Abstract
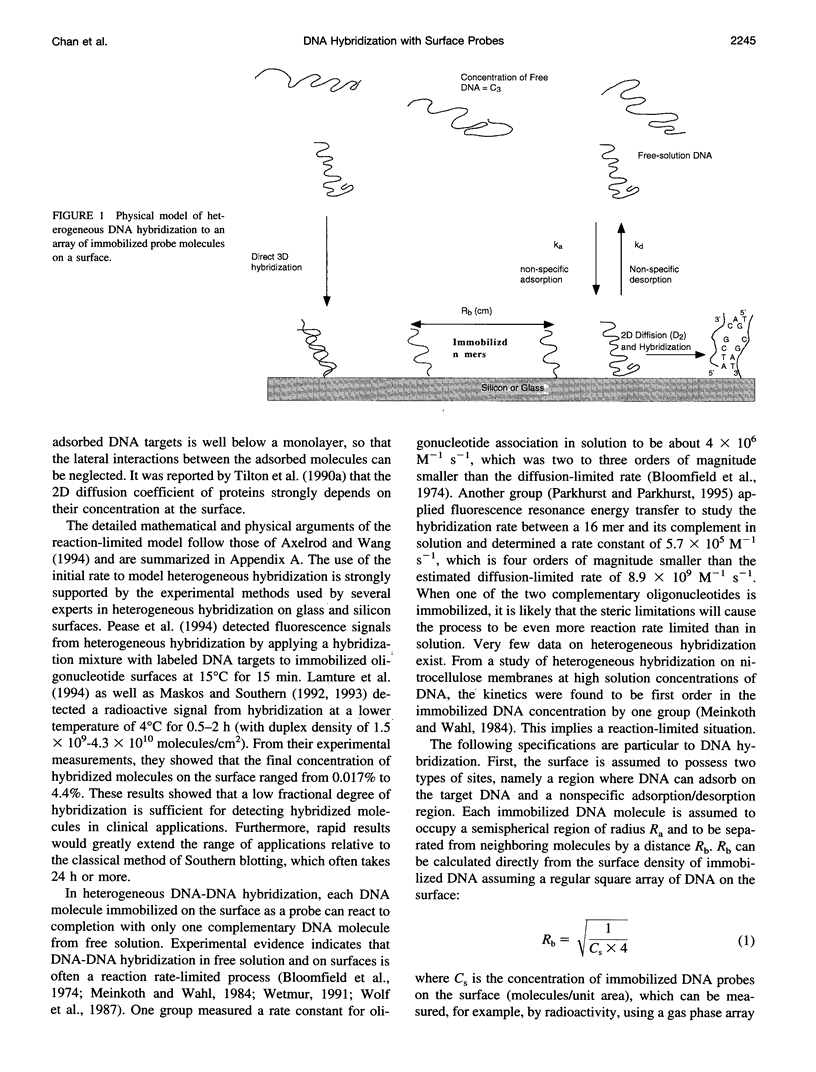
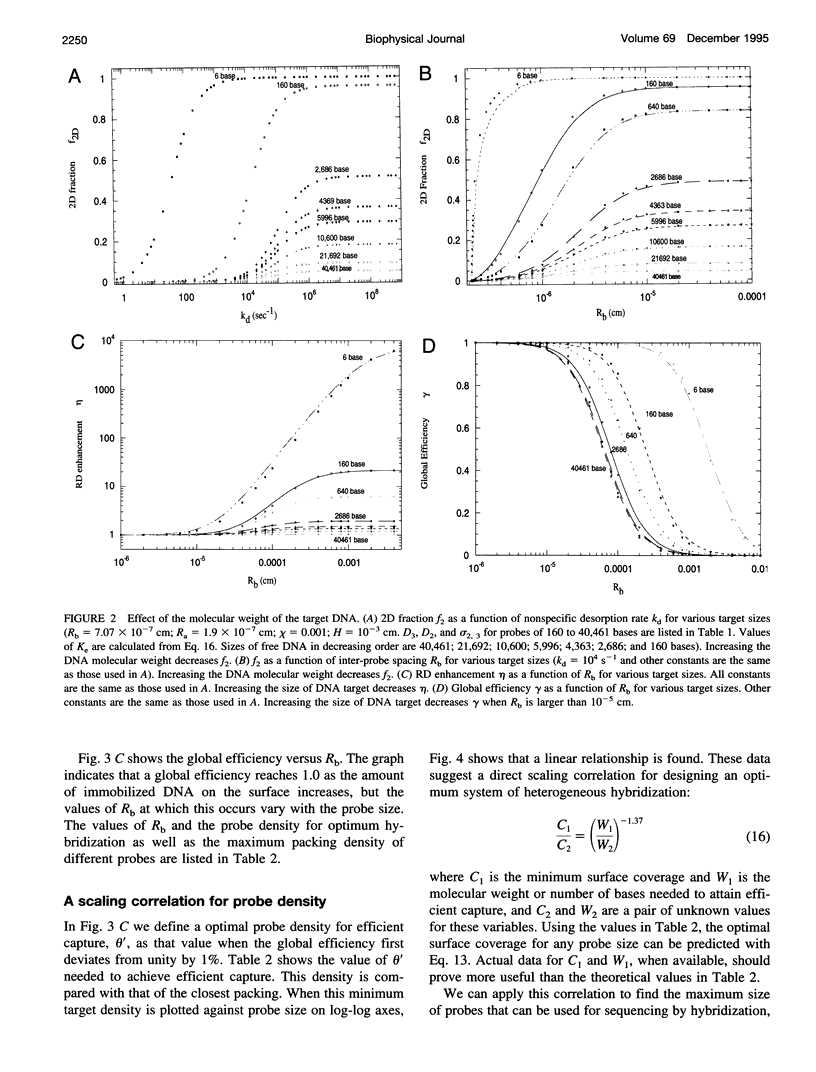
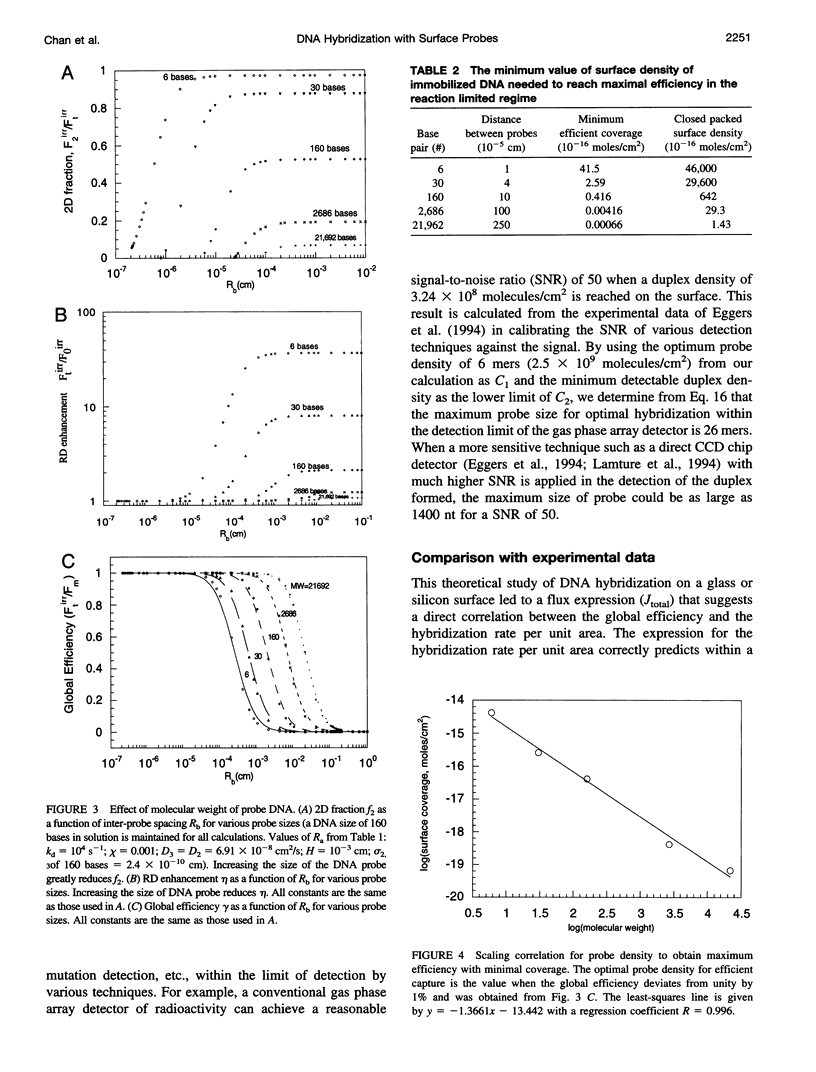
A mathematical model based on receptor-ligand interactions at a cell surface has been modified and further developed to represent heterogeneous DNA-DNA hybridization on a solid surface. The immobilized DNA molecules with known sequences are called probes, and the DNA molecules in solution with unknown sequences are called targets in this model. Capture of the perfectly complementary target is modeled as a combined reaction-diffusion limited irreversible reaction. In the model, there are two different mechanisms by which targets can hybridize with the complementary probes: direct hybridization from the solution and hybridization by molecules that adsorb nonspecifically and then surface diffuse to the probe. The results indicate that nonspecific adsorption of single-stranded DNA on the surface and subsequent two-dimensional diffusion can significantly enhance the overall reaction rate. Heterogeneous hybridization depends strongly on the rate constants for DNA adsorption/desorption in the non-probe-covered regions of the surface, the two-dimensional (2D) diffusion coefficient, and the size of probes and targets. The model shows that the overall kinetics of DNA hybridization to DNA on a solid support may be an extremely efficient process for physically realistic 2D diffusion coefficients, target concentrations, and surface probe densities. The implication for design and operation of a DNA hybridization surface is that there is an optimal surface probe density when 2D diffusion occurs; values above that optimum do not increase the capture rate. Our model predicts capture rates in agreement with those from recent experimental literature. The results of our analysis predict that several things can be done to improve heterogeneous hybridization: 1) the solution phase target molecules should be about 100 bases or less in size to speed solution-phase and surface diffusion; 2) conditions should be created such that reversible adsorption and two-dimensional diffusion occur in the surface regions between DNA probe molecules; 3) provided that 2) is satisfied, one can achieve results with a sparse probe coverage that are equal to or better than those obtained with a surface totally covered with DNA probes.