, 5). In the present study, a more detailed structure was obtained (Fig. 2) by combining the data from five tubular crystals falling into four different helical symmetries (Table 1). The density corresponding to a single E. coli core RNAP molecule (Fig. 2) was delineated by comparison with the Taq core RNAP x-ray structure (Fig. 3).
Figure 3.
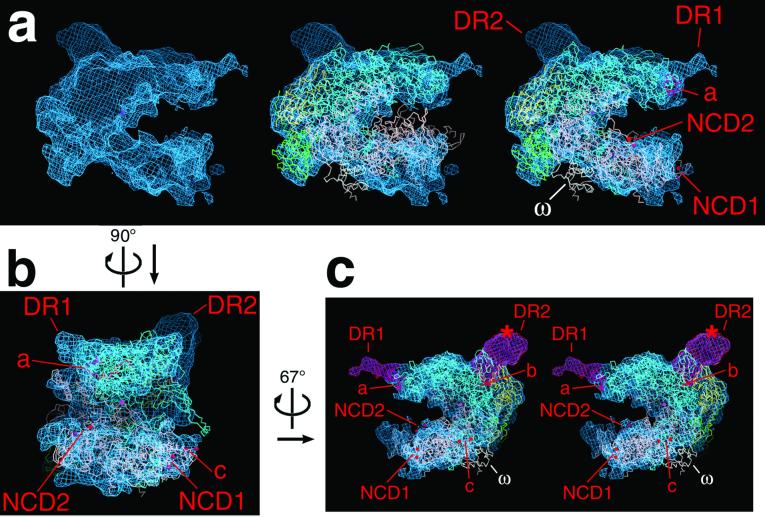
Fit of the Taq x-ray structure (αI, yellow; αII, green; β, cyan; β′, pink; ω, white) into the E. coli cryo-EM map (blue net). (a) One view of a single E. coli core RNAP molecule extracted from the cryo-EM map, with (Left) the cryo-EM map alone, (Center) the original (not flexed) Taq core RNAP x-ray structure (6 7) superimposed, showing the less than ideal fit of the β′ subunit, and (Right) the flexed Taq x-ray structure superimposed. (b) View (oriented with respect to a as indicated) of the E. coli core RNAP cryo-EM map with the flexed Taq core RNAP x-ray structure superimposed. (c) Stereo view (oriented with respect to b as indicated) of the E. coli core RNAP cryo-EM map with the flexed Taq core RNAP x-ray structure superimposed. The difference densities attributed to E. coli βDR1 and βDR2 (labeled DR1′ and DR2′ in all of the views) are shown in magenta. The red * in the lower-right view denotes the position of the positive difference peak determined by cryo-EM analysis of a mutant E. coli core RNAP containing a 30-kDa domain inserted in the middle of βDR2 (13). In all of the views, the active site Mg2+ is denoted by a magenta sphere. Differences between the E. coli cryo-EM map and the flexed Taq x-ray structure are labeled. These are E. coli βDR1 and βDR2, located near their insertion points with respect to Taq β208–233 (colored red and labeled a) and Taq β803–806 (colored red and labeled b). The red atoms labeled c denote a gap in the Taq β′ chain (from Taq β′ 32–68) that includes the Zn2+-binding motif universally conserved among prokaryotes. The red atoms labeled NCD1′ denote the gap in the Taq β′ chain (Taq β′ 156–451) caused by β′NCD1 (6, 7). The red atoms labeled NCD2 denote a gap in the Taq β′ chain (Taq β′ 1242–1249) where E. coli β′NCD2 is inserted (see Fig. 1). The ω subunit is also labeled. Figure was generated by using the program o (33).
Examination of the E. coli cryo-EM structure and the Taq x-ray structure together revealed a clear correspondence of most structural features. Direct superposition of the structures, however, resulted in a less than satisfactory fit (Fig. 3a) that could be optimized only by altering the Taq x-ray structure. An alignment procedure was developed (20) based on difference mapping, in combination with intermediate rigid-body and flexible fitting, to bring the deviating features of the Taq x-ray structure in register with the E. coli structure (see Materials and Methods). The resulting fit between the E. coli core RNAP cryo-EM structure and the “flexed” Taq core RNAP x-ray structure showed a remarkable correspondence of most structural features (Fig. 3), with the exceptions noted below.
