Abstract
The patch clamp-liposome technique was used to examine the stretch sensitivity of a model membrane ion channel, gramicidin A, in membrane patches of different bilayer thickness. We found that small changes in phospholipid acyl chain length (i.e., PC-20 to PC-18) can switch gramicidin A from a stretch-activated to a stretch-inactivated channel. The demonstration that subnanometer changes in bilayer thickness can reverse the response polarity of a model channel has implications for other signaling proteins that may experience local changes in bilayer thickness as a consequence of dynamic targeting to lipid microdomains, electrocompression, or chemical modification of the bilayer.
Gated membrane ion channels are designed to act as molecular transducers of electrical, chemical, or mechanical signals. They share the basic feature of being embedded in a lipid bilayer that is often assumed to behave as a passive substrate with the transducer's properties mainly dictated by membrane protein structure. However, recent studies indicate that certain membrane proteins can undergo dynamic targeting to lipid microdomains within the cell membrane and in doing so may change their signaling properties (1–6). Although, new interactions with other proteins within the microdomains may contribute to any changes, differences in the physical properties of the lipid domain (e.g., bilayer thickness and local curvature) may also play a role (7).
For organisms to survive and grow they need to transduce a variety of mechanical forces generated by osmotic pressure gradients, fluid shear, gravity, touch, substrate vibrations, and cytoskeleton reorganization. Putative mechanotransducers involving mechanosensitive membrane trafficking, ATP release, membrane receptors, and signaling pathways have been identified that may contribute to the integrated mechanosensitive response of living cells (8–11). Another class of molecular mechanotransducer, studied intensely over the last 20 years, is the mechanically gated (MG) membrane ion channel that has been broadly classified as stretch-activated or stretch-inactivated depending on whether membrane stretch opens or closes the channel (10, 11). In specific cell types, stretch activation (SA) and stretch inactivation (SI) channels have been shown to coexist within small membrane patches (12), and most recently it has been reported that a Shaker K+ channel can display either SA or SI depending on membrane potential (13). Although the voltage-gated Shaker channel may normally be expressed in cells that are protected from changes in bilayer tension (14, 15), the dual behavior seen in a single membrane ion channel is intriguing and represents a challenge for any model of mechanical gating.
Two general classes of models might account for the ability of a membrane channel to reverse its response to membrane stretch. In one model, the channel protein in switching between several conformational states undergoes changes in its membrane occupied area and/or external hydrophobic length. Membrane stretch by tending to both expand the area and reduce the thickness of the volumetrically incompressible bilayer (16) will shift the equilibrium distribution toward channel states of larger area (17) and shorter hydrophobic length (18). Other forms of stimulation (e.g., depolarization) by also shifting the state distribution may thereby alter the response to stretch (11). Support for this type of model requires information on the conformation of the channel states, which at this time is limited to the closed state of one type of bacterial MG channel (19, 20).
In another model, differences in stretch sensitivity arise because specific channel proteins (including a Shaker-like K+ channel) are dynamically targeted to lipid microdomains enriched in sphinogolipids and cholesterol (1–6). One feature of sphingolipids is that their acyl chains range between 20 and 26 carbon atoms, which is generally longer than the C-16 to C-22 of natural phospholipids (1). Thus, a channel protein in shifting between microdomains could experience different types of hydrophobic mismatch with its surrounding bilayer (i.e., positive, negative, or neutral) so that stretch-induced bilayer thinning by either decreasing or increasing the mismatch could produce SA or SI, respectively. To test this model in biological membranes is difficult because of the many classes of phospholipids that differ in the size and charge of their head group and the length and saturation of their acyl chains. Therefore, we have examined the stretch sensitivity of gramicidin A (gA) channels incorporated in liposomes composed of pure phospholipids that differ only in their acyl chain length. gA is a simple hydrophobic peptide that forms cation channels in lipid bilayers by the transmembrane association of one monomer from each monolayer (21, 22) and has proven to be a popular model for studying the effects of protein inclusions on lipid acyl chain order and dynamics (23). Although experiments carried out over the last 30 years indicate that gA channel gating varies with bilayer thickness and tension (18, 24–28), most studies have focused on changes in open channel properties (i.e., open channel lifetime and conductance) rather than the number of channels. Furthermore, the effect of bilayer thickness on tension sensitivity has not been determined. Here we use the patch clamp-liposome recording technique (29, 30) that allows tension to be reversibly increased in bilayer patches of specified acyl chain length.