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axelrod D., Wang M. D. Reduction-of-dimensionality kinetics at reaction-limited cell surface receptors. Biophys J. 1994 Mar;66(3 Pt 1):588–600. doi: 10.1016/s0006-3495(94)80834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H. C., Purcell E. M. Physics of chemoreception. Biophys J. 1977 Nov;20(2):193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle J. S., Lew A. M. An inexpensive alternative to glassmilk for DNA purification. Trends Genet. 1995 Jan;11(1):8–8. doi: 10.1016/s0168-9525(00)88977-5. [DOI] [PubMed] [Google Scholar]
- Burghardt T. P., Axelrod D. Total internal reflection/fluorescence photobleaching recovery study of serum albumin adsorption dynamics. Biophys J. 1981 Mar;33(3):455–467. doi: 10.1016/S0006-3495(81)84906-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverin A. B., Kramer F. R. Oligonucleotide arrays: new concepts and possibilities. Biotechnology (N Y) 1994 Nov;12(11):1093–1099. doi: 10.1038/nbt1194-1093. [DOI] [PubMed] [Google Scholar]
- DeLisi C. The biophysics of ligand-receptor interactions. Q Rev Biophys. 1980 May;13(2):201–230. doi: 10.1017/s0033583500001657. [DOI] [PubMed] [Google Scholar]
- Dwyer J. D., Bloomfield V. A. Brownian dynamics simulations of probe and self-diffusion in concentrated protein and DNA solutions. Biophys J. 1993 Nov;65(5):1810–1816. doi: 10.1016/S0006-3495(93)81235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers M., Hogan M., Reich R. K., Lamture J., Ehrlich D., Hollis M., Kosicki B., Powdrill T., Beattie K., Smith S. A microchip for quantitative detection of molecules utilizing luminescent and radioisotope reporter groups. Biotechniques. 1994 Sep;17(3):516–525. [PubMed] [Google Scholar]
- Garcia de la Torre J., Navarro S., Lopez Martinez M. C. Hydrodynamic properties of a double-helical model for DNA. Biophys J. 1994 May;66(5):1573–1579. doi: 10.1016/S0006-3495(94)80949-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Pearce K. H., Thompson N. L. Translational diffusion of bovine prothrombin fragment 1 weakly bound to supported planar membranes: measurement by total internal reflection with fluorescence pattern photobleaching recovery. Biophys J. 1994 Oct;67(4):1754–1766. doi: 10.1016/S0006-3495(94)80650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J. W., Fodor S. P. Combinatorial chemistry--applications of light-directed chemical synthesis. Trends Biotechnol. 