Within the β subunit, a 26-residue span of the Taq subunit (208–233, colored red and labeled a in Fig. 3) is replaced with a 141-residue segment in E. coli (221–361, DR1), a difference of 115 residues. The site of insertion of DR1 mapped onto the Taq subunit corresponds well with density present in the E. coli cryo-EM map that is not accounted for by the Taq x-ray structure, identifying this density as βDR1 (Fig. 3). The volume of the density in the cryo-EM map attributed to βDR1 (magenta in Fig. 3c) could accommodate only about 35 aa residues, indicating that this domain is largely disordered.
Similarly, a four-residue span of Taq β (803–806, labeled b in Fig. 3c) is replaced with a 103-residue segment in E. coli (931–1033, DR2) a difference of 99 residues. The site of insertion of DR2 mapped onto the Taq subunit corresponds with density present in the E. coli cryo-EM map not accounted for by the Taq x-ray structure. Furthermore, this extra density corresponds exactly to the location of an extra domain inserted into the middle of βDR2 that was localized previously by cryo-EM (red * in Fig. 3c; ref. 13). These two observations identify this density as βDR2. The volume of the density in the cryo-EM map attributed to βDR2 (magenta in Fig. 3c) is sufficient to accommodate the entire 99-residue insertion.
Within β′, there are two large differences between the E. coli and Taq subunits, NCD1 and NCD2. NCD1 is not modeled in the Taq x-ray structure (6, 7) and is missing in E. coli β′. Its point of insertion on the RNAP structure is indicated in Fig. 3.
Immediately following β′ conserved region G, the 188-residue NCD2 is found in E. coli but is missing in Taq. Surprisingly, there is little discrepancy between the E. coli cryo-EM map and the Taq x-ray structure in this region (Fig. 3), indicating that this domain is likely to be very flexible and disordered so as not to give rise to significant density in the Fourier-averaged map.
In the Taq x-ray structure, β′ residues 32–68 are not modeled because of flexibility and disorder within the crystals (6). The disordered segment includes four Cys residues, absolutely conserved among prokaryotes, that are presumed to participate in binding a Zn2+ ion (22). In the E. coli cryo-EM structure, a small region of density is present that we attribute to this segment, which may be visible because of the lower resolution of the map (labeled c in Fig. 3).
Finally, a recent study demonstrated that the bacterial RNAP ω subunit is a structural and functional homolog of an essential subunit of archaebacterial and eukaryotic RNAPs (Rpb6 of RNAP II), and that it plays a role in maintaining the proper fold of β′ for assembly into the RNAP (3). Surprisingly, density for ω, present at apparently full occupancy in the Taq RNAP crystal structure, is missing in the E. coli cryo-EM structure. Subunit ω appears to be present in the E. coli core RNAP preparations used for crystallization, based on SDS/PAGE analysis (data not shown), although the stoichiometry with respect to the other core subunits is difficult to assess because of its small size, which is less than 3% of the total core RNAP mass.
RNAP Conformational Change.
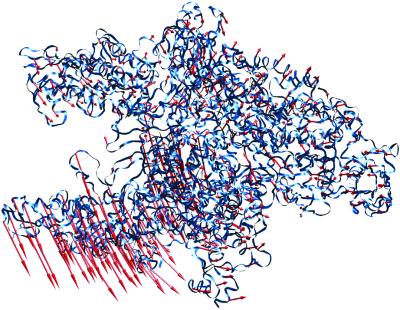
As described above, a large alteration of the Taq RNAP x-ray structure was required for an optimal fit to the E. coli cryo-EM structure. Partitioning of the structure into domains that move as rigid bodies was performed with the Hingefind algorithm (23). Using a maximum tolerance of 4 Å, the algorithm found six rigid domains that exhibited internal motions of 1.5 Å to 2.3 Å. Compared with the global conformational change (rms deviation of 6.6 Å), these internal motions were small, as expected for predominantly hinge-type movements. The structures (the original x-ray structure and the flexed structure fit to the cryo-EM map) were best compared by a least-squares alignment of the α subunits, which hardly changed during the flexing procedure. The conformational change is detailed in Fig. 4. After this alignment, the β subunits of the original and flexed structures also showed only small movements. The conformational change was dominated by a nearly 20° hinge-like rotation of a large domain containing components of both the β and β′ subunits, resulting in the opening of the main RNAP channel at its mouth by nearly 25 Å (Fig. 4).
Figure 4.
Conformational change of the Taq core RNAP x-ray structure. Shown is the α-carbon backbone of the Taq core RNAP x-ray structure (blue ribbon). Superimposed on every fifth α-carbon is a vector (red) denoting the magnitude and direction of displacement to the same α-carbon of the flexed structure. The flexed structure (not shown) is available for download at ftp://ftp.scripps.edu/pub/wriggers/dars02. Figure was generated with the program vmd (37).