Materials and Methods
Valine-gA was a gift of Roger Koeppe II, Univ. of Arkansas. Liposomes were formed from each of the phosphatidylcholines (PCs), dipalmitoleoyl (PC-16), dioleoyl (PC-18), dieicosenoyl (PC-20), dierucoyl (PC-22), 1,2-diacyl-sn-glycero-3-phosphocholines (unsaturated series) (Avanti Polar Lipids), or from asolectin (soybean lecithin; Sigma P-3644) according to a modified procedure (29, 30). The asolectin phospholipids contained the carbon chains C16:0, C18:0, C18:2, and C18:3 of which the 18:2 comprised 60%. Choline, ethanolamine, and inositol were the main phospholipid head groups. In brief, after rinsing a glass tube (50 ml) with chloroform, 2 mg of the appropriate PC was dissolved in chloroform that was then evaporated and dried by a N2 jet (for ≈15 min). The thin layer of lipid on the bottom of the glass tube was then resuspended in dehydration/rehydration (D/R) buffer (200 mM KCl/5 mM Hepes/KOH, pH 7.2) to achieve a final concentration of 10 mg/ml. The lipid solution was vortexed (≈1 min) to give a cloudy liquid that was sonicated for ≈5 min to give a clear liquid. Two microliters of gA in ethanol (10 μg/ml) was added to the lipid (i.e., 10 ng gA/1 mg lipid) in a centrifuge tube and placed on a platform rocker for 1 h at 23°C and then spun for 30 min at 90,000 rpm (TL-100 Ultracentrifuge Beckman) at 4°C. The pellet was then resuspended in 40 μl of D/R buffer, and 20-μl aliquots were then spotted onto an ethanol-cleaned slide and dehydrated for 6 h in a vacuum dessicator at 4°C. The spots were then rehydrated with 20 μl D/R buffer and left overnight. For patch clamping, 2 μl of the liposomes was placed in a chamber (≈1-ml volume) containing the recording solution (200 ml KCl, 40 mM MgCl2, 10 mM Hepes/KOH, pH 7.0). Within 15 min of plating clear blisters were seen to form under phase-contrast optics. Blister formation and patch stability depended on the PC type. Our main comparison was made between gA activity in PC-20 and PC-18 patches because success rate in forming tight seals and stable patches was high in both cases (>80% of attempts). However, only limited recordings were made from PC-22 patches because of the scarcity of suitable blebs for initial sealing or from PC-16 patches because patches often ruptured during or soon after seal formation. Patch stability, especially in response to suction, also decreased with increasing gA/lipid ratio and patch polarization (i.e., ≥100 mV) possibly because of gA-induced bilayer thinning and electrocompression, respectively. For this reason, we compared gA activities at the same low gA/lipid weight ratio (10−5) and at the same patch potential of 60 mV. Patches isolated from control liposomes (i.e., with no gA) showed no channel activity.
Single gA channel currents were recorded by using borosilicate (Sigma) patch pipettes that had a bubble number of ≈4 when measured in absolute ethanol (31), which corresponds to a resistance of ≈8 MΩ with recording solution in the pipette and bath. A tight seal formed on pipette tip contact with the liposome or after the brief application of suction. After seal formation the pipette tip was briefly passed through the solution-air interface to rupture the outer face of the closed vesicle (32). The patch current signal was amplified and filtered (100–200 Hz) with an Axopatch 1D (Axon Instruments, Foster City, CA) and digitized at 5 kHz with a computer using PCLAMP6 acquisition software. To stretch the membrane patch, a syringe arrangement was used to apply suction to the pipette holder, which was monitored with a piezoelectric pressure transducer (Omega Engineering, Stamford, CT). The suctions steps had rise times of ≈0.5 s, which were adequate to follow the relatively slow activation and deactivation of gA channels. We did not measure patch curvature changes during suction pulses, which would be required to directly convert pressure into a tension by using Laplace's law. However, Sukaharev et al. (33) have shown that with pressures greater than ≈60 mmHg changes in patch curvature saturate, as the membrane becomes inextensible. At this point membrane tension changes proportionally with pressure. The tension required to half maximally activate the Escherichia coli MscL is ≈12 dyn/cm (33), which corresponds to a suction of ≈80 mmHg with our pipettes (11). Accordingly, a suction pulse of 160 mmHg may transiently increase tension to 24 dyn/cm. Note sustained suctions of this magnitude invariably caused patch rupture.