1994 Jan;12(1):19–26. doi: 10.1016/0167-7799(94)90007-8. [DOI] [PubMed] [Google Scholar]
- Kubitscheck U., Wedekind P., Peters R. Lateral diffusion measurement at high spatial resolution by scanning microphotolysis in a confocal microscope. Biophys J. 1994 Sep;67(3):948–956. doi: 10.1016/S0006-3495(94)80596-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamture J. B., Beattie K. L., Burke B. E., Eggers M. D., Ehrlich D. J., Fowler R., Hollis M. A., Kosicki B. B., Reich R. K., Smith S. R. Direct detection of nucleic acid hybridization on the surface of a charge coupled device. Nucleic Acids Res. 1994 Jun 11;22(11):2121–2125. doi: 10.1093/nar/22.11.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livshits M. A., Florentiev V. L., Mirzabekov A. D. Dissociation of duplexes formed by hybridization of DNA with gel-immobilized oligonucleotides. J Biomol Struct Dyn. 1994 Feb;11(4):783–795. doi: 10.1080/07391102.1994.10508032. [DOI] [PubMed] [Google Scholar]
- Maskos U., Southern E. M. A study of oligonucleotide reassociation using large arrays of oligonucleotides synthesised on a glass support. Nucleic Acids Res. 1993 Oct 11;21(20):4663–4669. doi: 10.1093/nar/21.20.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskos U., Southern E. M. Oligonucleotide hybridizations on glass supports: a novel linker for oligonucleotide synthesis and hybridization properties of oligonucleotides synthesised in situ. Nucleic Acids Res. 1992 Apr 11;20(7):1679–1684. doi: 10.1093/nar/20.7.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Parkhurst K. M., Parkhurst L. J. Kinetic studies by fluorescence resonance energy transfer employing a double-labeled oligonucleotide: hybridization to the oligonucleotide complement and to single-stranded DNA. Biochemistry. 1995 Jan 10;34(1):285–292. doi: 10.1021/bi00001a035. [DOI] [PubMed] [Google Scholar]