Discussion
The structure of E. coli core RNAP was determined by cryo-EM and image processing of helical crystals to a nominal resolution of 15 Å. A recently described method for averaging Fourier–Bessel coefficients (19) was instrumental in including the data from five helical tubes, which fell into four different helical symmetries. The improved detail in the cryo-EM structure allowed an unambiguous fit of the Taq core RNAP x-ray structure into the E. coli cryo-EM map. The optimal fit required a large conformational change of the x-ray structure.
The comparison of the cryo-EM map and the altered x-ray structure revealed a close correspondence of structural features, with only a few exceptions that can be mostly explained by large insertions or deletions in the β subunits between E. coli and Taq. In particular, two significant densities present in the E. coli cryo-EM map (DR1 and DR2, Fig. 3) are not accounted for by the flexed Taq RNAP x-ray structure but are positioned near points in the Taq β subunit where insertions of about 100 aa (in each case) occur in E. coli β. Furthermore, one of these densities was identified previously to be associated with βDR2 from the results of a cryo-EM/labeling study (13).
DR1 and DR2 of the β subunit are poorly conserved or even missing in β homologs of other organisms, and E. coli RNAP can tolerate large deletions (more than 200 aa in some cases) in these regions without loss of function in vitro (11, 12). These properties suggest that the DRs must be structurally autonomous to accommodate such large deletions or insertions without disturbing the central functions of the enzyme. Thus, both DRs should comprise separate, isolated domains on the RNAP surface. The identification of βDR1 and βDR2 in the E. coli cryo-EM map shows that this is indeed the case, both regions comprise isolated densities that protrude from the surface of the enzyme.
Although the βDRs do not play important roles in RNAP assembly or basic transcription activity, it is assumed that their presence points to a role in regulatory functions specific to E. coli yet to be identified. Indeed, DR1 is targeted by the bacteriophage T4-Alc protein, which selectively induces premature termination of E. coli RNAP transcription on E. coli DNA during infection (12), indicating that regulatory factors can impact transcription through the DRs. Thus, we are left with the interesting picture of the RNAP molecule as a central core structure, contained within an approximately 40-Å radius sphere centered at the active site Mg2+ ion, that is highly conserved in sequence and nearly identical in structure among all organisms (7, 9, 24, 25). Outside this sphere, the sequences and structures diverge, and inserted at various points on the surface of the bacterial RNAPs are structurally autonomous domains that can serve as interaction modules for regulatory factors that modulate the various phases of the transcription cycle.
Although the ω subunit has recently been shown to be a structural and functional homolog of essential subunits in archaebacterial and eukaryotic RNAPs (3), density corresponding to ω is missing in the E. coli cryo-EM map. While ω was shown to play a role in preparing the β′ subunit for proper assembly with the α2β subassembly, this role was revealed only in the context of a temperature-sensitive assembly mutant of E. coli β′ (3) because the rpoZ gene (encoding ω) is not essential for E. coli viability under standard laboratory conditions (26). The interesting possibility exists that the ω subunit of the mesophilic E. coli RNAP is not tightly bound under normal laboratory conditions, being important only under stressful conditions that could encourage misfolding of the β′ subunit. In contrast, the RNAP from the thermophile Taq has evolved to assemble and function under conditions of high temperature (the optimal growth temperature for Taq is around 72°C), and, based on the x-ray crystallographic data, in Taq RNAP the ω subunit is tightly bound and may play a more important, and possibly essential, role in RNAP assembly. It would be interesting to test whether rpoZ is essential in extremophiles such as Taq.
The most striking finding of this study is the large conformation change required to fit the Taq x-ray structure into the E. coli cryo-EM map (Fig. 4). The very high conservation of sequence, structure, and function between E. coli and Taq (Fig. 1), and the fact that related conformational changes of a similar scale have been observed in different crystal forms and different functional complexes of yeast RNAP II (24, 27–30), leads to the conclusion that the observed conformational change reflects the normal flexibility of the RNAP rather than differences between E. coli and Taq RNAPs or crystallization artifacts.