Results
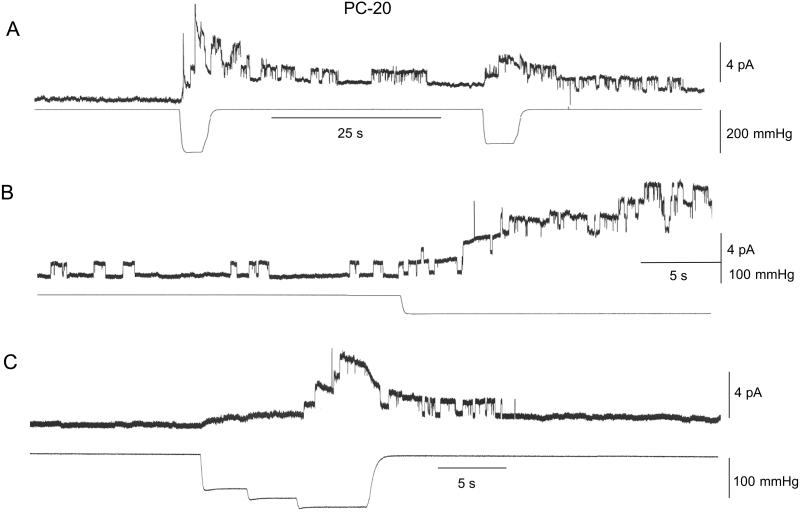
Fig. 1 shows current traces from three patches isolated from PC-20 liposomes. In the absence of stimulation, the large majority of PC-20 patches (28 of 33 patches) showed no prestretch channel activity whereas the other patches displayed a low frequency of openings (<1 s−1, see Fig. 1B). The single gA channel currents had an amplitude of ≈1 pA (at 60 mV) and an average duration of ≈1 s. In Fig. 1A, the initial suction step of 200 mmHg (note 1 mmHg = 133 Pa) transiently activated 6–8 channels (i.e., in <5 s). A second smaller suction step (150 mmHg) activated two channels. In Fig. 1B application of a suction step (100 mmHg) caused a slow, progressive activation of up to five channels before patch rupture. Fig. 1C illustrates the activation and deactivation of gA channel activity to a staircase increase in suction. Similar reversible SA of gA channels was seen in 30 of 33 patches. In general, minimal suction pulses of 80–100 mmHg were required to activate any gA channel currents, and current responses did not saturate with larger suction pulses (i.e., ≈200 mmHg) that often caused patch rupture. Qualitatively similar gA channel behavior was seen in patches from PC-22 liposomes (i.e., zero prestretch activity and SA with suctions pulses ≥100 mmHg) (three of three patches, data not presented).
Figure 1.
SA of gA channels measured in three different inside-out patches isolated from PC-20 liposomes. The suction pulse waveform for each panel is shown in the bottom trace. (A) In this patch there was no channel activity 100 s before suction stimulation. Application of a 5-s suction step of 200 mmHg activated ≈8 gA channels currents (≈1.1 pA) of which six deactivated after removal of the suction. Application of a 160-mmHg step reactivated three channels. (B) A different patch that displayed a spontaneous channel activity of ≈0.3 s−1 for the 100 s before stimulation with an average channel duration of ≈0.8 s (28 events). Stimulation with a suction step of 95 mmHg activated five channels. (C) In the third patch no channel activity was recorded in the 30 s before stimulation. A stair case increase in suction up to 125 mmHg activated four channel currents that deactivated within 10 s of removing the suction. In A and B the patch potential was 60 mV and in C it was 80 mV. In all cases the patch currents were recorded in symmetrical 200 mM KCl, 40 mM MgCl2, 10 mM Hepes/KOH, pH 7.0.