- Pease A. C., Solas D., Sullivan E. J., Cronin M. T., Holmes C. P., Fodor S. P. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci U S A. 1994 May 24;91(11):5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton M. J. Lateral diffusion in an archipelago. Effects of impermeable patches on diffusion in a cell membrane. Biophys J. 1982 Aug;39(2):165–173. doi: 10.1016/S0006-3495(82)84504-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M., Case-Green S. C., Elder J. K., Johnson M., Mir K. U., Wang L., Williams J. C. Arrays of complementary oligonucleotides for analysing the hybridisation behaviour of nucleic acids. Nucleic Acids Res. 1994 Apr 25;22(8):1368–1373. doi: 10.1093/nar/22.8.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassburger J., Reinert K. E. Diffusion of DNA at very low concentrations. Biopolymers. 1971;10(2):263–273. doi: 10.1002/bip.360100204. [DOI] [PubMed] [Google Scholar]
- Thompson N. L., Burghardt T. P., Axelrod D. Measuring surface dynamics of biomolecules by total internal reflection fluorescence with photobleaching recovery or correlation spectroscopy. Biophys J. 1981 Mar;33(3):435–454. doi: 10.1016/S0006-3495(81)84905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson N. L., Pearce K. H., Hsieh H. V. Total internal reflection fluorescence microscopy: application to substrate-supported planar membranes. Eur Biophys J. 1993;22(5):367–378. doi: 10.1007/BF00213560. [DOI] [PubMed] [Google Scholar]
- Tilton R. D., Gast A. P., Robertson C. R. Surface diffusion of interacting proteins. Effect of concentration on the lateral mobility of adsorbed bovine serum albumin. Biophys J. 1990 Nov;58(5):1321–1326. doi: 10.1016/S0006-3495(90)82473-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Gou S. Y., Axelrod D. Reaction rate enhancement by surface diffusion of adsorbates. Biophys Chem. 1992 Jun;43(2):117–137. doi: 10.1016/0301-4622(92)80027-3. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G. DNA probes: applications of the principles of nucleic acid hybridization. Crit Rev Biochem Mol Biol. 1991;26(3-4):227–259. doi: 10.3109/10409239109114069. [DOI] [PubMed] [Google Scholar]
- Wolf S. F., Haines L., Fisch J., Kremsky J. N., Dougherty J. P., Jacobs K. Rapid hybridization kinetics of DNA attached to submicron latex particles. Nucleic Acids Res. 1987 Apr 10;15(7):2911–2926. doi: 10.1093/nar/15.7.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]