From a comparison of two crystal forms of yeast RNAP II, a highly mobile domain termed the “clamp” was identified (24, 28, 29), comprising primarily the N terminus of Rpb1, and the C terminus of Rpb2 (the homologous structure in Taq RNAP corresponds to residues 1–624 of β′ and 1054–1115 of β). In the different structures of RNAP II, the clamp undergoes a swinging motion, resulting in opening or closing of the main DNA/RNA cleft, and this motion was proposed to be important for allowing entry of DNA into the cleft for the initiation of transcription, and for closing on the DNA and the DNA/RNA hybrid to provide processivity during transcription elongation. Closure of the RNAP jaws around the DNA and DNA/RNA hybrid was also proposed based on modeling of a ternary elongation complex of Taq RNAP (31). This idea was confirmed in a crystal structure of an elongation complex of yeast RNAP II (30), in which the clamp domain closes onto the DNA and the DNA/RNA hybrid within the cleft.
Our comparison of the Taq core RNAP crystal structure and the flexed structure fit to the E. coli core RNAP cryo-EM map reveals a consistent picture of RNAP conformational flexibility. As in yeast RNAP II, the bulk of the enzyme lies in a core module, comprising both α subunits, most of β (except for the C-terminal segment belonging to the clamp), and parts of β′ that form the active center and interact with β in the upper claw of the RNAP. Although domains within the core module move between the two structures, these changes are relatively small. The conformational change is dominated by a nearly 20° hinge-like rotation of the bottom claw (as seen in Fig. 4) that includes the clamp domain described above, but also includes the C terminus of β′ following the conserved region F bridging helix (from roughly residue 805 to the C terminus). The conformational change results in the displacement of some α-carbons by nearly 25 Å, opening the mouth of the main DNA/RNA channel of the RNAP.
The similarity of the conformational changes observed in eukaryotic and bacterial RNAPs underscores the likely functional significance. The high processivity of RNAP dictates that the claws remain closed during transcript elongation, as observed in the yeast RNAP II ternary complex structure (30). However, relatively small motions of the RNAP claws during elongation are not discounted and may occur in response to regulatory signals and factors (32), probably along the pathway of the larger conformational change observed here. The key functional relevance of the open-claw state of RNAP is likely to relate to the initiation phase of transcription, in which about 25 nt of DNA (from about +14 to −12) must be loaded inside the main channel before transcription can ensue.
The different conformational states of yeast RNAP II and bacterial core RNAPs are affected by differences in crystal packing forces. In both prokaryotes and eukaryotes, protein factors in addition to the core RNAP are required for promoter-specific initiation. In prokaryotes, it is a single polypeptide, the σ factor, that binds the core RNAP to create the initiation-competent holoenzyme. We suggest that, in addition to its central role in promoter recognition and melting, σ may play a role in controlling the opening and closing of the RNAP claws during different stages of the initiation process (14).
Acknowledgments
We are grateful to D. Stokes for use of the CM200 field emission gun at the Skirball Institute (New York), R. Beroukhim, W. Rice, and N. Unwin for helical processing software and assistance with its use, and C. Toyoshima for additional software. N.O. was supported by funds granted by the Norman and Rosita Winston Foundation and the Philippe Foundation. P.C. was supported by funds from the La Jolla Interfaces in Science Program/Burroughs Wellcome Fund. A.P. and G.Z. were supported by National Research Service Awards GM 17708–01 and 19441–01, respectively. This work was supported by National Institutes of Health Grant 1R01GM62968–01 (to W.W.) and a Burroughs Wellcome Travel Grant and National Institutes of Health Grant GM58020 (to S.A.D.).