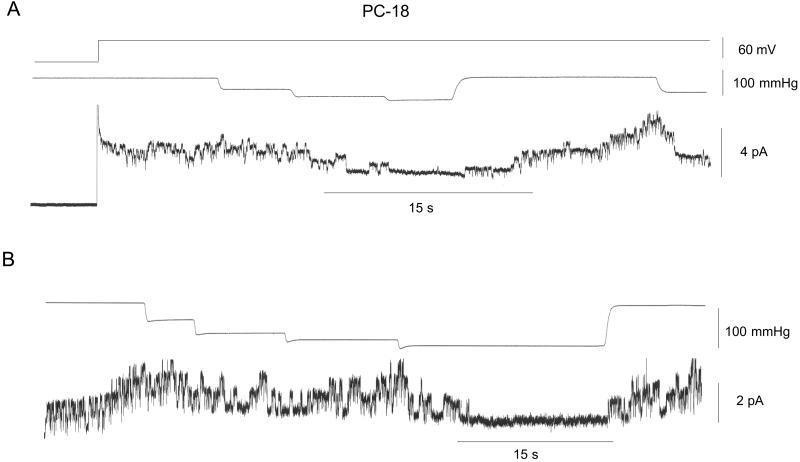
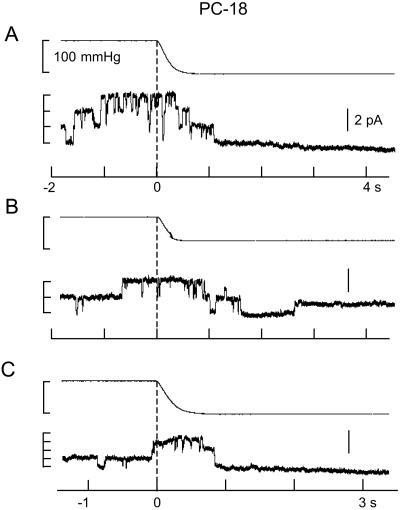
Fig. 2 shows current recordings from two patches isolated from PC-18 liposomes recorded under the same conditions as in Fig. 1. The gA channels had similar amplitude (≈1 pA) as in PC-20 patches with the major difference being the high level of prestretch (spontaneous) gA channel activity. This finding was despite a lower gA/lipid mol ratio in PC-18 (4.2 × 10−6, molecular weight 786) than in PC-20 (4.5 × 10−6, molecular weight 842). The similar number of gA channels (i.e., up to ≈8) that form in unstretched PC-18 (Fig. 2B) and stretched PC-20 patches (Fig. 1A) indicates there was no major difference in the number of monomers available for channel formation. Fig. 2 indicates staircase increases in suction produced no evidence of SA. However, after reaching a suction of ≈100 mmHg, channels were closed until the suction was removed. Fig. 3 shows that suction steps of ≈100 mmHg can close gA channels in 1–2 s. Similar SI of gA channel activity was seen in 12 of 14 PC-18 patches. We also saw reversible SI of prestretch gA activity in patches isolated from azolectin (soybean lecithin) liposomes (i.e., in which PC-18 is the major phopholipid) (eight of 10 patches) and in patches isolated from PC-16 liposomes (three of three patches) (data not presented).
Figure 2.
SI of gA channels in patches isolated from PC-18 liposomes. (A) The top trace indicates the patch potential that was stepped to 60 mV after seal formation. The middle trace shows the staircase increased in pressure (with steps to 45, 75, and 95 mmHg) that caused a deactivation of channels that was maintained for the duration of the 95-mmHg step. On removal of the suction, activity showed full recovery and reapplication of suction near the end of the current trace again closed the channels. (B) Similar protocol to A.
Figure 3.
gA channel turn-off in response to suction steps applied to an inside-out patch isolated from PC-18 liposomes. The top trace in each panel is the pressure trace. Before the suction step channel activity fluctuated between 2–3 open channels that in A and C closed within 0.5 s of the 100-mmHg suction step. In B there was a single channel reopened during the 75-mmHg suction step.
Discussion
Our main finding is that the response of a model MG channel can be reversed from SA to SI by small changes in lipid acyl chain length (i.e., PC-20 to PC-18). This result indicates that mechanosensitivity is not dictated solely by the channel structure but is a dynamic property that can vary with the physical properties of the lipid bilayer. Whether this is a feature shared by other channels will depend on the underlying mechanism. One mechanism that may explain our findings derives from the proposal of Haydon and colleagues based on planar bilayer studies (refs. 24 and 28; see also refs. 34 and 35). They suggested that if the hydrocarbon region of the bilayer is thicker than the hydrophobic length of the gA dimer (i.e., negative hydrophobic mismatch) there will be a local thinning of the bilayer associated with channel formation. On the other hand, if the length of the gA dimer is greater than the hydrocarbon thickness (i.e., positive mismatch) then the bilayer will be locally distorted to hide the exposed hydrophobic region of the gA molecule. Both bilayer distortions will result in an unfavorable energy increase because the lowest energy state will be when the bilayer thickness equals the dimer length. This model may account for our observation of higher prestretch gA channel activity in PC-18 compared with PC-20 patches (i.e., assuming the gA channel matches the PC-18 bilayer). It would also explain why ≈10 times less gA is required in PC-18 compared with PC-20 bilayers to achieve the same channel activity (36). Furthermore, if membrane stretch acts to thin as well as expand the bilayer (i.e., ΔA/A0 = −Δh/h0, where ΔA and Δh represent changes in area and thickness, respectively, and A0 and h0 are the original values, see refs. 11, 16, and 18), then stretching a relatively thick bilayer (PC-20) may reduce negative mismatch, thereby favoring the dimer (i.e., SA), whereas stretching a thinner bilayer (PC-18) may increase positive mismatch, thereby favoring the monomer (i.e., SI).
Several lines of evidence support the above model. First, x-ray diffraction studies of gA crystals indicate the ion bound channel has a maximum length of 26 Å (37). This finding compares with thickness estimates for the different bilayers of 26 Å (PC-16), 27 Å (PC-18), 30 Å (PC-20), and 33 Å (PC-22) (38, 39). Note these estimates, based on x-ray and neutron diffraction data, may overestimate the thickness of the fully hydrated bilayers (39). Second, Elliot et al. (28) reported that gA channel lifetime tended to increase in bilayers of decreasing thickness (≈29 Å to 22 Å) but in bilayers thinner than 22 Å, lifetime tended to decrease. In this study, bilayer thickness was calculated from electrical capacitance measurements but it was not determined how well this method correlates with estimates based on diffraction measurements. Thus, it remains possible that the nonmonotonic change in channel lifetime and reversal in stretch sensitivity occur at the same bilayer thickness. Elliot et al. (28) also reported that gA channel duration increased at higher bilayer tensions. However, in planar bilayers it is not possible to directly manipulate bilayer tension. Instead, the bilayers measured with higher tensions were relatively thicker (see figure 2 in ref. 28). As a consequence, tension and thickness effects on gA channels could not be separated. In a more recent study, Goulian et al. (18) used the aspiration technique (16) to alter tension isotropically in liposomes while monitoring GA activity in the aspirated membrane. They reported that gA channel opening rate and channel duration decreased as the tension in PC-18 liposomes relaxed from a pre-elevated level. There are several explanations that may account for the discrepancy with our finding that stretching PC-18 patches decreased the number of open gA channels. First, Goulian et al. recorded gA channels in acidic solution (pH 2.6) whereas we recorded at normal pH. Our patch studies indicate gA channel activity changes dramatically with pH. In particular, the prestretch gA channel activity evident at pH 7.0 in PC-18 patches is abolished at pH 2.6 and even strong suction pulses (i.e., ≥150 mmHg) fail to activate gA channels (unpublished results). Second, the protocol of Goulian et al. involved measuring channel activity as the liposome tension was reduced from 4 dyn/cm to zero. However, our patch recordings indicate that higher patch tensions (i.e., ≥10 dyn/cm) are required to cause SI of gA channels (i.e., based on a minimal pressure of ≈80 mmHg for SI, see Materials and Methods). The higher tensions would be consistent with the requirement that the PC-18 bilayer undergo near maximum thinning (e.g., ≈5%) to reduce gA dimer formation.
Other evidence consistent with the basic mechanism comes from studies of mini-gA channels synthesized with 11 instead of 15 aa so that their hydrophobic length was reduced by ≈5 Å (40). It was found that mini-gA channels form readily (i.e., spontaneously) in PC-14 and PC-16 bilayers but not in PC-18 bilayers. Presumably, the mini-gA channel suffers a similar degree of negative mismatch with PC-18 as the full-length gA channel suffers with PC-22. In this case, one would predict that mini-gA channels in PC-18 patches should be SA rather than SI. Finally, Watnick et al. (41), using 2H-NMR measurements, demonstrated that gA causes the largest increase in chain order in the PC-14 bilayer, somewhat less in PC-16, and has no affect on chain order in PC-18 bilayers. Again this finding is consistent with the idea that the full-sized gA dimer is well matched with the hydrocarbon interior of the PC-18 bilayer.