Abbreviations
- DR
dispensable region
- EM
electron microscopy
- NCD
nonconserved domain
- RNAP
RNA polymerase
- Taq
Thermus aquaticus
References
- 1.Archambault J, Friesen J D. Microbiol Rev. 1993;57:703–724. doi: 10.1128/mr.57.3.703-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang G, Darst S A. Science. 1998;281:262–266. doi: 10.1126/science.281.5374.262. [DOI] [PubMed] [Google Scholar]
- 3.Minakhin L, Bhagat S, Brunning A, Campbell E A, Darst S A, Ebright R H, Severinov K. Proc Natl Acad Sci USA. 2001;98:892–897. doi: 10.1073/pnas.98.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darst S A, Polyakov A, Richter C, Zhang G. J Struct Biol. 1998;124:115–122. doi: 10.1006/jsbi.1998.4057. [DOI] [PubMed] [Google Scholar]
- 5.Darst S A, Polyakov A, Richter C, Zhang G. Cold Spring Harbor Symp Quant Biol. 1998;63:269–276. doi: 10.1101/sqb.1998.63.269. [DOI] [PubMed] [Google Scholar]
- 6.Campbell E A, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst S A. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 7.Zhang G, Campbell E A, Minakhin L, Richter C, Severinov K, Darst S A. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 8.Mooney R A, Artsimovitch I, Landick R. J Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darst S A. Curr Opin Struct Biol. 2001;11:155–162. doi: 10.1016/s0959-440x(00)00185-8. [DOI] [PubMed] [Google Scholar]
- 10.Severinov K. Curr Opin Microbiol. 2000;3:118–125. doi: 10.1016/s1369-5274(00)00062-x. [DOI] [PubMed] [Google Scholar]
- 11.Borukhov S, Severinov K, Kashlev M, Lebedev A, Bass I, Rowland G C, Lim P-P, Glass R E, Nikiforov V, Goldfarb A. J Biol Chem. 1991;266:23921–23926. [PubMed] [Google Scholar]
- 12.Severinov K, Kashlev M, Severinova E, Bass I, McWilliams K, Kutter E, Nikiforov V, Snyder L, Goldfarb A. J Biol Chem. 1994;269:14254–14259. [PubMed] [Google Scholar]
- 13.Opalka N, Mooney R A, Richter C, Severinov K, Landick R, Darst S A. Proc Natl Acad Sci USA. 1999;97:617–622. doi: 10.1073/pnas.97.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polyakov A, Severinova E, Darst S A. Cell. 1995;83:365–373. doi: 10.1016/0092-8674(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 15.Beroukhim R, Unwin N. Neuron. 1995;15:323–331. doi: 10.1016/0896-6273(95)90037-3. [DOI] [PubMed] [Google Scholar]
- 16.Toyoshima C, Unwin N. J Cell Biol. 1990;111:2623–2635. doi: 10.1083/jcb.111.6.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice W J, Young H S, Martin D W, Sachs J R, Stokes D L. Biophys J. 2001;80:2187–2197. doi: 10.1016/S0006-3495(01)76191-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mimori Y, Yamashita I, Murata K, Fujiyoshi Y, Yonekura K, Toyoshima C, Namba K. J Mol Biol. 1995;249:69–87. doi: 10.1006/jmbi.1995.0281. [DOI] [PubMed] [Google Scholar]
- 19.DeRosier D, Stokes D L, Darst S A. J Mol Biol. 1998;289:159–165. doi: 10.1006/jmbi.1999.2677. [DOI] [PubMed] [Google Scholar]
- 20.Wriggers W, Chacon P. Structure (London) 2001;9:779–788. doi: 10.1016/s0969-2126(01)00648-7. [DOI] [PubMed] [Google Scholar]
- 21. Chacon, P. & Wriggers, W. (2002) J. Mol. Biol., in press. [DOI] [PubMed]
- 22.Treich I, Riva M, Sentenac A. J Biol Chem. 1991;266:21971–21972. [PubMed] [Google Scholar]
- 23.Wriggers W, Schulten K. Proteins Struct Funct Genet. 1997;29:1–14. [PubMed] [Google Scholar]
- 24.Cramer P, Bushnell D A, Kornberg R D. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 25.Ebright R H. J Mol Biol. 2000;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 26.Gentry D, Xiao H, Burgess R, Cashel M. J Bacteriol. 1991;173:3901–3903. doi: 10.1128/jb.173.12.3901-3903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asturias F J, Meredith G D, Poglitsh C L, Kornberg R D. J Mol Biol. 1997;272:536–540. doi: 10.1006/jmbi.1997.1273. [DOI] [PubMed] [Google Scholar]
- 28.Cramer P, Bushnell D A, Fu J, Gnatt A L, Maier-Davis B, Thompson N E, Burgess R R, Edwards A M, David P R, Kornberg R D. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 29.Fu J, Gnatt A L, Bushnell D A, Jensen G J, Thompson N E, Burgess R R, David P R, Kornberg R D. Cell. 1999;98:799–810. doi: 10.1016/s0092-8674(00)81514-7. [DOI] [PubMed] [Google Scholar]
- 30.Gnatt A L, Cramer P, Fu J, Bushnell D A, Kornberg R D. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 31.Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst S A. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 32.Landick R. Cell. 2001;105:567–570. doi: 10.1016/s0092-8674(01)00381-6. [DOI] [PubMed] [Google Scholar]
- 33.Jones T A, Zou J-Y, Cowan S, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Jokerst R S, Weeks J R, Zehring W A, Greenleaf A L. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- 35.Sweetser D, Nonet M, Young R A. Proc Natl Acad Sci USA. 1987;84:1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebright R H, Busby S. Curr Opin Genet Dev. 1995;5:197–203. doi: 10.1016/0959-437x(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 37.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;1:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]