A further consideration concerns the expected bilayer thinning caused by increased tension and how this compares with the bilayer thickness at which gA open channel probability (Po) should be maximal. It is usually assumed that the maximum dilatation/thinning tolerated by a bilayer is ≤5%, as determined by dividing the rupture tension (10–20 dyn/cm) by the area expansion modulus (200–500 dyn/cm) (ref. 42, see also ref. 43). In this case, stretching a PC-20 bilayer (30 Å) may thin it by ≈1.5 Å, which compares with 4 Å required to achieve neutral mismatch with gA (26 Å) and presumably maximum Po. However, this discrepancy is consistent with our inability to achieve saturated responses (maximum Po) with suction pulses of 150 mmHg (see Fig. 1A) that we estimate would increase tension to ≈20 dyn/cm. A sustained suction at this level invariably resulted in patch rupture, indicating patches can tolerate higher transient tensions. This finding is also consistent with the idea that patch rupture is a stochastic event that requires a finite time to occur. A related consideration concerns the observation that full-sized gA channels form readily not only in PC-16 but also in PC-14 bilayers (40). Furthermore, CD measurements indicate gA dimers form in bilayers with acyl chain lengths as short as 10 (44). These results would appear inconsistent with the idea that thinning the PC-18 bilayer by only ≈1.5 Å inactivates the gA channel. A possible explanation is that in very thin bilayers (i.e., <PC-16) the gA dimer is able to tilt in relation to the plane of the membrane and thereby relieve positive mismatch (23, 24). In this case, one might expect that membrane stretch would be less effective in causing SI in these thinner bilayers.
Although gA forms a channel that in many respects behaves like no other known channel, increasing evidence indicates that sensitivity to hydrophobic surface matching may be a general feature of MG channels. For example, hydrophobic matching has been evoked to explain the modulation by amphipaths and lysophopholipids of a variety of membrane ion channels (ref. 45 and references therein), including prokaryotic (46, 47) and eukaryotic MG channels (ref. 48 and references therein). These agents were originally proposed to act on gA channel activity by changing the energetic cost of bilayer deformation caused by hydrophobic mismatch (45). In the case of MscL, the currently favored model of mechanical gating assumes the closed-open gating transition involves a significant increase in membrane occupied area (≈600 Å2) so that bilayer tension can do significant work on the channel (33). However, recent modeling studies also indicate a reduction in the open channel length (49, 50). Furthermore, less tension is required to activate MscL in thinner compared with thicker bilayers, consistent with the open channel having a shorter hydrophobic length (B.M., unpublished work). A similar dependence of stretch sensitivity on bilayer thickness has been reported for MG channels in the archaeon Thermoplasma volcanium (47). In terms of the energetics of bilayer deformation (51), the hydrophobic mismatch model may also provide a physically more plausible basis for the tension sensitivity of channels in animal cells (e.g., refs. 13 and 52). Although it seems unlikely that these ion-selective channels with much smaller pore diameters than MscL derive their tension sensitivity from similar or even larger changes in membrane occupied area (i.e., ≥600 Å2), only relatively small changes in hydrophobic mismatch (<1 Å) are required to generate tension sensitivity (51).
In the case of the Shaker K+ channel that also reverses its stretch sensitivity (13), several possible explanations may apply. However, one relates to the observation that Shaker-like channels (Kv2.1) undergo selective targeting to lipid rafts (6). Furthermore, depletion of cellular cholesterol not only alters the Kv2.1-associated raft buoyancy but also shifts the Kv2.1 inactivation curve by ≈40 mV without affecting peak current density or channel activation (6). In this case, if hydrophobic surface matching contributes to lipid raft recruitment (23), then the channel in changing conformation (i.e., caused by depolarization) may partition into a new lipid microdomain and as a consequence change its signaling properties, including stretch sensitivity.
The demonstration here of a bilayer-controlled switch in signaling by a mechanotransducer channel emphasizes that the bilayer is much more than a neutral solvent. Instead, it may actively modulate the specificity and fidelity of signaling by membrane proteins. This feature, in combination with protein-related factors (e.g., oligomerization state and cytoskeleton association) that determine not only protein recruitment into lipid microdomains but also the dynamic organization of the bilayer itself (53), indicates a dynamic reciprocity in lipid–protein interactions that is presumably necessary for the higher-order spatial and temporal control of signaling, presently unique to complex living cells.
Acknowledgments
We thank the Raine Medical Research Foundation and the Australian Research Council for their support, Albert Raso for preparing the liposomes, and Dr. Rosario Maroto for critical comments on the manuscript. This study was performed while O.P.H. was a Raine Visiting Professor at the University of Western Australia.
Abbreviations
- gA
gramicidin A
- SA
stretch activation
- SI
stretch inactivation
- MG
mechanically gated
- PC
phosphatidylcholine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Harder T, Simons K. Curr Opin Cell Biol. 1997;9:534–542. doi: 10.1016/s0955-0674(97)80030-0. [DOI] [PubMed] [Google Scholar]
- 2.Brown D A, London E. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 3.Ferron O, Smith T W, Michell T, Kelly R A. J Biol Chem. 1997;272:17744–17748. doi: 10.1074/jbc.272.28.17744. [DOI] [PubMed] [Google Scholar]
- 4.Bravo-Zehnder M, Orio P, Norambuena A, Wallner M, Meera P, Toro L, Gonzalez A. Proc Natl Acad Sci USA. 2000;97:13114–13119. doi: 10.1073/pnas.240455697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lockwich T P, Liu X, Singh B B, Jadlowiec J, Weiland S, Ambudkar I S. J Biol Chem. 2000;275:11934–11942. doi: 10.1074/jbc.275.16.11934. [DOI] [PubMed] [Google Scholar]
- 6.Martens J R, Navarro-Polanco R, Coppock E A, Nishiyama A, Parshley L, Grobaski T D, Tamkun M M. J Biol Chem. 2000;275:7443–7446. doi: 10.1074/jbc.275.11.7443. [DOI] [PubMed] [Google Scholar]
- 7.Andersen O S, Sawyer D B, Koeppe R E. In: Biomembrane Structure and Function. Garber B P, Earswaran K R K, editors. New York: Adenine; 1992. pp. 227–243. [Google Scholar]
- 8.Dai J, Sheetz M P. Cold Spring Harbor Symp Quant Biol. 1995;XL:567–571. doi: 10.1101/sqb.1995.060.01.060. [DOI] [PubMed] [Google Scholar]
- 9.Maroto R, Hamill O P. J Biol Chem. 2001;276:23867–23872. doi: 10.1074/jbc.M101500200. [DOI] [PubMed] [Google Scholar]
- 10.Sachs F, Morris C E. Rev Physiol Biochem Pharmacol. 1998;132:1–77. doi: 10.1007/BFb0004985. [DOI] [PubMed] [Google Scholar]
- 11.Hamill O P, Martinac B. Physiol Rev. 2001;81:686–740. doi: 10.1152/physrev.2001.81.2.685. [DOI] [PubMed] [Google Scholar]
- 12.Morris C E, Sigurdson W J. Science. 1989;243:807–809. doi: 10.1126/science.2536958. [DOI] [PubMed] [Google Scholar]
- 13.Gu X, Juranka P F, Morris C E. Biophys J. 2001;80:2678–2693. doi: 10.1016/S0006-3495(01)76237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Hamill O P. J Physiol (London) 2000;523:101–115. doi: 10.1111/j.1469-7793.2000.00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris C E, Homann U. J Membr Biol. 2001;179:79–102. doi: 10.1007/s002320010040. [DOI] [PubMed] [Google Scholar]
- 16.Evans E A, Hochmuth R M. In: Topics in Membrane and Transport. Kleinzeller A, Bronner F, editors. Vol. 10. New York: Academic; 1978. pp. 1–64. [Google Scholar]
- 17.Opsahl L, Webb W W. Biophys J. 1994;66:71–74. doi: 10.1016/S0006-3495(94)80751-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goulian M, Mesquita O N, Fygenson D K, Nielsen C, Andersen O S, Libchaber A. Biophys J. 1998;74:328–337. doi: 10.1016/S0006-3495(98)77790-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang G, Spencer R, Lee A, Barclay M, Rees C. Science. 1998;282:2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 20.Perozo E, Kloda A, Cortes D M, Martinac B. J Gen Physiol. 2001;118:193–205. doi: 10.1085/jgp.118.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urry D W, Goodall M C, Glickson J D, Mayers D F. Proc Natl Acad Sci USA. 1971;68:1907–1911. doi: 10.1073/pnas.68.8.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koeppe R E, II, Andersen O S. Annu Rev Biophys Biomol Struct. 1996;25:231–258. doi: 10.1146/annurev.bb.25.060196.001311. [DOI] [PubMed] [Google Scholar]
- 23.Killian J A. Biochim Biophys Acta. 1992;1113:391–425. doi: 10.1016/0304-4157(92)90008-x. [DOI] [PubMed] [Google Scholar]
- 24.Hladky S B, Haydon D A. Biochim Biophys Acta. 1972;274:294–313. doi: 10.1016/0005-2736(72)90178-2. [DOI] [PubMed] [Google Scholar]
- 25.Veatch W R, Mathies R, Eisenberg M, Stryer L. J Mol Biol. 1975;99:75–92. doi: 10.1016/s0022-2836(75)80160-4. [DOI] [PubMed] [Google Scholar]
- 26.Neher E, Eibl H J. Biochim Biophys Acta. 1977;464:37–44. doi: 10.1016/0005-2736(77)90368-6. [DOI] [PubMed] [Google Scholar]
- 27.Kolb H-A, Bamberg E. Biochim Biophys Acta. 1977;464:127–141. doi: 10.1016/0005-2736(77)90376-5. [DOI] [PubMed] [Google Scholar]
- 28.Elliot J R, Needham D, Dilger J P, Haydon D A. Biochim Biophys Acta. 1983;735:95–103. doi: 10.1016/0005-2736(83)90264-x. [DOI] [PubMed] [Google Scholar]
- 29.Delcour A H, Martinac B, Adler J, Kung C. Biophys J. 1989;56:631–635. doi: 10.1016/S0006-3495(89)82710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Häse C C, Le Dain A C, Martinac B. J Biol Chem. 1995;270:18329–18334. doi: 10.1074/jbc.270.31.18329. [DOI] [PubMed] [Google Scholar]
- 31.Corey D P, Stevens C F. In: Single Channel Recording. Sakmann B, Neher E, editors. New York: Plenum; 1983. pp. 53–68. [Google Scholar]
- 32.Hamill O P, Marty A, Neher E, Sakmann B, Sigworth F J. Pflügers Archiv. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 33.Sukahrev S I, Sigurdson W J, Kung C, Sachs F. J Gen Physiol. 1998;113:525–539. doi: 10.1085/jgp.113.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang H W. Biophys J. 1986;50:1061–1070. doi: 10.1016/S0006-3495(86)83550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helfrich P, Jakobsson E. Biophys J. 1990;57:1075–1084. doi: 10.1016/S0006-3495(90)82625-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mobashery N, Nielsen C, Andersen O S. FEBS Lett. 1997;412:15–20. doi: 10.1016/s0014-5793(97)00709-6. [DOI] [PubMed] [Google Scholar]
- 37.Koeppe R E, Berg J M, Hodgson K O, Stryer L. Nature (London) 1979;279:723–725. doi: 10.1038/279723a0. [DOI] [PubMed] [Google Scholar]
- 38.Wiener M C, White S H. Biophys J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis B A, Engelman D M. J Mol Biol. 1983;166:211–217. doi: 10.1016/s0022-2836(83)80007-2. [DOI] [PubMed] [Google Scholar]
- 40.Arndt H-D, Knoll A, Koert U. Chembiochemistry. 2001;3:221–223. doi: 10.1002/1439-7633(20010302)2:3<221::AID-CBIC221>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 41.Watnick P I, Chan S I, Dea P. Biochemistry. 1990;29:6215–6221. doi: 10.1021/bi00478a015. [DOI] [PubMed] [Google Scholar]
- 42.Bloom M, Evans E, Mouritsen O G. Q Rev Biophys. 1991;24:293–397. doi: 10.1017/s0033583500003735. [DOI] [PubMed] [Google Scholar]
- 43.Nichol J A, Hutter O F. J Physiol (London) 1996;493:187–198. doi: 10.1113/jphysiol.1996.sp021374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greathouse D V, Hinton J F, Kim K S, Koeppe R E. Biochemistry. 1994;33:4291–4299. doi: 10.1021/bi00180a025. [DOI] [PubMed] [Google Scholar]
- 45.Lundbæk J A, Andersen O S. J Gen Physiol. 1994;104:645–673. doi: 10.1085/jgp.104.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinac B, Adler J, Kung C. Nature (London) 1990;348:261–263. doi: 10.1038/348261a0. [DOI] [PubMed] [Google Scholar]
- 47.Kloda A, Martinac B. Cell Biochem Biophys. 2001;34:321–347. doi: 10.1385/CBB:34:3:321. [DOI] [PubMed] [Google Scholar]
- 48.Hamill O P, McBride D W. Pharmacol Rev. 1996;48:231–252. [PubMed] [Google Scholar]
- 49.Gullingsrud J, Kosztin D, Schulten K. Biophys J. 2001;80:2074–2081. doi: 10.1016/S0006-3495(01)76181-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sukharev S, Betanzos M, Chiang C-S, Guy H R. Nature (London) 2001;409:720–724. doi: 10.1038/35055559. [DOI] [PubMed] [Google Scholar]
- 51.Nielsen C, Goulian M, Andersen O S. Biophys J. 1998;74:1966–1983. doi: 10.1016/S0006-3495(98)77904-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Gao F, Popov V L, Wen J L, Hamill O P. J Physiol (London) 2000;523:117–130. doi: 10.1111/j.1469-7793.2000.t01-1-00117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simons K, Toome D. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]